Abstract
Hutchinson–Gilford progeria syndrome (HGPS), a progeroid syndrome in children, is caused by mutations in LMNA (the gene for prelamin A and lamin C) that result in the deletion of 50 aa within prelamin A. In normal cells, prelamin A is a “CAAX protein” that is farnesylated and then processed further to generate mature lamin A, which is a structural protein of the nuclear lamina. The mutant prelamin A in HGPS, which is commonly called progerin, retains the CAAX motif that triggers farnesylation, but the 50-aa deletion prevents the subsequent processing to mature lamin A. The presence of progerin adversely affects the integrity of the nuclear lamina, resulting in misshapen nuclei and nuclear blebs. We hypothesized that interfering with protein farnesylation would block the targeting of progerin to the nuclear envelope, and we further hypothesized that the mislocalization of progerin away from the nuclear envelope would improve the nuclear blebbing phenotype. To approach this hypothesis, we created a gene-targeted mouse model of HGPS, generated genetically identical primary mouse embryonic fibroblasts, and we then examined the effect of a farnesyltransferase inhibitor on nuclear blebbing. The farnesyltransferase inhibitor mislocalized progerin away from the nuclear envelope to the nucleoplasm, as determined by immunofluoresence microscopy, and resulted in a striking improvement in nuclear blebbing (P < 0.0001 by χ2 statistic). These studies suggest a possible treatment strategy for HGPS.
Keywords: aging, lamin A/C, laminopathy
Hutchinson–Gilford progeria syndrome (HGPS) is a progeroid syndrome characterized by a host of aging-like phenotypes, including a wizened appearance of the skin, osteoporosis, alopecia, and premature atherosclerosis (1). Children with HGPS die at the mean age of 13, generally from myocardial infarctions or strokes (1). This disease is caused by the accumulation of a mutant form of prelamin A that cannot be processed to mature lamin A (1). In normal cells, wild-type prelamin A is virtually undetectable because it is fully converted to mature lamin A, a structural protein of the nuclear lamina (2, 3). The nuclear lamina is an intermediate filament meshwork adjacent to the inner nuclear membrane that provides structural support for the nucleus (2, 3).
Prelamin A contains a nuclear localization signal and terminates with a CAAX motif (2), in which C is a cysteine, A residues are usually aliphatic amino acids, and X can be one of many different residues. CAAX motifs are also found on lamin B1, lamin B2, the Ras family of proteins, and many other cellular proteins. The CAAX motif triggers three sequential enzymatic posttranslational modifications, beginning with protein prenylation. In the case of prelamin A, the first processing step is carried out by protein farnesyltransferase (FTase) and involves the addition of a 15-carbon farnesyl lipid to the thiol group of the cysteine within the CAAX motif. Second, the last 3 aa of the protein (i.e., -AAX) are removed by a prenylprotein-specific endoprotease. For prelamin A, this step is likely to be a redundant function of two endoplasmic reticulum membrane endoproteases, Zmpste24 and Rce1 (4). Third, the newly exposed farnesylcysteine is methylated by Icmt, a prenylprotein-specific membrane methyltransferase of the endoplasmic reticulum (5). After these CAAX-box modifications have been completed, prelamin A (in contrast to other CAAX proteins) undergoes an additional processing step. The last 15 aa of the protein (including the farnesylcysteine methyl ester) are clipped off by Zmpste24 and then degraded, leaving behind mature lamin A (4, 6, 7).
The farnesylation of prelamin A is important for its targeting to the nuclear envelope (8–10). Each of the three CAAX motif modifications of prelamin A render the C terminus of the protein more hydrophobic, facilitating its association with the inner nuclear membrane, where the protein is cleaved, releasing mature lamin A (9, 11). In the absence of farnesylation (for example, in mevinolin-treated cells), prelamin A accumulates in the nucleoplasm and does not reach the nuclear envelope (9, 11). In the setting of Zmpste24 deficiency, farnesyl prelamin A accumulates at the nuclear envelope (6, 12) and adversely affects the integrity of the nuclear envelope. The nuclei of Zmpste24-deficient fibroblasts are misshapen, containing numerous nuclear blebs (6, 12).
HGPS is most commonly caused by a de novo point mutation in exon 11 of LMNA (1). This mutation, which occurs in codon 608, activates a cryptic splice site and leads to the in-frame deletion of 50 aa within prelamin A. This deletion leaves the CAAX motif intact; hence, the mutant prelamin A (progerin) is predicted to undergo farnesylation, release of the -AAX, and carboxyl methylation. However, the site for the second endoproteolytic cleavage step is eliminated by the deletion (1). Thus, progerin cannot be processed to lamin A and likely retains a farnesylcysteine methyl ester at its C terminus. Like Zmpste24-deficient cells, HGPS fibroblasts contain misshapen nuclei with numerous blebs of the nuclear envelope (1). In human HGPS cells, the severity of nuclear blebbing is variable, depending in part on the number of times the cells have been passaged (13). A GFP-tagged progerin has been reported to accumulate along the nuclear envelope of HeLa cells and cause nuclear shape abnormalities (13).
