Abstract
The UV components of sunlight (UVA and UVB) are implicated in the etiology of human skin cancer. The underlying mechanism of action for UVB carcinogenicity is well defined; however, the mechanistic involvement of UVA in carcinogenesis is not fully delineated. We investigated the genotoxicity of UVA1 versus UVB in the overall genome and in the p53 tumor suppressor gene in normal human skin fibroblasts. Immuno-dot blot analysis identified the cis-syn cyclobutane pyrimidine-dimer (CPD) as a distinctive UVB-induced lesion and confirmed its formation in the genomic DNA of UVA1-irradiated cells dependent on radiation dose. HPLC/tandem MS analysis showed an induction of 8-oxo-7,8-dihydro-2′-deoxyguanosine in the genomic DNA of UVA1-irradiated cells only. Mapping of DNA damages by terminal transferase-dependent PCR revealed preferential, but not identical, formation of polymerase-blocking lesions and/or strand breaks along exons 5-8 of the p53 gene in UVB- and UVA1-irradiated cells. The UVB-induced lesions detected by terminal transferase-PCR were almost exclusively mapped to pyrimidine-rich sequences; however, the UVA1-induced lesions were mapped to purine- and pyrimidine-containing sequences along the p53 gene. Cleavage assays with lesion-specific DNA repair enzymes coupled to ligation-mediated PCR showed preferential, but not identical, formation of CPDs along the p53 gene in UVB- and UVA1-irradiated cells. Additionally, dose-dependent formation of oxidized and ring-opened purines and abasic sites was established in the p53 gene in only UVA1-irradiated cells. We conclude that UVA1 induces promutagenic CPDs and oxidative DNA damage at both the genomic and nucleotide resolution level in normal human skin fibroblasts.
Ultraviolet (UV) irradiation from the sun is linked to basal and squamous cell carcinomas of the skin and cutaneous malignant melanoma in humans (1-3). The etiologically relevant UV wavelengths for development of these diseases are UVA (320-400 nm) and UVB (280-320 nm) (4, 5). UVA constitutes the major proportion of the solar UV reaching the surface of the Earth (≈95%). The remaining fraction is the residual UVB, which is not absorbed by passage through the stratospheric ozone layer (6, 7). Biologically, however, UVB is a few orders of magnitude more potent than UVA in inducing photocarcinogenesis (8).
The carcinogenic effect of UVB is irrefutably ascribed to its ability to produce promutagenic DNA lesions including cis-syn cyclobutane pyrimidine-dimers (CPDs) and to a lesser extent pyrimidine (6-4) pyrimidone photoproducts [(6-4)PPs]. The carcinogenicity of UVA is partly attributed to its mutagenicity; however, the mechanistic involvement of UVA in mutagenesis remains controversial. The latter is caused by the ambiguous DNA damaging effects of this wavelength (4, 5).
Generally, the poor absorption of UVA by DNA supports the notion that UVA indirectly triggers mutagenesis via photosensitization reactions. Presumably, UVA sensitizes intracellular chromophores, thereby generating reactive oxygen species, which in turn may cause promutagenic DNA lesions, e.g., 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxo-dG) and DNA single-strand breaks (4, 5). We have recently shown the formation of 8-oxo-dG followed by induction of signature mutations of this lesion, i.e., G → T transversions, in mouse fibroblasts irradiated with UVA1 (340-400 nm) (9) or treated with a UVA1-activated sensitizer, δ-aminolevulinic acid (10). There is also a second notion that UVA might induce CPDs as a consequence of weak, yet direct, absorption of UVA photons by DNA (11, 12) and/or through triplet photosensitization reactions (5). In fact, the genomic DNA of rodent cells irradiated with UVA is shown to contain detectable levels of CPDs (13-15). Yet, in normal human cells, the plausibility of these two scenarios, i.e., the mechanism through which UVA induces mutagenesis, has not been investigated.
Here, we analyze the DNA damaging properties of UVA1 versus UVB at both the genomic and nucleotide resolution level, specifically in the p53 tumor suppressor gene, in normal human skin fibroblasts.
Materials and Methods
Cell Culture and Irradiation. A strain of human fibroblasts was developed from a normal foreskin biopsy according to standard protocols (16). The cells were cultured in DMEM (Irvine Scientific) supplemented with 10% FBS. The monolayer ≈70% confluent cell cultures were irradiated with UVA1 or UVB for different durations of time under uniformly controlled conditions. Briefly, after removing the media, the cultures were washed thoroughly with PBS, filled with a 1-cm layer of PBS, placed on ice, and irradiated with UVA1 (216, 432, and 648 J/cm2) or UVB (0.26, 0.52, and 0.78 J/cm2). The UVA source was a Sellas Sunlight System (Medizinische Geräte, Gevelsberg, Germany) with an average fluence rate of 60 mW/cm2 emitting almost exclusively wavelengths of 340-400 nm (see figure 1 of ref. 9). The UVB source consisted of three fluorescent tubes (TL 20 W/12R, Philips, Eindhoven, The Netherlands) filtered through a cellulose acetate sheet, which cuts off wavelengths <295 nm. The source had a peak spectral emission at 312 nm and an average fluence rate of 1.15 mW/cm2 determined by a UVX radiometer (Ultraviolet Products). Immediately after irradiation, all cells including dead and alive ones were harvested by trypsinization and subjected to DNA isolation. A set of representative cultures was processed similarly and used for determining cell viability by means of trypan blue dye exclusion assay. All experiments were run in triplicate.
Genomic DNA Isolation. For all analyses except HPLC-tandem MS (MS/MS) genomic DNA was isolated by using a standard phenol/chloroform extraction and ethanol precipitation protocol (17). The DNA was dissolved in Tris-EDTA buffer and preserved at -80°C until further analysis.
