Abstract
There is an increasing demand for easy, high-throughput (HTP) methods for protein engineering to support advances in the development of structural biology, bioinformatics and drug design. Here, we describe an N- and C-terminal cloning method utilizing Gateway cloning technology that we have adopted for chimeric and mutant genes production as well as domain shuffling. This method involves only three steps: PCR, in vitro recombination and transformation. All three processes consist of simple handling, mixing and incubation steps. We have characterized this novel HTP method on 96 targets with >90% success. Here, we also discuss an N- and C-terminal cloning method for domain shuffling and a combination of mutation and chimeragenesis with two types of plasmid vectors.
INTRODUCTION
As structural and in silico biology have advanced, systematic and robust high-throughput (HTP) methods for protein engineering are required to generate mutants that will allow us to model and analyze protein function. Production of protein drugs, including chimeric antibodies, requires a method for mutagenesis and chimeragenesis. While many methods for chimeragenesis, such as domain shuffling (1,2), have been developed, the majority utilize complex processes to obtain the desired protein and are not amenable to HTP automation.
Recently, the cloning systems with in vitro recombination were developed (3,4) and are amenable to HTP cloning. Gateway cloning technology, one of such systems, utilizes the bacteriophage λ recombination system in vitro (4). We characterized this recombination system and succeeded in introducing N- and C-terminal fragments on each recombination site by in vitro recombination and cloning in an HTP manner. This cloning strategy represents an easy HTP method for site-directed chimeragenesis, protein engineering and mutagenesis.
In the Gateway cloning system, a fragment carrying attB1 and attB2 recombination sites on both ends is prepared by PCR and introduced into a Donor vector carrying attP1 and attP2 recombination sites by an in vitro recombination reaction driven by BP clonase. attB1, attB2, attP1 and attP2 sequences are designed from natural attB and attP sequences, respectively, to obtain specificity without reduced recombination efficiency. There are two processes in Gateway cloning: the first is intermolecular recombination between a fragment and a plasmid at attB1 and attP1 or attB2 and attP2 and the second is intramolecular recombination. We have developed a novel strategy for Gateway cloning, named ‘N- and C-term’ (N- and C-terminal) Gateway cloning, in which two independent fragments, each carrying attB1 or attB2 on their ends, are combined with a single plasmid containing both attP1 and attP2 sites. The resulting linear constructs carrying both N- and C-terminal fragments are circularized and the Gateway clone is constructed. It was reported that homologous recombination in Escherichia coli was available for DNA cloning (5,6). We adopted homologous recombination in E.coli to circularize a plasmid. This cloning technique offers a rapid, automatable way to construct chimeric genes and introduce point mutations, deletions, insertions and substitutions at any place in the target genes as required.
MATERIALS AND METHODS
PCR conditions
An aliquot of 20 pmol of primers and 50 ng of template DNA were used to obtain N- and C-terminal fragments by PCR with 0.5 U Pfx polymerase (Invitrogen Corp., Carlsbad, CA) in 25 μl reaction mixture. Cycling parameters for PCR were 15 cycles, consisting of 1 min at 95°C for denaturing, 2 min at 55°C for annealing, 3 min at 68°C for extension and 5 min at 68°C for final extension.
BP in vitro recombination
BP recombination was according to the industrial manual with a half scale. Our condition is as follows: PCR products containing N- and C-terminal fragments (2.5 μl each) and 150 ng pDONR 201 were combined in a 10 μl reaction mixture containing 1 μl BP clonase (Invitrogen Corp.). It is not necessary to purify PCR products. The mixture was incubated at 25°C for 16 h.
Transformation
After the reaction was stopped by proteinase K, 2.5 μl of reaction mixture was mixed with 25 μl DH5α competent cell (Invitrogen Corp.) for transformation. The procedure was according to the industrial manual. All cells were spread on Luria–Bertani (LB) plate containing 50 μg/ml Kanamycin or 25 μg/ml Zeocin.