We hypothesized that the farnesylation of progerin targets the protein to the nuclear envelope, where it might weaken the nuclear lamina and cause nuclear blebbing. We further hypothesized that blocking farnesylation with an FTase inhibitor (FTI) would mislocalize progerin away from the nuclear envelope and reduce nuclear blebbing. Some researchers might argue that the latter hypothesis is unattractive, given that the FTI would also block the posttranslational processing of lamin B1 and lamin B2, potentially weakening the lamina further. However, we hypothesized that the salutary effects of blocking farnesylation of progerin would “trump” any deleterious effects of the FTI on the lamina and lead to an overall improvement in nuclear blebbing.
To test the impact of blocking the farnesylation of progerin on nuclear shape, we reasoned that it would be helpful to create gene-targeted “HGPS mice” that express progerin at levels sufficient to cause nuclear blebbing. The existence of a gene-targeted mouse model would make it possible to study the impact of an FTI on nuclear shape in independent lines of low-passage, genetically identical mouse embryonic fibroblasts (MEFs). In this study, we adopted exactly this strategy, first by generating a mouse model of HGPS and then by using primary MEFs to define the impact of an FTI on the nuclear blebbing phenotype.
Materials and Methods
Gene-Targeted Mouse Model of HGPS. The DNA fragments for the arms of the gene-targeting vector were generated by long-range PCR of genomic DNA from 129/Ola ES cells. A 6-kb 5′ fragment, which spanned from the end of intron 5 to sequences 1.9 kb downstream from the 3′ UTR (encoded by exon 12) was amplified with 5′-GGCTTCCTGGTCACTGGATA-3′ and 5′-GATCTGCCTGGAAGCTGAGT-3′ and cloned into pCR2.1-TOPO-XL (Invitrogen). Next, a 5-kb EcoRI fragment (spanning from a polylinker EcoRI site in pCR2.1-TOPO-XL to an EcoRI site 0.9 kb downstream from the 3′ UTR) was cleaved from the vector and cloned into pBSK (Stratagene). To create the 5′ arm of a sequence-replacement vector, this EcoRI fragment was subjected to two sequential site-directed mutagenesis reactions (QuikChange, Stratagene). First, intron 10 was deleted with the primer 5′-GATGGAGAAGAGCTCCTCCATCACCACCGTGGTTCCCACTGCAGCGGCTCGGGGGACCCC-3′ and the reversed and complemented primer. Second, the last 150 nt of exon 11 and intron 11 were deleted with the primer 5′-GACAAGGCTGCCGGTGGAGCGGGAGCCCAGAGCTCCCAGAACTGCAGCATCATGTAATCT-3′ and the reversed and complemented primer.
To create the gene-targeting vector, the mutant EcoRI fragment was cloned into the polylinker EcoRI site of pKSloxPNT-mod. The 3′ arm, consisting of sequences immediately downstream of those in the 5′ arm, was amplified with the primers 5′-GACAGCCACCTGGTCAGTTT-3′ and 5′-GTAACTCTGGCTGCCCTCAA-3′ and then cloned into pCR2.1-TOPO-XL. To complete the gene-targeting vector, this fragment was cloned into the polylinker AscI site of pKSloxPNTmod. The integrity of the vector (≈17 kb in length) was verified by DNA sequencing and restriction mapping.
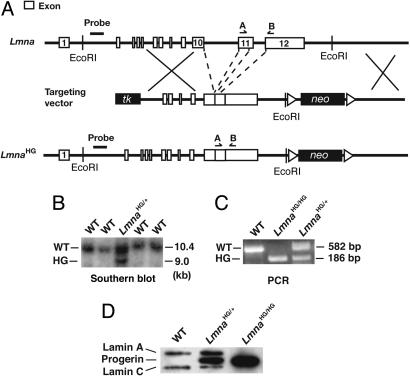
The vector was linearized with NotI and electroporated into strain 129/Ola ES cells. To identify clones carrying the targeted LmnaHG allele, we performed Southern blot analyses of EcoRI-digested genomic DNA with a 348-bp 5′-flanking probe. The probe was generated by PCR from mouse genomic DNA with the following primers: 5′-CAAGGAGCTCGGATTCTGTC-3′ and 5′-GTCAGGGAAGAGTGCAGAGG-3′. The probe detected a 10.4-kb band in the wild-type Lmna allele and a 9.3-kb band in the LmnaHG allele. Genotyping was also performed on genomic DNA by PCR with the following primers: 5′-TGAGTACAACCTGCGCTCAC-3′ and 5′-CAGACAGGAGGTGGCATGT-3′. The PCR fragment, spanning from exon 11 to exon 12, was 582 bp in the wild-type Lmna allele and 186 bp in the LmnaHG allele.