DNA Isolation and Nucleoside Preparation for HPLC-MS/MS. Genomic DNA was isolated by using a salt-based extraction method as described (9). Detailed descriptions of this methodology are provided in Supporting Text, which is published as supporting information on the PNAS web site.
HPLC-MS/MS for Quantifying 8-Oxo-dG. Quantification of 8-oxo-dG was done by using the method of Singh et al. (18). Detailed descriptions of this methodology are provided in Supporting Text.
Immuno-Dot Blot Assay for Detecting CPDs and (6-4)PPs. The immuno-dot blot assay was performed by using TD-M-2 and 64-M-2 mAbs specific for CPDs and (6-4)PPs, respectively (19, 20) (kindly provided by T. Mori, O. Nikaido, and A. Yasui, Tohoku University, Sendai, Japan). The assay was performed as described (21). Detailed descriptions of this methodology are provided in Supporting Text. To quantitate the immuno-dot blot assay results, we prepared standard controls with known quantities of photodimers, and subsequently established a calibration curve for each run of the assay. The standard controls were made from the genomic DNAs of normal human skin fibroblasts irradiated with increasing doses of UVC (i.e., 1, 10, 25, 50, 100, 150, 200, and 250 J/m2) emitted from a germicidal lamp and UVB (i.e., 200, 600, 1,200, and 2,400 J/m2). The standard controls for CPD determination were subjected to T4 endonuclease V digestion, and then run on a 1.5% alkaline/agarose electrophoresis gel (9) with specific ladder markers in the range of 100 to 10,000 bp (New England Biolabs). The standard controls for (6-4)PPs determination were subjected to two consecutive treatments with (i) CPD photolyase and (ii) UV damage endonuclease (Trevigen, Gaithersburg, MD) (22), and subsequently run on an electrophoresis gel as described above. The UV damage endonuclease is known to cleave both CPDs and (6-4)PPs (23-25). Therefore, a pretreatment with CPD photolyase, which rids DNA of all CPDs, followed by digestion with UV damage endonuclease can specifically identify the remaining (6-4)PPs in each sample.
Terminal Transferase-Dependent PCR (TD-PCR) for Mapping of Photodimers and/or DNA Strand Breaks in the p53 Gene. TD-PCR is a highly sensitive polymerase arrest-based assay, which involves an initial gene-specific primer extension followed by PCR amplification and labeling of the extended product, and finally resolution of the products by gel electrophoresis (26). During primer extension, all DNA lesions that stop polymerase progression, i.e., bulky lesions, e.g., CPDs or (6-4)PPs, and DNA strand breaks, produce prematurely terminated single-stranded DNA fragments. After undergoing exponential amplification by PCR, these fragments can be labeled, and subsequently visualized on a sequencing gel. The fragments are identified as specific bands along the sequencing gel whose intensities reflect the frequency of the lesions (27). Technically, however, 8-oxo-dG is a nonbulky DNA lesion and cannot be detected by TD-PCR. Detailed descriptions of the methodology for TD-PCR are provided in Supporting Text and Table 1, which is published as supporting information on the PNAS web site.
Ligation-Mediated PCR (LM-PCR) for Mapping of Specific Types of DNA Damage in the p53 Gene. The principles of LM-PCR are similar to TD-PCR with the only differences being the primer extension and ligation steps (17). In LM-PCR, enzymatically or chemically cleaved single-strand DNA breaks having 5′ phosphate termini undergo primer extension to produce blunt-ended fragments to which a linker is subsequently ligated (28). To cleave each specific type of DNA damage we used the following enzymes: T4 endonuclease V combined with CPD-photolyase for CPDs, formamidopyrimidine DNA glycosylase (Fpg) for oxidized and ring-opened purines, and thymine glycol DNA glycosylase for oxidized and ring-opened pyrimidines. Detailed descriptions of this methodology are provided in Supporting Text.
Results
Cytotoxicity Examination for UVA1 and UVB. To mimic a real-life situation, we deliberately chose high irradiation doses corresponding to 4-12 and 1-3 minimal erythema doses of UVA and UVB, respectively. These doses are equivalent to several hours of exposure to midday summer sunlight at 45° latitude (29, 30). The relatively instantaneous consequence of acute sun exposure is erythema ensued by cutaneous cell lethality (31, 32). However, UV-induced DNA damage and mutagenesis are likely to occur consequent to chronic (accumulative) sun exposure (31, 32). For experimental purposes, therefore, it is justified to administer high doses of UV to initiate DNA damage, at the price of compromising cell viability. In the present study, the choice of dose was determined by examining the cytotoxicity of UVA1 and UVB to ensure that the highest administered doses of each of these two wavelengths yielded a minimal viability of ≈1-5% in the irradiated cells.