RESULTS
Experimental design
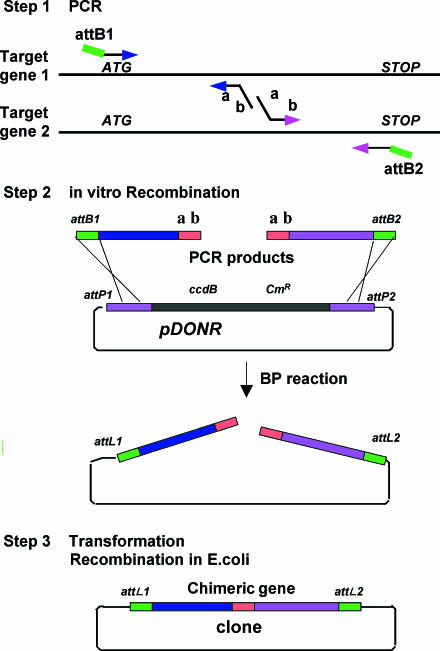
The general outline of N- and C-terminal Gateway cloning method for chimeragenesis is illustrated in Figure 1. N- and C-terminal fragments were obtained by PCR with attB attached primers and internal gene primers. Internal gene primers were designed to create a homologous region at the 3′ end of N-terminal and 5′ of C-terminal fragments. Symbols a and b in Figure 1 indicate gene specific sequences of joining sites of the 3′ end of N- and 5′ end of C-terminal fragments, respectively. These fragments were then introduced into a Donor vector (pDONR) by BP recombination (without any purification steps). E.coli was then transformed with the linear plasmid product. The desired, mutated clone was obtained after the linear plasmid was circularized in the E.coli cell utilizing the homologous sequences at its ends.
Figure 1.
Outline of chimeragenesis with Gateway technology. For details see text.
The length of homologous sequences required for plasmid circularization in E.coli
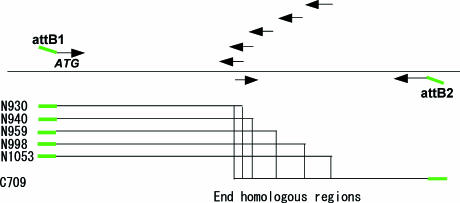
To address the question of the minimal length of homologous sequences required for this cloning method, we prepared five attB1-N-terminal fragments containing various lengths of homology when paired with a single attB2-C-terminal. Five attB1-N-terminal fragments, N930, N940, N959, N998 and N1053, and one attB2-C-terminal fragment, C709, were produced by PCR. The length of each attB1-N- and attB2-C-terminal fragment was 930, 940, 959, 998, 1053 and 709 bp, respectively. Five attB1-N-terminal fragments had 10, 20, 39, 78 and 133 bp homologous sequences to attB2-C-terminal fragment at their ends. The design of PCR and products used for the BP reaction are shown in Figure 2.
Figure 2.
Preparation of N- and C-terminal fragments with different lengths of homologous sequences on the ends. Five N-terminal and one C-terminal fragments were prepared by PCR. The combinations of N- and C-terminal fragments gave 10, 20, 39, 78 and 133 bp homologous sequences on the ends of each fragment.
Each attB1-N-terminal fragment, attB2-C-terminal fragment and pDONR201 were mixed at a molar ratio of 5:5:1 and BP recombination was carried out. After stopping the reaction with proteinase K, linear DNA samples were directly introduced into E.coli DH5α (recA strain). The number of colonies obtained from 37.5 ng vector DNA and the efficiency of clone recovery for each condition are shown in Table 1. Even 10 bp homologous region gave 25 colonies. We analyzed the sequences of the junction region between the N- and C-terminal fragment to check the fidelity of homologous recombination in the recA strain. None of the six clones sequenced from each condition showed unexpected sequences in the junction region. This indicated that a 10 bp homologous sequence was sufficient for correct homologous recombination at the ends of the plasmid, although the number of clones obtained was smaller than with other conditions.
Table 1.
Relation between the length of homologous region and efficiency of cloning
| Length of homologous region | 10 bp | 20 bp | 39 bp | 78 bp | 133 bp |
|---|---|---|---|---|---|
| Number of colonies | 25 | 110 | 334 | 367 | 406 |
| Rate of right clone | 6/6 | 6/6 | 6/6 | 6/6 | 6/6 |
The effect of homologous sequence in the target gene
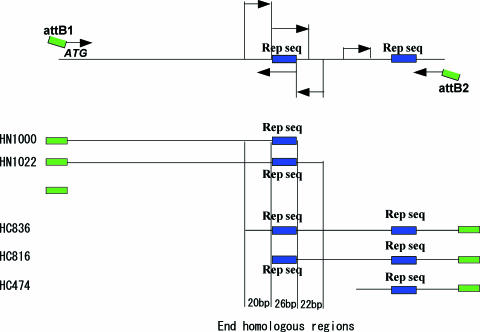
Some genes contain tandem repeats or short homologous sequences. We wanted to determine the effect of such homologous sequences when using the N-term C-term cloning method. To address this question we constructed a clone containing a tandem repeat. We chose a 1732 bp cDNA for the test gene. This cDNA carried two 26 bp identical sequences, named ‘rep seq’ sites, and several homologous regions, whose size and homology were 10–30 bp and 70–90%. The ‘rep seq’ sites are indicated with blue boxes in Figure 3. We prepared two N-terminal, HN1000 and HN1022, and three C-terminal fragments, HC836, HC816 and HC474, by PCR. HN1000 and HN1022 fragments carried ‘rep seq’ sites at 3′ end and 22 bp inside of the end, respectively. Both HC816 and HC836 had two ‘rep seq’ sites, one proximal and one distal. The proximal ‘rep seq’ sites of HC816 and HC836 were located at 5′ end and 22 bp inside of the end, respectively. The distance between the end and distal ‘rep seq’ site was several hundred base pairs. HC474 had only a distal ‘rep seq’ site.