Creating LmnaHG MEFs. Targeted 129/Ola ES cells were microinjected into C57BL/6 blastocysts to produce chimeric mice. To generate MEFs, chimeras were bred with C57BL/6 females. Wild-type primary MEFs (Lmna+/+) and LmnaHG/+ MEFs were prepared from the same litter of day 13.5 embryos and genotyped by PCR. Because the MEFs were generated from embryos of chimera matings, they were genetically identical (one 129/Ola chromosome and one C57BL/6 chromosome). Cells homozygous for the LmnaHG mutation were generated by subjecting LmnaHG/+ cells to several months of selection in increasing concentrations of G418 (14). The LmnaHG/HG cells were euploid, but many of the cells were differentiated, as determined by morphology.
FTIs. For all experiments, we used PB-43, which is a member of the tetrahydroquinoline family of protein FTIs (15) that displays potent inhibition of rat FTase in vitro (IC50 = 1.7 nM). PB-43 readily crosses cell membranes, as demonstrated by its ability to kill Plasmodium falciparum in human red blood cells (M.H.G., unpublished data). PB-43 was synthesized by described methods (15) and shown to be pure by HPLC on a reverse-phase column. The compound was dissolved in DMSO at a concentration of 10 mM and stored in aliquots at -80°C.
Treatment of Cells with the FTI and Western Blot Analyses. Adherent early-passage MEFs in six-well tissue culture plates were incubated with the vehicle control (DMSO) or the indicated concentrations of PB-43 diluted in culture medium at 37°C for 48 h. The cells were washed with PBS, and urea-soluble extracts were prepared as described in ref. 11. Cell pellets solubilized with SDS-containing buffers were also prepared and yielded results indistinguishable from those with urea extraction. Proteins were size-separated on 4–12% gradient polyacrylamide Bis-Tris gels (Invitrogen) and then electrophoretically transferred to nitrocellulose membranes for Western blotting. The following antibody dilutions were used: 1:400 anti-lamin A/C goat IgG (sc-6215, Santa Cruz Biotechnology), 1:400 anti-lamin B (sc-6217, Santa Cruz Biotechnology), 1:6,000 anti-mouse prelamin A rabbit antiserum (12, 13), 1:500 anti-Hdj-2 mouse IgG (LabVision, Fremont, CA), 1:1,000 anti-actin goat IgG (sc-1616, Santa Cruz Biotechnology), 1:6,000 horseradish peroxidase (HRP)-labeled anti-goat IgG (sc-2020, Santa Cruz Biotechnology), 1:4,000 HRP-labeled anti-mouse IgG (Amersham Biosciences), and 1:6,000 HRP-labeled anti-rabbit IgG (Amersham Biosciences). Antibody binding was detected with the ECL Plus chemiluminescence system (Amersham Biosciences) and exposure to x-ray film.
Immunofluoresence Microscopy. Primary cells of different genotypes were grown on coverslips, fixed in 3% paraformaldehyde, permeabilized with 0.2% Triton X-100, and blocked with BSA (12). Cells were incubated for 60 min with antibodies against lamin A (sc-20680) or lamin B (sc-6217) (Santa Cruz Biotechnology). After washing, cells were stained with species-specific Cy3-conjugated secondary antibodies (Jackson Immuno-Research) and DAPI to visualize DNA. Images were obtained on an Axiovert 40CFL microscope (Zeiss) with a ×63/1.25 oil-immersion objective and processed with axovision software (version 4.2; Zeiss). Nuclear shape abnormalities were scored by two independent observers, who were blinded to genotype or treatment group.