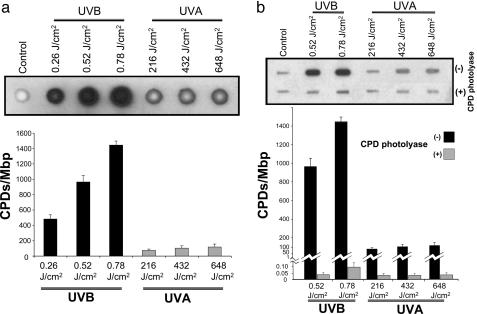
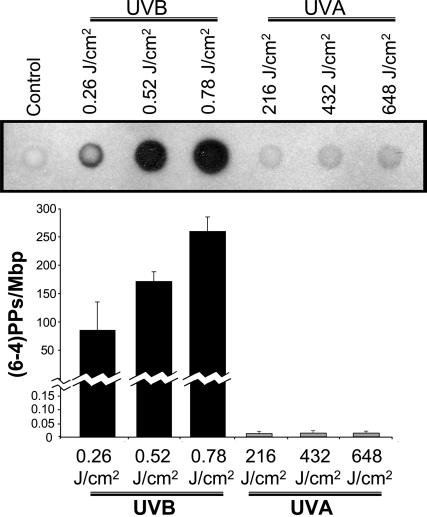
Quantifying CPDs and (6-4)PPs in the Genome by Immuno-Dot Blot Assay. Genomic DNA of the UVB-irradiated human fibroblasts contained substantial levels of CPDs dependent on radiation dose. The UVA1-irradiated cells showed less, yet significant, levels of CPDs in the genomic DNA (Fig. 1a). To verify the specificity of CPD detection, we pretreated the DNAs of UVB- and UVA1-irradiated cells with CPD-photolyase (33), and subsequently performed the immuno-dot blot assay. The levels of CPDs in the pretreated DNAs returned closely to the baseline level established in the genomic DNA of nonirradiated cells (Fig. 1b). The genomic DNA of UVB-irradiated cells showed a radiation dose-dependent formation of (6-4)PPs. However, there was no appreciable formation of (6-4)PPs in the genomic DNA of UVA1-irradiated cells (Fig. 2).
Fig. 1.
Quantifying CPDs in the genome by immuno-dot blot assay. (a) Genomic DNAs of UVB-irradiated (0.26, 0.52, and 0.78 J/cm2) or UVA1-irradiated (216, 432, and 648 J/cm2) human fibroblasts were analyzed with immuno-dot blot assay by using TD-M-2 mAb specific for CPDs (19, 20), as described in Materials and Methods. (b) To verify the specificity of CPD detection, the DNAs of UVB- and UVA1-irradiated cells were pretreated with CPD-photolyase, and subsequently subjected to immuno-dot blot analysis. To quantitate the results, we prepared standard controls with known quantities of CPDs, and subsequently established a calibration curve for luminescence signals specific for each assay run (see Materials and Methods). Results are expressed as means of three independent experiments, with each experiment run in triplicate (nine measurements). Error bar, SD. Mbp, megabase pair DNA.
Fig. 2.
Quantifying (6-4)PPs in the genome by immuno-dot blot assay. Genomic DNAs of UVB-irradiated (0.26, 0.52, and 0.78 J/cm2) or UVA1-irradiated (216, 432, and 648 J/cm2) human fibroblasts were analyzed with immuno-dot blot assay by using 64-M-2 mAb specific for (6-4)PPs (19, 20), as described in Materials and Methods. To quantitate the results, we prepared standard controls with known quantities of (6-4)PPs, and subsequently established a calibration curve for luminescence signals specific for each assay run (see Materials and Methods). Results are expressed as means of three independent experiments, with each experiment run in triplicate (nine measurements). Error bar, SD. Mbp, megabase pair DNA.
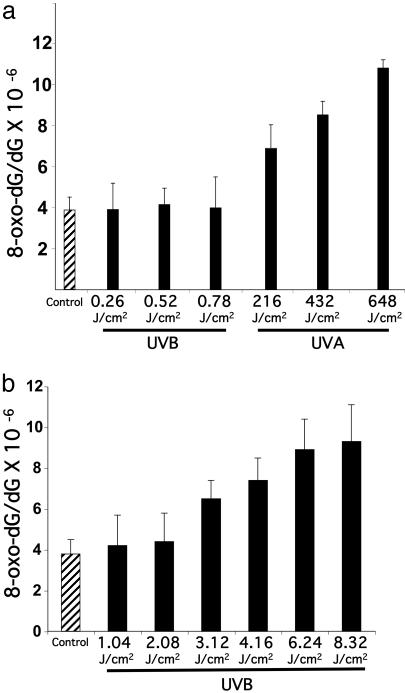
Quantifying 8-Oxo-dG in the Genome by HPLC-MS/MS. Genomic DNA of the UVA1-irradiated human fibroblasts contained elevated levels of 8-oxo-dG relative to control, i.e., genomic DNA of nonirradiated cells (Fig. 3a). The extent of 8-oxo-dG generation depended on UVA1 dose, reaching a maximum of ≈3-fold over background (P < 0.01). There was no increase in 8-oxo-dG levels in the genomic DNA of UVB-irradiated cells relative to control. To investigate whether high-dose UVB irradiation of cells can lead to 8-oxo-dG formation, a set of representative cultures was irradiated with UVB at increasing doses of 1.04, 2.08, 3.12, 4.16, 6.24, and 8.32 J/cm2. As shown in Fig. 3b, a minimum dose of 4.16 J/cm2 of UVB was required to cause an ≈2-fold increase in the background level of 8-oxo-dG in the irradiated cells. At this dose, however, UVB was overwhelmingly lethal (cell survival < 0.1%).
Fig. 3.
Quantifying 8-oxo-dG in the genome by HPLC-MS/MS. (a) Genomic DNAs of UVB-irradiated (0.26, 0.52, and 0.78 J/cm2) or UVA1-irradiated (216, 432, and 648 J/cm2) human fibroblasts were assayed with HPLC-MS/MS, as described in Materials and Methods.(b) Effects of high-dose UVB irradiation on the formation of 8-oxo-dG in human fibroblasts. Results are expressed as means of three independent experiments, with each experiment run in triplicate (nine measurements). Error bar, SD.