Figure 3.
Preparation of N- and C-terminal fragments from a template gene carrying identical sequences. The target gene had two identical sequences named ‘rep seq’ indicated by blue boxes. Two N-terminal fragments, HN1000 and HN1022, and three C-terminal fragments, HC836, HC816 and HC474, were prepared by PCR. HC836 and HC816 had two ‘rep seq’ on their 5′ ends and inside of the fragments. The other fragments had only one rep seq'.
We measured the cloning efficiency and reliability with various combinations of N and C fragments. The combinations HN1000/HC836, HN1000/HC816 and HN1022/HC816 carried 46, 26 and 48 bp homologous sequence at their ends and a 26 bp homologous sequence of ‘rep seq’ sites, respectively. The combinations HN1000/HC474 and HN1022/HC474 had no end homologous sequence but a 26 bp homologous sequence of ‘rep seq’ sites. We randomly selected 16 colonies from each experiment for characterization. The number of colonies obtained from 37.5 ng vector DNA and the ratio of correct clones are shown in Table 2. Sequence analysis of the resultant clones from the combinations HN1000/HC474 and HN1022/HC474 showed that recombination had occurred between ‘rep seq’ sites at the end of the N-terminal fragment and the mid region of the C-terminal fragment. The ratios of homologous recombination between two ‘rep seq’ sites obtained by the combinations HN1000/HC474 and HN1022/HC474 were 12/16 and 8/16, respectively. The remaining clones in these experiments showed recombination between the other homologous regions.
Table 2.
Relation between the length of homologous region and cloning efficiency for the target containing tandem repeats
| Combination of fragments | NH1000 × HC474 | NH1022 × HC474 | NH1000 × HC816 | NH1000 × HC836 | NH1022 × HC816 |
|---|---|---|---|---|---|
| End homologous region (bp) | 0 | 0 | 26 | 46 | 48 |
| Number of colonies | 132 | 37 | 124 | 76 | 195 |
| Recombination between the end of N-terminal and C-terminal fragments | – | – | 6/16 | 15/16 | 14/16 |
| Recombination between the end of N-terminal fragment and inside ‘rep seq’ of C-terminal fragment | 12/16 | 8/16 | 5/16 | 1/16 | 1/16 |
| Others | 4/16 | 8/16 | 5/16 | 0/16 | 1/16 |
The combination HN1000/HC816, which had a 26 bp homologous region at both ends of N- and C-terminal fragments, was tested for efficiency to produce specific designed clones by end recombination. The ratio of designed clone obtained by the end recombination was 6/16. The ratio of the recombination between ‘rep seq’ of the N-terminal fragment and the mid C-terminal fragment was 5/16. The ratio of the recombination among other homologous sequence was also 5/16. The combinations HN1000/HC836 and HN1022/HC816 showed 46 and 48 bp homologous region at the ends, respectively, and the frequency of the end recombination of these combinations was very high at 15/16 and 14/16, respectively. These results suggested that the combination of N- and C-terminal fragments with long, (∼40–50 bp), homologous regions at their ends produced the desired clones with high reliability even if the gene had short homologous regions, such as ∼30 bp identical sequence.