Results
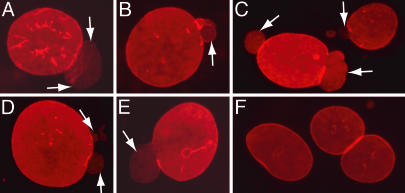
We created a mouse model of HGPS that expressed large amounts of progerin. To accomplish that goal, we created a mutant Lmna allele, LmnaHG, that exclusively yields progerin (Fig. 1A). We deleted intron 10 of Lmna, thereby eliminating lamin C synthesis; we also deleted the last 150 nt of exon 11 and intron 11, which results in the synthesis of progerin (and precludes wild-type prelamin A synthesis). Two targeted ES cell clones (from 192 G418- and FIAU-resistant clones) were identified by Southern blotting (Fig. 1B) and used to create 22 high-percentage chimeric mice. Those mice were bred with C57BL/6 mice to produce the LmnaHG/+ mice for this study (Fig. 1C). In addition, LmnaHG/HG cell lines were created from LmnaHG/+ ES cells by high-G418 selection (Fig. 1D). As predicted, primary MEFs from LmnaHG/+ embryos produced progerin, along with lamin A and lamin C from the wild-type Lmna allele (Figs. 1D and 3), whereas the LmnaHG/HG cells yielded only progerin (Fig. 1D). Lmna expression in wild-type ES cells is low (2, 3, 16). However, the LmnaHG/HG cells, which had undergone ≈2 months of high G418 selection and appeared to be differentiated, expressed progerin at levels comparable with those in MEFs (Fig. 1D). Not surprisingly, the nuclei of many LmnaHG/+ MEFs were misshapen and contained large blebs (Fig. 2).
Fig. 1.
Production of a mutant Lmna allele, LmnaHG, that yields progerin. (A) Gene-targeting strategy, which involves deleting intron 10, intron 11, and the last 150 nt of exon 11 of Lmna. (B) Southern blot analysis detecting the LmnaHG allele in mouse ES cells, with EcoRI-cleaved genomic DNA and the indicated 5′ flanking probe. (C) PCR identification of the LmnaHG allele. Results with wild-type MEFs (WT), heterozygous MEFs (LmnaHG/+), homozygous ES cells (LmnaHG/HG), and heterozygous MEFs (LmnaHG/+) are shown. (D) Western blotting identification of progerin with a lamin A/C-specific monoclonal antibody. Wild-type cells, LmnaHG/+ MEFs, and LmnaHG/HG cells are shown. On SDS/PAGE gels, the electrophoretic migration of progerin in mouse LmnaHG/+ MEFs and human HGPS fibroblasts was identical (data not shown).
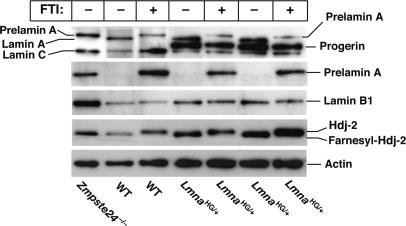
Fig. 3.
Western blot analysis of wild-type, LmnaHG/+, and Zmpste24-/- MEFs. Cells were grown in the presence and absence of an FTI (10 μM PB-43), and Western blot analyses of cell extracts were performed with antibodies specific for prelamin A (specific for the extreme C terminus), lamin A/C (binds to both lamin A and lamin C), lamin B1, Hdj-2, and actin.
Fig. 2.
Immunofluorescence microscopy of wild-type and LmnaHG/+ fibroblasts. (A–E) Immunostaining showing nuclear blebs in LmnaHG/+ MEFs. Blebs are indicated by white arrows. (F) Lmna+/+ MEFs. In this experiment, cells were stained for lamin B1 (red).
We sought to define the impact of an FTI, PB-43, on nuclear shape. To document that this compound was effective in blocking farnesylation, we treated LmnaHG/+ MEFs with the PB-43 FTI and then examined the electrophoretic migration of Hdj-2, a farnesylated protein, with Western blot analyses of SDS/PAGE gels (Fig. 3). In FTI-treated MEFs, nearly all of the Hdj-2 migrated slowly and almost none migrated normally, indicating that the FTI had been effective in blocking protein farnesylation.
We also performed Western blot analyses of wild-type and LmnaHG/+ cell extracts with an antibody specific for the C terminus of prelamin A (Fig. 3). The prelamin A-specific antibody does not normally detect prelamin A in cells because prelamin A is farnesylated and rapidly converted to mature lamin A. However, in the presence of the FTI, prelamin A processing is blocked, and prelamin A is easily detectable in both wild-type and LmnaHG/+ MEFs (Fig. 3). Western blot analyses with an antibody against the N-terminal portion of lamin A detected mature lamin A in wild-type MEFs and in LmnaHG/+ MEFs but detected the slightly larger prelamin A (and virtually no mature lamin A) in MEFs that had been treated with the FTI. The FTI did not yield clear-cut or consistent alterations in the amount of either progerin or lamin B1 in LmnaHG/+ MEFs (Fig. 3).