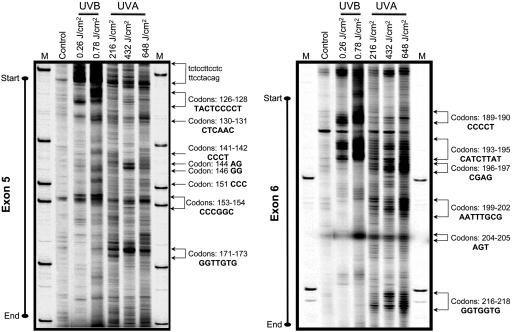
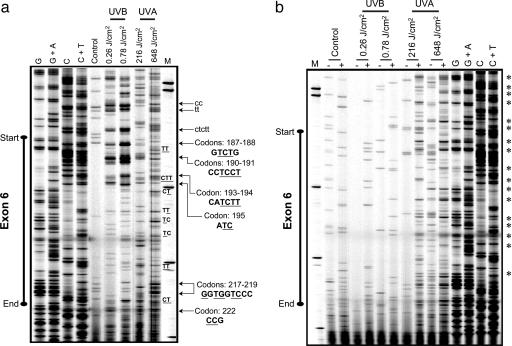
Mapping of Polymerase-Blocking DNA Lesions in the p53 Gene by TD-PCR. The UVB- and UVA1-irradiated human fibroblasts showed preferential, but not identical, formation of polymerase-blocking DNA lesions along the p53 gene (Fig. 4). The lesions were mapped to precise locations in introns and in exons 5-8 of the p53. Representative lesion-mapping data for exons 5 and 6 of the p53 gene are shown in Fig. 4. The UVB-induced lesions were almost exclusively formed at pyrimidine-rich sequences in the p53 gene. However, the UVA1-induced lesions were positioned at purine/pyrimidine sequences along the p53 gene. The former is highlighted in a stretch of pyrimidines at the intron/exon 5 junction of the p53 gene, which was a hotspot of UVB-induced lesions (Fig. 4). The latter is exemplified by the formation of UVA1-induced lesions at multiple purine/pyrimidine sequences along exon 6 of the p53 gene (Fig. 4). Mapping of Specific Types of DNA Damage in the p53 Gene by LM-PCR. T4 endonuclease V-cleaved DNAs of UVB- and UVA1-irradiated human fibroblasts showed preferential, but not identical, formation of CPDs along the p53 gene (Fig. 5a and Figs. 6a and 7a, which are published as supporting information on the PNAS web site). The hotspots and dose dependency of CPD formation were overtly pronounced in the DNA of UVB-irradiated cells relative to UVA1-irradiated cells. Representative CPD mapping data for exons 5, 6, and 8 of the p53 gene are shown in Figs. 7a, 5a, and 6a, respectively. For the most part, mapping of CPDs in the p53 gene by LM-PCR corresponded well with the mapping of polymerase-blocking DNA lesions in this gene determined by TD-PCR (for comparison, see Figs. 7a and 5a versus Fig. 4). Methodologically, each LM-PCR lesion-derived band represents a “single” nucleotide position, whereas the respective TD-PCR band may not be as precise. That is because the ribotailing step of TD-PCR adds a few residues (on average 3 nt) to the 3′ end of the polymerase-extended fragment (17, 26, 27). This fact might explain minor discrepancies observed between the locations of lesions detected by the respective assays.
Fig. 4.
Mapping of polymerase-blocking DNA lesions in the p53 gene (nontranscribed strand) by TD-PCR. Genomic DNAs of UVB-irradiated (0.26 and 0.78 J/cm2) or UVA1-irradiated (216, 432, and 648 J/cm2) human fibroblasts were subjected to TD-PCR, as described in Materials and Methods. Representative lesion-mapping data for exons 5 (Left) and 6 (Right) of the p53 gene are shown. Hotspots of lesion formation are indicated by arrows, and the corresponding nucleotide positions, e.g., respective codons, are specified. Sequence contexts of the lesions in introns and exons are written in lowercase and uppercase fonts, respectively. M, sizing standard.
Fig. 5.
Mapping of CPDs and Fpg-sensitive sites in exon 6 of the p53 gene (nontranscribed strand) by LM-PCR. Genomic DNAs of UVB-irradiated (0.26 and 0.78 J/cm2) or UVA1-irradiated (216 and 648 J/cm2) human fibroblasts were subjected to T4 endonuclease V cleavage and CPD-photolyase reactivation (to create ligatable ends) (a), or treatment with Fpg (+) or digestion buffer only (-), and subsequently assayed by LM-PCR (b), as described in Materials and Methods. Hotspots of lesion formation are indicated by arrows, and the corresponding nucleotide positions, e.g., respective codons, are specified. Sequence contexts of the lesions in introns and exons are written in lowercase and uppercase, respectively. Underlined bases are the exact positions where the lesions are formed. Major Fpg-sensitive sites are identified by *. To specifically identify G, G + A, C, and C + T, a Maxam/Gilbert sequencing ladder (61) was included in all runs. All samples were processed in parallel and run on the same gel but were separated for better illustration. M, sizing standard.
Fpg protein-cleaved DNA of UVA1- but not UVB-irradiated human fibroblasts showed specific lesion formation along the p53 gene dependent on radiation dose (Figs. 5b, 6b, and 7b). The lesions were almost exclusively mapped to purine-containing sequences, mostly guanines, in introns as well as in exons 5-8 of the p53 gene. A similar, yet less extensive, lesion formation was observed in non-Fpg-cleaved DNA of the cells irradiated with 648 J/cm2 of UVA1. This finding implies that the administered dose of UVA1 generates sufficient numbers of DNA strand breaks that are readily detectable by LM-PCR (Figs. 5b, 6b, and 7b). There was no appreciable lesion formation in the DNA of UVA1- or UVB-irradiated cells cleaved with thymine glycol DNA glycosylase protein (data not shown).