The amount of N- and C-terminal fragments required for cloning
We analyzed the range and suitable amount of DNA fragments for cloning. We selected the combination N959/C709, whose length of end homologous region was 39 bp, for this analysis. We measured colony forming ability and reliability of cloning with a combination of different molar ratios of fragments and 150 ng pDONR vector, from 1/32:1/32:1 to 32:32:1. The amount of PCR fragment was from 1.56 to 1636 ng. After the BP reaction, we introduced linear plasmid, which had 37.5 ng vector DNA, into E.coli and counted the number of colonies. We selected eight colonies from each experiment and sequence analyzed these clones. The results are summarized in Table 3. For cloning of this 1.5 kb gene, the optimal ratio of fragments and a vector was 2:1. Using this condition, 425 colonies were obtained with all eight sequenced clones having the desired sequence. This experiment showed that the desired clone was obtained with high reliability using a wide variation in the relative quantity. Even when using <2 ng of insert fragment, 3% of vector DNA gave the right clone although the rate of the right clone was only 1/8.
Table 3.
Relation between the amount of N- and C-terminal fragments and cloning efficiency
| Ratio between vector and fragment | 1/32 | 1/16 | 1/8 | 1/4 | 1/2 | 1/1 | 2 | 4 | 8 | 16 | 32 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of colonies | 15 | 14 | 58 | 177 | 203 | 362 | 425 | 397 | 273 | 157 | 77 |
| Rate of right clone | 1/8 | 6/8 | 6/8 | 5/8 | 7/8 | 6/8 | 8/8 | 8/8 | 8/8 | 7/8 | 7/8 |
HTP Gateway cloning with N- and C-terminal fragments
HTP methods require simple procedures, which will work for a wide range of amount of substrates to reliably complete each reaction. The results of all experiments described above showed that the N- and C-terminal cloning method using the Gateway system incorporate not only easy handling procedures and high cloning efficiency but also that the reliability of this method was very high when the ends of N- and C-terminal fragments were designed to carry 40 bp or longer homologous sequences for recombination in E.coli. These results demonstrate that our N- and C-terminal cloning method was suitable for HTP cloning and chimeragenesis.
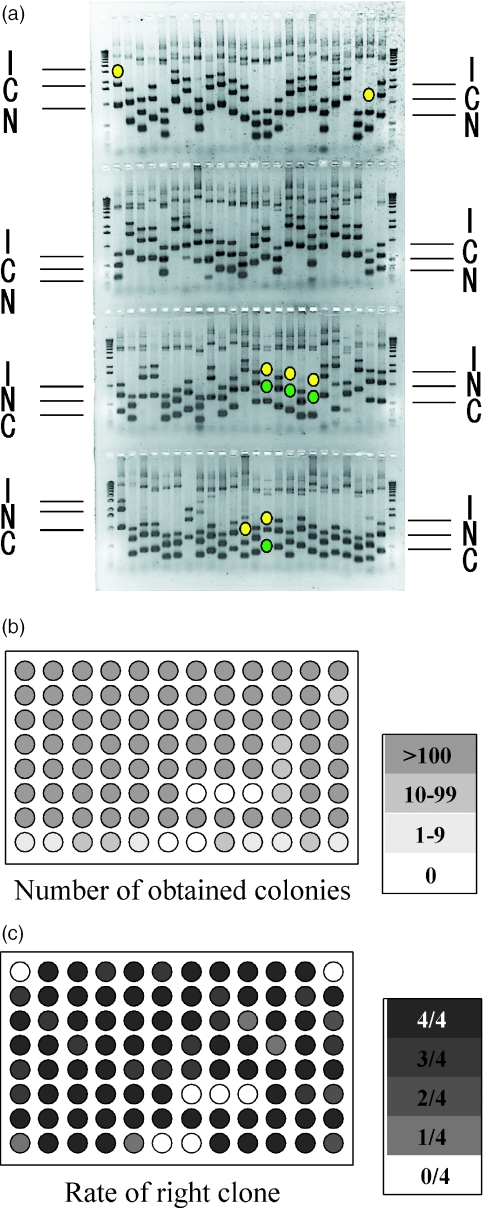
We measured the success rate of this method for HTP handling with 96 target genes. These samples were randomly chosen from human full-length cDNA library. The sizes of their open reading fragment were from 603 to 3381 bp. Among these clones, 37 clones had 603–1000 bp, 39 clones had 1001–2000 bp and 20 clones had more than 2000 bp long target regions for cloning. The primers for amplification of N- and C-terminal fragments were designed to produce 30–60% full-length fragments containing 40 bp end homologous sequences. Ninety-six combinations of N- and C-terminal fragments were mixed with 150 ng pDNOR201 vector and in vitro BP recombination reactions were carried out. One-fourth of each reaction mixture was used to transform competent E.coli and the cells were plated on LB containing 50 μg/ml of Kanamycin. After the colonies were counted, four colonies from each plate were randomly selected, and the length of inserts and their sequences were analyzed. The amplified N- and C-terminal fragments and inserts of clones are shown in Figure 4a. Cloning efficiency and rates of the desired clones are illustrated in Figure 4b and c, respectively. No colonies were obtained from five targets, four of which did not produce N- or C-terminal fragments by PCR (indicated by green dots in Figure 4a). A total of 66 out of 96 reactions (69%) gave only correct clones and 16 out of 96 (17%) gave only one wrong clones when four clones from each point were analyzed. More than 85% (82 out of 96) gave the desired clones with high reliability. Cloning failed completely for only 7 out of 96 targets using this system equating to a 93% success rate when four clones from each reaction were checked.