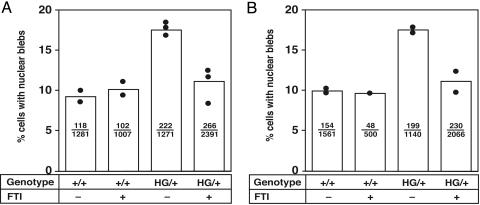
To judge the impact of the FTI on nuclear blebbing in LmnaHG/+ MEFs, we examined both Lmna+/+ and LmnaHG/+ MEFs in a blinded fashion by immunofluorescence microscopy. LmnaHG/+ MEFs had more nuclear blebs than Lmna+/+ cells (P < 0.0001, χ2 test) (Fig. 4). The FTI did not affect nuclear shape in Lmna+/+ MEFs. However, FTI treatment of the LmnaHG/+ MEFs reduced the frequency of nuclear blebbing (P < 0.0001). This result was consistent in several independently isolated LmnaHG/+ cell lines and in several independent experiments (Fig. 4). Of note, the frequency of blebbing in FTI-treated LmnaHG/+ MEFs was not different, as determined by the χ2 statistic, from the frequency of blebbing in FTI-treated or untreated Lmna+/+ MEFs.
Fig. 4.
Bar graph showing increased frequency of nuclear blebbing in LmnaHG/+ MEFs and a reduction in nuclear blebbing in LmnaHG/+ MEFs treated with an FTI (10 μM PB-43). A and B represent two independent experiments. Each black circle shows the frequency of nuclear blebbing with an independently isolated fibroblast cell line. Bars indicate the mean frequency of blebbing. The number of cells with nuclear blebs and the total number of cells examined are recorded within each bar. In both experiments, the FTI did not change the frequency of blebbing in Lmna+/+ MEFs, as determined by the χ2 test. LmnaHG/+ MEFs exhibited more blebbing than Lmna+/+ cells (P < 0.0001, χ2 test). An FTI reduced the frequency of blebbing in LmnaHG/+ MEFs (P < 0.0001, χ2 test). The frequency of blebbing in FTI-treated LmnaHG/+ MEFs was not different from the frequency of blebbing in the treated or untreated Lmna+/+ MEFs. Very similar results, with identical levels of statistical significance, were obtained when the microscopic slides were scored by a second blinded observer.
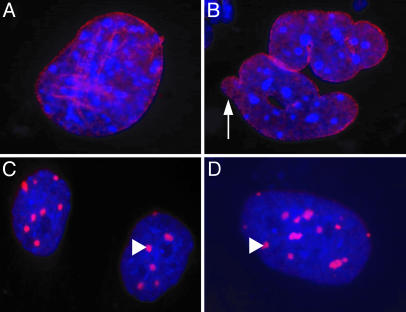
Because LmnaHG/+ MEFs synthesize lamin A and lamin C in addition to progerin, it was impossible to use immunofluorescence microscopy of LmnaHG/+ MEFs to define the location of progerin within those cells. However, the intracellular location of progerin could be examined in experiments with LmnaHG/HG cells, which synthesize exclusively progerin. In those cells, progerin was located mainly at the nuclear envelope (Fig. 5 A and B). However, in the presence of the FTI, virtually all of the progerin was mislocalized to intensely staining aggregates within the nucleoplasm, and none of it was detectable at the nuclear envelope (Fig. 5 C and D).
Fig. 5.
Immunofluoresence images showing the distribution of progerin in untreated and FTI-treated LmnaHG/HG cells. DNA was visualized with DAPI (blue), and progerin was visualized with an antibody against lamin A (red). (A and B) Untreated LmnaHG/HG cells, showing progerin along the nuclear envelope. Misshapen nuclei were common (arrow). (C and D) FTI-treated LmnaHG/HG cells, revealing intensely staining progerin aggregates (arrow-heads) in the nucleoplasm.
Discussion
FTIs were developed initially as anticancer agents (17). The concept was straightforward: to eliminate the farnesyl lipid from mutationally activated Ras proteins, thereby mislocalizing these signaling proteins away from the plasma membrane, where they “cause trouble” by stimulating uncontrolled cell division. In this study, we assessed an analogous concept with HGPS, which is a disease in which another farnesylated protein causes trouble in the cell. The synthesis of progerin leads to misshapen nuclei and frequent nuclear blebs (1). We hypothesized that blocking farnesylation would mislocalize progerin away from the nuclear envelope and ameliorate the nuclear blebbing phenotype. We generated a gene-targeted mouse model of HGPS, created genetically identical primary LmnaHG/+ MEFs as well as LmnaHG/HG cells, and tested the impact of an FTI on both nuclear blebbing and progerin localization. In untreated cells, progerin was located mainly along the nuclear envelope, whereas in FTI-treated cells the protein was mislocalized to the nucleoplasm. In the LmnaHG/+ MEFs, the FTI reduced nuclear blebbing to a baseline level observed in untreated wild-type cells. These studies visualize progerin localization independently of lamin A and lamin C, and they show that mislocalization of progerin is associated with a change in nuclear shape. Interestingly, the FTI did not adversely affect nuclear shape in wild-type fibroblasts.