Discussion
The present study is a comprehensive investigation of DNA damaging effects of UVA1 versus UVB in the overall genome and in the p53 tumor suppressor gene in normal human skin fibroblasts. Our immuno-dot blot analysis verified the CPD as a major UVB-induced lesion and confirmed its formation in the genomic DNA of UVA1-irradiated cells dependent on radiation dose (Fig. 1). In addition, a dose-dependent formation of (6-4)PPs was observed in the genomic DNA of UVB-irradiated cells. However, the formation of (6-4)PPs could not be established in the genomic DNA of UVA1-irradiated cells (Fig. 2). Another class of photolesions, i.e., Dewar valence photoisomers, are known to be formed through photoisomerization of (6-4)PPs by radiation at 320 nm (15, 34, 35). Given the fact that our UVA source has no emission at or below 320 nm and only negligibly emits wavelengths between 330 and 340 nm, we could essentially rule out the possibility of formation of Dewar valence photoisomers in the genomic DNA of UVA1-irradiated cells. Using a similar methodology, Perdiz et al. (15) have reported comparable results on the formation of photodimeric lesions and the nondetectability of Dewar valence photoisomers in UVA-irradiated Chinese hamster ovary cells.
Our HPLC-MS/MS analysis showed an induction of 8-oxo-dG in the genomic DNA of UVA1-irradiated cells dependent on radiation dose. There was an ≈3-fold increase in the baseline level of 8-oxo-dG consequent to the highest UVA1 irradiation dose (P < 0.01) (Fig. 3a). It is likely that oxidative damages other than 8-oxo-dG may also be produced via UVA-derived photosensitization reactions. Possible candidates are the consequential products of singlet oxygen induction triggered by photosensitization reactions (36-38), including intrastrand base/base crosslinks and cyclo products, secondary oxidation products of 8-oxo-dG, e.g., diastereomeric spiroiminodihydantoins, lipid peroxidation product-induced DNA lesions, e.g., malondialdehyde- and 4-hydroxy-2-nonenal-derived DNA adducts, and DNA-protein crosslinks (39-44). Furthermore, only at lethal doses of UVB did we observe an appreciable formation of 8-oxo-dG in the genomic DNA of the irradiated cells (Fig. 3b). Presumably, 8-oxo-dG generation at extreme doses of UVB is overshadowed by the excessive production of cytotoxic dimeric photolesions. Using an alkaline elution assay, Kielbassa et al. (45) have demonstrated the formation of Fpg-sensitive sites (indicative of combined 8-oxo-dGs and formamidopyrimidines) in Chinese hamster ovary cells irradiated with 20 and 30 kJ/m2 of UVB at wavelengths of 310 and 320 nm, respectively. In agreement with our findings, no discernible induction of Fpg-sensitive sites was, however, detected at radiation doses of 250 J/m2 and 3 kJ/m2 at the respective wavelengths (figure 4 of ref. 45). Kielbassa et al. did not report any cytotoxicity examination for the administered doses of UVB. Using HPLC coupled with electrochemical detection technique, Zhang et al. (46) have shown an induction of 8-oxo-dG in calf thymus DNA and HeLa cells irradiated with 50-300 kJ/m2 of UVB. Obviously, the possibility of artifactual generation of 8-oxo-dG during sample preparation and processing is a matter of concern (47). In the latter study (46), the background levels of 8-oxo-dG determined in calf thymus DNA and HeLa cells were ≈5- and 7-fold, respectively, higher than that established in normal human skin fibroblasts by our HPLC-MS/MS assay. Unlike our recently optimized salt-based DNA isolation technique, however, a chloroform/isoamyl alcohol extraction method for isolating DNA was used by Zhang et al. (46).
Mapping of DNA lesions by TD-PCR revealed preferential, but not identical, formation of photodimers and/or DNA strand breaks along the p53 gene in UVB- and UVA1-irradiated cells. The UVB-induced lesions were almost exclusively formed at pyrimidine-rich sequences in the p53 gene (Fig. 4). These findings confirm the preferential formation of photodimers in the genomic DNA of UVB-irradiated cells (Figs. 1 and 2). On the other hand, the UVA1-induced lesions were mapped to purine/pyrimidine sequences along the p53 gene (Fig. 4). The latter is to be expected in light of our observation that UVA1 gave rise to CPDs together with 8-oxo-dGs and presumably other oxidative lesions in the overall genome (Figs. 1 and 3a).
LM-PCR analysis showed preferential, but not identical, formation of CPDs along the p53 gene in UVB- and UVA1-irradiated cells. Many of the UVB-induced CPD hotspots found in the p53 gene were hardly detectable in the respective positions in the DNA of UVA1-irradiated cells (Figs. 7a, 5a, and 6a). This finding indicates that the underlying mechanism of CPD formation by UVB may somehow differ from that by UVA. It is known that CPDs are formed through direct absorption of UVB photons by DNA (4, 5). However, triplet photosensitization reactions are thought to be partly responsible for UVA-induced CPD formation (5). If true, thymine would be the main target because it has the lowest triplet state energy among all four DNA bases (48). Accordingly, sequence contexts of 5′TT, 5′TC, and 5′CT, respectively, are to be hit by UVA-derived triplet photosensitization reactions (49). In confirmation, most UVA1-induced CPDs along the p53 gene were formed at these sequence contexts, especially consequent to the highest irradiation dose. Nonetheless, the possibility of CPD formation caused by a marginal, yet direct, absorption of UVA by DNA cannot be entirely ruled out (4, 5). This idea is supported by our observation that there were some similarities between the patterns of CPD formation induced by UVA1 and UVB, respectively, in the p53 gene (Figs. 7a, 5a, and 6a). Rochette et al. (14) had previously reported a preferential formation of CPDs at 5′TT sequences in the adenine phosphoribosyl transferase gene in UV-irradiated Chinese hamster ovary cells.