Figure 4.
Summary of HTP N- and C-terminal cloning. (a) Gel showed N- and C-terminal fragments used for cloning and inserted fragments of clones. Symbols N and C indicate N- and C-terminal fragments, and I indicates insert fragments amplified by PCR. The dots indicate the position of missing fragments; green for N- or C-terminal; and yellow for right insert fragments. (b and c) Number of obtained colonies and the rate of right clone.
Introduction of mutations
The N- and C-terminal Gateway cloning method was also applicable to site-directed mutagenesis, introducing point mutation, deletion, insertion or substitution at any position in the gene according to the primer design. When two priming sites were separated from each other, the resultant clone by the N- and C-terminal method was a deletion clone that lacked the sequence between the two priming sites. Using mutated primer and primers containing additional sequences at the 5′ end of oligo, DNA produced a point mutant and insertion clone, respectively. The combination of deletion and insertion procedures gave a substitution clone. We constructed a point mutation, 120 bp deletion, 80 bp insertion and 80 bp substitution clones with 1.5 kb cDNA and evaluated these mutant clones by sequencing. We checked six clones of each mutant, and all clones showed the designed sequence in the whole region of the gene.
DISCUSSION
There is an increasing requirement for HTP protein engineering methods to meet the demands of drug discovery and the development of chimeric antibodies for human healthcare. In an attempt to meet this demand, the Multisite Gateway technology was reported (7,8). This system assembled multiple DNA sequences and makes domain shuffling easily. However, this system required the recombination sequences, an att site, between assembled DNA segments and added several amino acid residues between domains. To improve this point, we developed an HTP N- and C-terminal cloning method using the Gateway system. The Gateway system uses in vitro site-specific recombination generating clones without requirement for restriction enzyme digestion and ligation. BP reactions are highly efficient and we obtained desired clones using <10 pg PCR fragments for 1 kb targets (data not shown).
HTP methods for cloning should allow the handling of numerous samples and generate the desired clones in a short period. We demonstrated that our N- and C-terminal cloning system showed not only reliability, ease of production and screening, but also ability to utilize a wide range of substrates, and that high success rate was shown when 96 samples were handled. Our N- and C-terminal cloning method contained only three steps: PCR, BP in vitro recombination and transformation. The simplicity of the PCR process (mixing primer set, template DNA, reaction solution and enzyme) facilitates establishment of this HTP method. The BP reaction requires simple mixing of PCR products, pDONR vector and reaction mixture without any purification step. The transformation step also requires only mixing of samples after the BP reaction and competent cells without any purification step. Therefore, all steps can easily be automated.
We usually use 50–100 ng template plasmid DNA for 15 cycles PCR with Pfx polymerase (Invitrogen Corp.) and obtained 10–100 ng PCR fragments. In the case of our test gene in this paper, which was 1.5 kb, we obtained the desired clones efficiently with 3 ng to 1.6 μg PCR fragments. Fifteen cycles of PCR with high fidelity PCR enzyme, such as Pfx or KOD, gave 1/10 kb error rate from our analysis of more than 10 000 clones. The error rate of synthetic oligo DNA for cloning primer (synthesized by Invitrogen Corp., Japan) was 1/2000 bp according to the analysis of 2500 clones. The test experiment for the HTP method described in this paper failed to gain colonies against five templates. We characterized 364 clones, 91 target genes × 4 sub-clones, and observed 42 incorrect clones. This showed that accuracy was 88.5%. Only 2 out of 42 incorrect clones showed a base substitution error. The other showed different sizes of inserts or vector background. Such size differences were easily observed by colony PCR and gel electrophoresis. This suggested that the accuracy of this HTP method was very high and colony PCR was sufficient for selection of desired clones. All of our results demonstrated that the N- and C-terminal cloning method described here was suitable for HTP chimeragenesis and site-specific mutagenesis.