Nuclear blebbing is the principal cellular phenotype in HGPS (13, 18), and the FTI clearly ameliorates this phenotype. The next obvious questions are as follows. Will the nuclear shape abnormalities in LmnaHG/+ cells be accompanied by any disease phenotypes at the “whole-animal” level? And, if so, would an FTI also ameliorate those disease phenotypes, including perhaps the atherosclerotic disease that claims the lives of most humans with the disease? With regard to the first question, the LmnaHG/+ mice clearly exhibit unequivocal disease phenotypes (S.H.Y., L.G.F., and S.G.Y., unpublished data). By 4 months of age, all of the LmnaHG/+ mice (n = 22) exhibit growth retardation and/or bone disease, akin to homozygous Zmpste24-deficient mice (4). Detailed radiographic and pathologic analyses (e.g., assessing bone density and defining atherosclerosis susceptibility in inbred backgrounds) will take many months to complete. However, the existence of tractable phenotypes in our heterozygous gene-targeted mice makes us confident that this model could be very useful for assessing therapeutic strategies.
To properly assess the impact of FTIs on disease phenotypes in mice, we first need to identify methods for the long-term oral delivery of the drugs and document that the drug is efficacious in blocking protein farnesylation in multiple tissues. Again, we anticipate that optimization of FTI delivery schemes will require considerable experimentation. In the meantime, we can only speculate about whether FTIs might be useful for treating the disease phenotypes in the LmnaHG/+ mice. Optimists would contend that it makes intuitive sense (i.e., in line with Occam's razor) that improvements in “whole-animal” disease phenotypes would parallel improvements in cellular phenotypes. Optimists would also point out that the nuclear blebbing phenotypes in cultured fibroblasts and the whole-animal disease phenotypes were clearly associated in a recent study of Zmpste24-deficient mice (12). Zmpste24-deficient fibroblasts, which accumulate wild-type farnesyl prelamin A, manifest nuclear blebbing (6, 12), and Zmpste24-deficient mice develop a variety of progeria-like phenotypes (4, 12). When prelamin A synthesis was reduced by 50% (by introducing a single copy of a Lmna knockout allele), the nuclear blebbing in fibroblasts and the disease phenotypes in mice were eliminated (12). That study was also intriguing because it proved that dramatic improvements in progeria-like disease phenotypes can occur with a 50% reduction in the amount of farnesyl prelamin A in the cell. Thus, one might easily imagine that an FTI could ameliorate the disease phenotypes even with incomplete inhibition of farnesylation and incomplete mislocalization of progerin (i.e., without pushing FTI doses to levels associated with side effects). However, drug toxicity may not be much of an issue because FTIs have been generally well tolerated in humans and in mice (19, 20). Genetic studies also support the idea that it should be possible to give FTIs safely on a long-term basis (21). Completely inactivating FTase in adult mice by using Cre/loxP approaches did not cause histopathological abnormalities or any noticeable disease phenotypes (21).
There are also reasons to be pessimistic about the proposition that an FTI would be a panacea for HGPS. Although an FTI mislocalizes progerin and improves nuclear blebbing, one could imagine that nonfarnesylated progerin could still be toxic in vivo. Nonfarnesylated progerin is still a structurally abnormal protein, and it is sobering to remember that even single amino acid substitutions in mature lamin A and lamin C (neither of which is farnesylated) can cause a host of different genetic diseases (e.g., several forms of muscular dystrophy, cardiomyopathy, partial lipodystrophy, and mandibuloacral dysplasia) (2, 3). Note in particular that some mutations in the carboxyl-terminal region of lamin A (i.e., the region not shared with lamin C) cause human genetic diseases (22, 23). Moreover, one could argue that improved nuclear blebbing in cultured cells might not be an accurate indicator of disease phenotypes at the whole-animal level, for the simple reason that some LMNA missense mutations have been reported to cause human disease without affecting nuclear shape in cultured fibroblasts (22). Last, a pessimist would certainly point out that HGPS is a dominant disease, caused by a single mutant chromosome. Treatment with an FTI interferes with the biogenesis of mature lamin A from the normal LMNA allele. To our knowledge, the short or long-term consequences of reducing lamin A biogenesis in humans are not known, although a single LMNA null allele, causing a 50% reduction in both lamin A and lamin C synthesis, causes muscular dystrophy (24).