Furthermore, LM-PCR analysis identified Fpg-sensitive sites along the p53 gene in UVA1- but not UVB-irradiated cells. The formation of Fpg-sensitive sites was dose-dependently related to UVA1. The Fpg-sensitive sites were mapped almost exclusively to purine-containing sequences, mainly guanines, throughout exons 5-8ofthe p53 gene (Figs. 5b, 6b, and 7b). The Fpg protein is specific for nicking of oxidized and ring-opened purines, although it also cleaves abasic sites (22). The detection of these lesions in the p53 gene by LM-PCR is consistent with the verified presence of 8-oxo-dGs in the overall genome consequent to UVA1 irradiation (Fig. 3a); other oxidized guanine lesions not quantified by HPLC-MS/MS can be detected by LM-PCR. This latter might explain the relatively low ratio of 8-oxo-dGs to CPDs at the genomic level (Figs. 1 versus 3) and the comparable levels of oxidized purines and CPDs at the nucleotide resolution level in UVA1-irradiated cells (Figs. 5, 6, 7). Our data also confirm a recent finding by Agar et al. (50) who demonstrated a striking bias in 8-oxo-dG formation localized to the basal epidermal layer of human squamous cell carcinoma and solar keratoses. Concurrently, the signature mutations of 8-oxo-dG, i.e., GC → TA was prevalent at the same localization (50). Agar et al. ascribed these observations to the fact that human epidermis attenuates UVA photons much less efficiently than those of UVB. As a result, UVA photons can effectively reach the basal layer of the skin wherein tumorigenesis is initiated consequent to the DNA damaging effects of this wavelength. Mechanistically, our observations favor the idea that singlet oxygen generated via photosensitization reactions is the major contributor to UVA-induced oxidative DNA damage (10, 51). At the same time, there may be some contribution of hydroxyl radicals produced via superoxide and Fenton reactions to this process, especially at extreme irradiation doses (52, 53). The latter is reflected by the significant induction of DNA strand breaks upon 648 J/cm2 of UVA1 irradiation, which was readily detectable by LM-PCR in the respective noncleaved DNAs (Figs. 7b, 5b, and 6b).
Previously, we have shown 8-oxo-dG-mediated induction of mutagenesis in mouse embryonic fibroblasts irradiated with UVA1 (9) or treated with UVA1 in presence of δ-aminolevulinic acid, a precursor of the active intracellular photosensitizer protoporphyrin IX (10). In both cases, there was no detectable formation of CPDs or induction of signature mutations of photodimeric DNA lesions, i.e., C → T and CC → TT at dipyrimidine sites. To have a low cytotoxicity, however, we did not administer UVA1 doses exceeding 36 J/cm2 (9). Obviously, viable and replicating cells are needed if the induced DNA damages are to be fixed and translated into mutations. Irradiating either mouse embryonic fibroblasts (9) or normal human fibroblasts (data not shown) with 36 J/cm2 of UVA1 caused readily detectable levels of oxidative DNA damages, while marginal CPD formation was observed. This finding emphasizes the importance of irradiation dose, and potentially cell type and contents of endogenous photosensitizers, in shaping the overall pattern of UVA-induced DNA damage and mutagenesis.
The p53 mutational spectrum in sunlight-associated human skin cancer is dominated by the signature mutations of photodimeric DNA lesions (4, 5, 31, 32). These mutations are likely to have arisen from exposure to the UVB fraction of sunlight, although some contribution of the predominant UVA fraction is not precluded [per joule basis UVB is up to 50,000 times more genotoxic than UVA (8)] (31, 32). Of concern, however, is the excessive artificial irradiation among frequent users of high-powered tanning beds/baths, which emit mainly far wavelength UV. In view of our observation that UVA at extreme doses induces promutagenic DNA lesions, it is important to educate the public on a possible health risk involved with the regular use of these tanning devices. Prospectively, it will be interesting to establish a mutational spectrum for the p53 gene in individuals with a history of frequent use of tanning beds/baths.
In conclusion, we have demonstrated a unique DNA damaging property of UVA1 both in the genome and the p53 tumor suppressor gene in normal human fibroblasts. The UVA1-induced DNA lesions at high irradiation doses include promutagenic CPDs and oxidative DNA damage. However, at lower irradiation doses, the latter lesions are more relevant and might explain the induction of G → T transversion mutations previously observed in UVA1 mutagenesis experiments in mouse cells (9, 10). In humans, however, the signature mutation of G → T transversion does not prevail in the p53 mutational spectrum in malignant melanomas associated with sunlight exposure. Nevertheless, a substantial number of mutations in this malignancy occurs at guanine residues, i.e., G:C → A:T, G:C → T:A, and G:C → C:G comprise 57.8%, 3.94%, and 3.94%, respectively, of all mutations (International Agency for Research on Cancer P53 Database, R9 Version, www-p53.iarc.fr/p53DataBase.htm). Taken together, the contribution of each class of lesions to the overall mutagenicity of solar UV in mammalian cells may depend on (i) irradiation dose, (ii) efficiency of nucleotide excision repair versus base excision repair, the respective systems handling CPDs (54-58) and oxidized lesions (59, 60), and (iii) the content of endogenous photosensitizers specific for each cell type.
Supplementary Material
Acknowledgments
We thank Dr. Timothy O'Connor for kindly providing DNA repair enzymes for LM-PCR, Seung-Gi Jin for technical assistance in the immuno-dot blot assay, and Steven Bates for help with cell culturing. This work was supported by National Institute of Environmental Health Sciences Grant ES06070 (to G.P.P.).
Author contributions: A.B., A.D.R., and G.P.P. designed research; A.B., T.W.S., H.-H.C., C.C., and B.X. performed research; A.B., A.D.R., and G.P.P. analyzed data; and A.B. and G.P.P. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: (6-4)PP, pyrimidine (6-4) pyrimidone photoproduct; 8-oxo-dG, 8-oxo-7,8-dihydro-2′-deoxyguanosine; CPD, cis-syn cyclobutane pyrimidine-dimer; Fpg, formamidopyrimidine DNA glycosylase; MS/MS, tandem MS; LM-PCR, ligation-mediated PCR; TD-PCR, terminal transferase-dependent PCR.