Since two types of pDONR vector carrying different antibiotics resistant genes, Kanamycin and Zeocin, exist, serial N- and C-terminal cloning for chimeragenesis and domain shuffling are possible. The length of insertion and substitution depends on the length of reliable oligo primers. Reasonable length for the reliable primers is 80mer. This length creates an 80 bp insertion because 40 bp is required for homologous recombination in E.coli and 20 bp is used as the priming site for PCR. We established one-step adapter PCR, using four primers in one tube PCR, as described previously (9). This method allowed a 200 bp insertion. Serial cloning also gives longer insertion and substitution.
Translocation of chromosomes produces chimeric proteins underlying many diseases, including hematological malignancies and solid tumors. Mitelman et al. (10) reported that 209 fusion genes on hematological malignancies and 271 fusion genes on solid tumors. The characterization of chimeric proteins is important to understand the mechanism of such diseases. However, one of the biggest problems is the difficulty to obtain full-length cDNA clones with fusion from patents. Recently, several projects produced huge number of human full-length cDNAs (11,12), which covered most genes of human. The method presented here can create all chimeric clones using the established full-length cDNAs and will open the cyclopedic study on functional analyses of fusion proteins in cancer.
In this paper, we introduced the method for protein engineering included chimera protein production. Our method was for assembling two DNA fragments and constructing plasmids by HTP way. This method allowed introducing any kinds of sequences, such as regulatory sequences, localization signals and control elements at any place of genes and had the wide application. We believe that this method helps the works of many research fields.
Acknowledgments
The authors thank Drs J. Chesnut and M. Jones for critical reading of the manuscript. This work was supported by a grant from the NEDO Project of Ministry of Economy, Trade and Industry. Funding to pay the Open Access publication charges for this article was provided by Invitrogen Japan, Invitrogen Corporation.
Conflict of interest statement. None declared.
REFERENCES
- 1.O'Maille P.E., Bakhtina M., Tsai M.-D. Structure-based combinatorial protein engineering (SCOPE) J. Mol. Biol. 2002;321:677–691. doi: 10.1016/s0022-2836(02)00675-7. [DOI] [PubMed] [Google Scholar]
- 2.Hiraga K., Arnold F.H. General method for sequence-independent site-directed chimeragenesis. J. Mol. Biol. 2003;330:287–296. doi: 10.1016/s0022-2836(03)00590-4. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R.W., Velappan N., Pavlik P., Chasteen L., Bradbury A. Recombinatorial cloning using heterologous lox sites. Genome Res. 2004;14:1119–1129.. doi: 10.1101/gr.1821804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartley J.L., Temple G.F., Brasch M.A. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y., Buchholz F., Muyrers J.P.P., Stewart A.F. A new logic for DNA engineering using recombination in Escherichia coli. Nature Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 6.Li M.Z., Elledge S.J. MAGIC, an in vivo genetic method for the rapid construction of recombinant DNA molecules. Nature Genet. 2005;37:311–319. doi: 10.1038/ng1505. [DOI] [PubMed] [Google Scholar]
- 7.Sasaki Y., Sone T., Yoshida S., Yahata K., Hotta J., Chesnut J.D., Honda T., Imamoto F. Evidence for high specificity and efficiency of multiple recombination signals in mixed DNA cloning by the Multisite Gateway system. J. Biotechnol. 2004;107:233–243. doi: 10.1016/j.jbiotec.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Cheo D.L., Titus S.A., Byrd D.R.N., Hartley J.L., Temple G.F., Brash M.A. Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: functional analysis of multi-segment expression clones. Genome Res. 2004;14:2111–2120. doi: 10.1101/gr.2512204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kagawa N., Kemmochi K., Tanaka S. One-step adapter PCR method for HTP Gateway technology cloning. Quest. 2004;1:53–55. [Google Scholar]
- 10.Mitelman F., Johansson B., Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nature Genet. 2004;36:331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- 11.Strausberg R.L., Feingold E.A., Grouse L.H., Derge J.G., Klausner R.D., Collins F.S., Wagner L., Shenmen C.M., Schuler G.D., Altschul S.F., et al. Generation and initial analysis of more than 15 000 full-length human and mouse cDNA sequences. Proc. Natl Acad. Sci. USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., et al. Complete sequencing and characterization of 21 243 full-length human cDNAs. Nature Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]