In this study, we sought to create a HGPS allele that yielded large amounts of progerin, and our strategy was successful. Although it may have been technically simpler to introduce the codon 608 point mutation identified in humans with HGPS (24), we did not follow this approach because we worried that that the “point mutation” approach would not yield sufficient amounts of progerin to elicit phenotypes in the mouse. In humans with HGPS, most of the transcripts from the mutant LMNA allele actually yield wild-type lamin C and wild-type lamin A rather than progerin (1). We worried about this type of an allele for mouse experimentation because experience with mouse Lmna mutations has suggested that mice may be “tougher” than humans when it comes to the dose of mutant lamin proteins required to elicit disease phenotypes. For example, humans carrying a single LMNA H222P mutation develop muscular dystrophy, whereas two copies of the H222P allele are required to elicit a muscle phenotype in mice (25). Similarly, humans heterozygous for a LMNA nonsense mutation develop muscular dystrophy (24), whereas mice with a single Lmna knockout allele are normal (26). Thus, we worried that the point mutation approach in the mouse might not yield sufficient levels of progerin to elicit phenotypes. Accordingly, we chose a gene-targeting strategy that would guarantee high levels of progerin expression. As it turned out, we generated the first Lmna mutant mice that exhibit a phenotype with a single copy of the mutant allele.
Progerin is clearly the “culprit molecule” in HGPS (1, 13, 18), and studies raise the hope that FTIs might ultimately prove to be useful for treating HGPS. The FTI strategy appears to be well suited for testing in children because the drugs have been studied extensively, are generally well tolerated, and can be given orally. However, FTIs are not the only potential hope for HGPS. Recent studies have indicated that the nuclear blebbing phenotype in HGPS fibroblasts can be ameliorated with morpholino antisense reagents (18) or by expressing short hairpin RNA constructs (RNA interference) (Junko Oshima, personal communication). These RNAi and antisense methods directly reduce the production of progerin transcripts, so the finding of reduced nuclear blebbing is probably not particularly surprising. Nevertheless, those studies were very important because they provided hope to patients affected by HGPS and because they will stimulate interest in overcoming the practical and pharmacological obstacles to delivering RNAi and morpholino oligonucleotides to humans.
Acknowledgments
We thank Dr. Luanne Peters for counting chromosomes in the LmnaHG/HG ES cells. This work was supported in part by National Institutes of Health Grants AI054384 (to M.H.G.), R01 CA099506, and R01 AR050200 and a grant from the Progeria Research Foundation (to S.G.Y.).
Author contributions: L.G.F. designed research; S.H.Y., M.O.B., J.I.T., X.Q., Y.H., S.S., and M.M. performed research; S.H.Y., S.G.Y., and L.G.F. analyzed data; P.B. and M.H.G. contributed new reagents/analytic tools; and S.H.Y., S.G.Y., and L.G.F. wrote the paper.
Abbreviations: HGPS, Hutchinson–Gilford progeria syndrome; FTase, protein farnesyltransferase; FTI, FTase inhibitor; MEF, mouse embryonic fibroblast.
References
- 1.Eriksson, M., Brown, W. T., Gordon, L. B., Glynn, M. W., Singer, J., Scott, L., Erdos, M. R., Robbins, C. M., Moses, T. Y., Berglund, P., et al. (2003) Nature 423, 293-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mounkes, L. C., Burke, B. & Stewart, C. L. (2001) Trends Cardiovasc. Med. 11, 280-285. [DOI] [PubMed] [Google Scholar]
- 3.Burke, B. & Stewart, C. L. (2002) Nat. Rev. Mol. Cell Biol. 3, 575-585. [DOI] [PubMed] [Google Scholar]