References
- 1.Woodhead, A. D., Setlow, R. B. & Tanaka, M. (1999) J. Epidemiol. 9, S102-S114. [DOI] [PubMed] [Google Scholar]
- 2.de Gruijl, F. R. (1999) Eur. J. Cancer 35, 2003-2009. [DOI] [PubMed] [Google Scholar]
- 3.Jhappan, C., Noonan, F. P. & Merlino, G. (2003) Oncogene 22, 3099-3112. [DOI] [PubMed] [Google Scholar]
- 4.Pfeifer, G. P., You, Y. H. & Besaratinia, A. (2005) Mutat. Res. 571, 19-31. [DOI] [PubMed] [Google Scholar]
- 5.Cadet, J., Sage, E. & Douki, T. (2005) Mutat. Res. 571, 3-17. [DOI] [PubMed] [Google Scholar]
- 6.Setlow, R. B. (1974) Proc. Natl. Acad. Sci. USA 71, 3363-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Gruijl, F. R. (2002) Skin Pharmacol. Appl. Skin Physiol. 15, 316-320. [DOI] [PubMed] [Google Scholar]
- 8.de Gruijl, F. R., Sterenborg, H. J., Forbes, P. D., Davies, R. E., Cole, C., Kelfkens, G., van Weelden, H., Slaper, H. & van der Leun, J. C. (1993) Cancer Res. 53, 53-60. [PubMed] [Google Scholar]
- 9.Besaratinia, A., Synold, T. W., Xi, B. & Pfeifer, G. P. (2004) Biochemistry 43, 8169-8177. [DOI] [PubMed] [Google Scholar]
- 10.Besaratinia, A., Bates, S. E., Synold, T. & Pfeifer, G. P. (2004) Biochemistry 43, 15557-15566. [DOI] [PubMed] [Google Scholar]
- 11.Sutherland, J. C. & Griffin, K. P. (1981) Radiat. Res. 86, 399-409. [PubMed] [Google Scholar]
- 12.Kuluncsics, Z., Perdiz, D., Brulay, E., Muel, B. & Sage, E. %(1999%) J. Photochem. Photobiol. B 49, 71-80. [DOI] [PubMed] [Google Scholar]
- 13.Douki, T., Reynaud-Angelin, A., Cadet, J. & Sage, E. (2003) Biochemistry 42, 9221-9226. [DOI] [PubMed] [Google Scholar]
- 14.Rochette, P. J., Therrien, J. P., Drouin, R., Perdiz, D., Bastien, N., Drobetsky, E. A. & Sage, E. (2003) Nucleic Acids Res. 31, 2786-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perdiz, D., Grof, P., Mezzina, M., Nikaido, O., Moustacchi, E. & Sage, E. (2000) J. Biol. Chem. 275, 26732-26742. [DOI] [PubMed] [Google Scholar]
- 16.Gray, B. A. & Gollin, S. M. (1987) Am. J. Med. Genet. 28, 521-526. [DOI] [PubMed] [Google Scholar]
- 17.Pfeifer, G. P., Chen, H. H., Komura, J. & Riggs, A. D. (1999) Methods Enzymol. 304, 548-571. [DOI] [PubMed] [Google Scholar]
- 18.Singh, R., McEwan, M., Lamb, J. H., Santella, R. M. & Farmer, P. B. (2003) Rapid Commun. Mass Spectrom. 17, 126-134. [DOI] [PubMed] [Google Scholar]
- 19.Mori, T., Nakane, M., Hattori, T., Matsunaga, T., Ihara, M. & Nikaido, O. (1991) Photochem. Photobiol. 54, 225-232. [DOI] [PubMed] [Google Scholar]
- 20.Chadwick, C. A., Potten, C. S., Nikaido, O., Matsunaga, T., Proby, C. & Young, A. R. (1995) J. Photochem. Photobiol. B 28, 163-170. [DOI] [PubMed] [Google Scholar]
- 21.You, Y. H., Lee, D. H., Yoon, J. H., Nakajima, S., Yasui, A. & Pfeifer, G. P. (2001) J. Biol. Chem. 276, 44688-44694. [DOI] [PubMed] [Google Scholar]
- 22.Yoon, J. H., Lee, C. S., O'Connor, T. R., Yasui, A. & Pfeifer, G. P. (2000) J. Mol. Biol. 299, 681-693. [DOI] [PubMed] [Google Scholar]
- 23.Freyer, G. A., Davey, S., Ferrer, J. V., Martin, A. M., Beach, D. & Doetsch, P. W. (1995) Mol. Cell. Biol. 15, 4572-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yajima, H., Takao, M., Yasuhira, S., Zhao, J. H., Ishii, C., Inoue, H. & Yasui, A. (1995) EMBO J. 14, 2393-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaur, B., Avery, A. M. & Doetsch, P. W. (1998) Biochemistry 37, 11599-11604. [DOI] [PubMed] [Google Scholar]
- 26.Komura, J. & Riggs, A. D. (1998) Nucleic Acids Res. 26, 1807-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen, H. H., Kontaraki, J., Bonifer, C. & Riggs, A. D. (2001) Sci. STKE, PL1 (1-17). [DOI] [PubMed]
- 28.Pfeifer, G. P., Steigerwald, S. D., Mueller, P. R., Wold, B. & Riggs, A. D. (1989) Science 246, 810-813. [DOI] [PubMed] [Google Scholar]
- 29.Parrish, J. A., Jaenicke, K. F. & Anderson, R. R. (1982) Photochem. Photobiol. 36, 187-191. [DOI] [PubMed] [Google Scholar]
- 30.Robert, C., Muel, B., Benoit, A., Dubertret, L., Sarasin, A. & Stary, A. (1996) J. Invest. Dermatol. 106, 721-728. [DOI] [PubMed] [Google Scholar]
- 31.Daya-Grosjean, L. & Sarasin, A. (2005) Mutat. Res. 571, 43-56. [DOI] [PubMed] [Google Scholar]
- 32.Melnikova, V. O. & Ananthaswamy, H. N. (2005) Mutat. Res. 571, 91-106. [DOI] [PubMed] [Google Scholar]
- 33.Kosmoski, J. V. & Smerdon, M. J. (1999) Biochemistry 38, 9485-9494. [DOI] [PubMed] [Google Scholar]
- 34.Rosenstein, B. S. & Mitchell, D. L. (1987) Photochem. Photobiol. 45, 775-780. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, J. S., Lu, H. F. & Kotyk, J. J. (1990) Photochem. Photobiol. 51, 161-167. [DOI] [PubMed] [Google Scholar]
- 36.Cadet, J., Douki, T., Pouget, J. P. & Ravanat, J. L. (2000) Methods Enzymol. 319, 143-153. [DOI] [PubMed] [Google Scholar]
- 37.Ravanat, J. L., Di Mascio, P., Martinez, G. R., Medeiros, M. H. & Cadet, J. (2000) J. Biol. Chem. 275, 40601-40604. [DOI] [PubMed] [Google Scholar]
- 38.Stary, A. & Sarasin, A. (2000) Methods Enzymol. 319, 153-165. [DOI] [PubMed] [Google Scholar]
- 39.Ravanat, J. L. & Cadet, J. (1995) Chem. Res. Toxicol. 8, 379-388. [DOI] [PubMed] [Google Scholar]
- 40.Randerath, K., Zhou, G. D., Somers, R. L., Robbins, J. H. & Brooks, P. J. (2001) J. Biol. Chem. 276, 36051-36057. [DOI] [PubMed] [Google Scholar]
- 41.Niles, J. C., Wishnok, J. S. & Tannenbaum, S. R. (2001) Org. Lett. 3, 963-966. [PubMed] [Google Scholar]
- 42.Bellon, S., Ravanat, J. L., Gasparutto, D. & Cadet, J. (2002) Chem. Res. Toxicol. 15, 598-606. [DOI] [PubMed] [Google Scholar]
- 43.Gu, C. & Wang, Y. (2004) Biochemistry 43, 6745-6750. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, Q. & Wang, Y. (2005) Nucleic Acids Res. 33, 1593-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kielbassa, C., Roza, L. & Epe, B. (1997) Carcinogenesis 18, 811-816. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, X., Rosenstein, B. S., Wang, Y., Lebwohl, M., Mitchell, D. M. & Wei, H. (1997) Photochem. Photobiol. 65, 119-124. [DOI] [PubMed] [Google Scholar]
- 47.Cadet, J., Bellon, S., Berger, M., Bourdat, A. G., Douki, T., Duarte, V., Frelon, S., Gasparutto, D., Muller, E., Ravanat, J. L. & Sauvaigo, S. (2002) Biol. Chem. 383, 933-943. [DOI] [PubMed] [Google Scholar]
- 48.Lamola, A. A. (1968) Photochem. Photobiol. 8, 601-616. [DOI] [PubMed] [Google Scholar]
- 49.Sauvaigo, S., Douki, T., Odin, F., Caillat, S., Ravanat, J. L. & Cadet, J. (2001) Photochem. Photobiol. 73, 230-237. [DOI] [PubMed] [Google Scholar]
- 50.Agar, N. S., Halliday, G. M., Barnetson, R. S., Ananthaswamy, H. N., Wheeler, M. & Jones, A. M. (2004) Proc. Natl. Acad. Sci. USA 101, 4954-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Epe, B. (1991) Chem. Biol. Interact. 80, 239-260. [DOI] [PubMed] [Google Scholar]
- 52.Epe, B., Pflaum, M. & Boiteux, S. (1993) Mutat. Res. 299, 135-145. [DOI] [PubMed] [Google Scholar]
- 53.Darr, D. & Fridovich, I. (1994) J. Invest. Dermatol. 102, 671-675. [DOI] [PubMed] [Google Scholar]
- 54.Svoboda, D. L., Smith, C. A., Taylor, J. S. & Sancar, A. (1993) J. Biol. Chem. 268, 10694-10700. [PubMed] [Google Scholar]
- 55.Fritz, L. K. & Smerdon, M. J. (1995) Biochemistry 34, 13117-13124. [DOI] [PubMed] [Google Scholar]
- 56.Cleaver, J. E., Cortes, F., Karentz, D., Lutze, L. H., Morgan, W. F., Player, A. N., Vuksanovic, L. & Mitchell, D. L. (1988) Photochem. Photobiol. 48, 41-49. [DOI] [PubMed] [Google Scholar]
- 57.Cleaver, J. E., Jen, J., Charles, W. C. & Mitchell, D. L. (1991) Photochem. Photobiol. 54, 393-402. [DOI] [PubMed] [Google Scholar]
- 58.Nakajima, S., Lan, L., Kanno, S., Takao, M., Yamamoto, K., Eker, A. P. & Yasui, A. (2004) J. Biol. Chem. 279, 46674-46677. [DOI] [PubMed] [Google Scholar]
- 59.Dizdaroglu, M. (2003) Mutat. Res. 531, 109-126. [DOI] [PubMed] [Google Scholar]
- 60.Rosenstein, B. S. & Mitchell, D. L. (1991) Radiat. Res. 126, 338-342. [PubMed] [Google Scholar]
- 61.Maxam, A. M. & Gilbert, W. (1980) Methods Enzymol. 65, 499-560. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.