- 4.Bergo, M. O., Gavino, B., Ross, J., Schmidt, W. K., Hong, C., Kendall, L. V., Mohr, A., Meta, M., Genant, H., Jiang, Y., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 13049-13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai, Q., Choy, E., Chiu, V., Romano, J., Slivka, S. R., Steitz, S. A., Michaelis, S. & Philips, M. R. (1998) J. Biol. Chem. 273, 15030-15034. [DOI] [PubMed] [Google Scholar]
- 6.Pendás, A. M., Zhou, Z., Cadiñanos, J., Freije, J. M. P., Wang, J., Hultenby, K., Astudillo, A., Wernerson, A., Rodríguez, F., Tryggvason, K. & Lopéz-Otín, C. (2002) Nat. Genet. 31, 94-99. [DOI] [PubMed] [Google Scholar]
- 7.Corrigan, D. P., Kuszczak, D., Rusinol, A. E., Thewke, D. P., Hrycyna, C. A., Michaelis, S. & Sinensky, M. S. (2005) Biochem. J. 387, 129-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hennekes, H. & Nigg, E. A. (1994) J. Cell Sci. 107, 1019-1029. [DOI] [PubMed] [Google Scholar]
- 9.Lutz, R. J., Trujillo, M. A., Denham, K. S., Wenger, L. & Sinensky, M. (1992) Proc. Natl. Acad. Sci. USA 89, 3000-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izumi, M., Vaughan, O. A., Hutchison, C. J. & Gilbert, D. M. (2000) Mol. Biol. Cell 11, 4323-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalton, M. & Sinensky, M. (1995) Methods Enzymol. 250, 134-148. [DOI] [PubMed] [Google Scholar]
- 12.Fong, L. G., Ng, J. K., Meta, M., Cote, N., Yang, S. H., Stewart, C. L., Sullivan, T., Burghardt, A., Majumdar, S., Reue, K., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 18111-18116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldman, R. D., Shumaker, D. K., Erdos, M. R., Eriksson, M., Goldman, A. E., Gordon, L. B., Gruenbaum, Y., Khuon, S., Mendez, M., Varga, R. & Collins, F. S. (2004) Proc. Natl. Acad. Sci. USA 101, 8963-8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mortensen, R. M., Conner, D. A., Chao, S., Geisterfer-Lowrance, A. A. T. & Seidman, J. G. (1992) Mol. Cell. Biol. 12, 2391-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nallan, L., Bauer, K. D., Bendale, P., Rivas, K., Yokoyama, K., Hornéy, C. P., Pendyala, P. R., Floyd, D., Lombardo, L. J., Williams, D. K., et al. (2005) J. Med. Chem. 48, 3704-3713. [DOI] [PubMed] [Google Scholar]
- 16.Raharjo, W. H., Enarson, P., Sullivan, T., Stewart, C. L. & Burke, B. (2001) J. Cell Sci. 114, 4447-4457. [DOI] [PubMed] [Google Scholar]
- 17.Reiss, Y., Goldstein, J. L., Seabra, M. C., Casey, P. J. & Brown, M. S. (1990) Cell 62, 81-88. [DOI] [PubMed] [Google Scholar]
- 18.Scaffidi, P. & Misteli, T. (2005) Nat. Med. 11, 440-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caraglia, M., D'Alessandro, A. M., Marra, M., Giuberti, G., Vitale, G., Viscomi, C., Colao, A., Prete, S. D., Tagliaferri, P., Tassone, P., et al. (2004) Oncogene 23, 6900-6913. [DOI] [PubMed] [Google Scholar]
- 20.Rogers, M. J. (2003) Curr. Pharm. Des. 9, 2643-2658. [DOI] [PubMed] [Google Scholar]
- 21.Mijimolle, N., Velasco, J., Dubus, P., Guerra, C., Weinbaum, C. A., Casey, P. J., Campuzano, V. & Barbacid, M. (2005) Cancer Cell 7, 313-324. [DOI] [PubMed] [Google Scholar]
- 22.Muchir, A., Medioni, J., Laluc, M., Massart, C., Arimura, T., van der Kooi, A. J., Desguerre, I., Mayer, M., Ferrer, X., Briault, S., et al. (2004) Muscle Nerve 30, 444-450. [DOI] [PubMed] [Google Scholar]
- 23.Csoka, A. B., Cao, H., Sammak, P. J., Constantinescu, D., Schatten, G. P. & Hegele, R. A. (2004) J. Med. Genet. 41, 304-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonne, G., Di Barletta, M. R., Varnous, S., Bécane, H.-M., Hammouda, E.-H., Merlini, L., Muntoni, F., Greenberg, C. R., Gary, F., Urtizberea, J.-A., et al. (1999) Nat. Genet. 21, 285-288. [DOI] [PubMed] [Google Scholar]
- 25.Arimura, T., Helbling-Leclerc, A., Massart, C., Varnous, S., Niel, F., Lacene, E., Fromes, Y., Toussaint, M., Mura, A. M., Keller, D. I., et al. (2005) Hum. Mol. Genet. 14, 155-169. [DOI] [PubMed] [Google Scholar]
- 26.Sullivan, T., Escalante-Alcalde, D., Bhatt, H., Anver, M., Bhat, N., Nagashima, K., Stewart, C. L. & Burke, B. (1999) J. Cell Biol. 147, 913-919. [DOI] [PMC free article] [PubMed] [Google Scholar]