Abstract
To investigate the function of transcription factor Sp1 in β-like globin gene activation, we analyzed the recruitment of Sp1, fetal Krüppel-like factor 2 (FKLF2), and related factors at the human β-globin locus in a human fetal liver and mouse erythroleukemia hybrid cell (A181γ cell) that contains a single copy of human chromosome 11. Sp1 binds at the GT boxes of the cis-elements throughout the β-locus, but it is phosphorylated and lost over DNase I hypersensitive site (HS)2, HS3, HS4, and the human β-globin gene promoter after A181γ cell differentiation. The binding of FKLF2 at HS2 and HS3 was unchanged. Histone deacetylase 1, which could be recruited by Sp1, is also lost over HS2 and HS3 after differentiation, resulting in the acetylation of histones 3 and 4 across the human β-globin locus. We previously detected in vivo GT footprints over the β-globin locus after A181γ differentiation. Here, we report that after differentiation, the p300/CREB-binding protein-associated factor is recruited by FKLF2 to the locus control region to acetylate histones 3 and 4 at the human β-globin gene locus. Our results suggest that Sp1 is an inhibitor of β-like globin gene transcription during erythroid terminal differentiation. Its phosphorylation and release allow the erythroid-specific FKLF2 or erythroid Krüppel-like factor to interact with other erythroid-specific transcription factors to initiate the transcription of β-like globin genes.
Keywords: β-globin gene regulation, erythroid-specific transcription factors, chromatin immunoprecipitation
The human α- and β-globin genes are arranged in the order of their expression during ontogeny and expressed in a tissue-specific manner in erythroid cells. They provide an interesting, although elaborate, model for the study of eukaryotic gene regulation (1). The expression of the β-like globin genes is under the influence of the locus control region (LCR), which consists of at least five DNase I hypersensitive sites (HS), HS1–5, upstream of the ε-globin gene (2). The LCR is rich in sequences that bind erythroid-specific transcription factors such as GATA-1 and NF-E2, as well as general transcriptional factors such as Sp1. Gene expression is regulated by the LCR at two levels: the formation of an active chromatin structure and the activation of transcription (3). Previous studies suggest that the formation of an active chromatin domain involves histone modifications and dynamic changes in transcriptional factor binding that result in changes in in vivo GT (GGTGTGGGG) and GC (GGGGCGGGG) footprint patterns in the human β-globin locus (4, 5).
The ubiquitously expressed Sp1 zinc-finger protein is the first described member of Krüppel-like factors known as Sp1/XKLF transcription factor family that binds to the consensus sequences of the GC and GT boxes (6, 7). The GC and GT boxes can be found in many tissue-specific regulatory sequences such as the β-globin gene promoter and hypersensitive sites (8), as well as ubiquitously expressed gene promoters and enhancers of house-keeping genes such as the human translation termination factor 1 gene (9). Although the binding of erythroid Krüppel-like factor (EKLF) and fetal Krüppel-like factor 2 (FKLF2) proteins at these sequences of the human β-globin locus has been demonstrated and their function in the expression of human β-like globin genes investigated (10, 11), the role of Sp1 during human β-globin gene expression is not yet understood. In this study, we used a mouse erythroleukemia cell line that contained human chromosome 11 (A181γ cells) and applied chromatin immunoprecipitation (ChIP) to assess the role of Sp1 binding in the induction of β-globin gene expression in the human β-globin locus (12, 13).
Materials and Methods
Cell Culture and Globin Gene Induction. Human–mouse erythroleukemia hybrid A181γ cells containing human chromosome 11 were maintained in RPMI medium 1640 supplemented with 10% FCS. For globin gene induction, hexamethylenebisacetamide was added at a final concentration of 4 mM, and the cells were cultured for 4 days.
RT-PCR Analysis of Globin Gene Expression. Total RNA was extracted from induced and uninduced A181γ cells by TRIzol reagent (Invitrogen). Five micrograms of total RNA was subjected to reverse transcription in a 25-μl volume containing dNTP, RNasin, oligo(dT)n, and M-MLV reverse transcriptase (Promega) at 42°C for 1 h. One microliter of the cDNA was then amplified by β- or γ-globin primers in a 25-μl PCR reaction (Table 1).
Table 1. Primer sequences for RT-PCR and ChIP.
| Prime names | Sequences | Length, bp |
|---|---|---|
| β-Forward for RT-PCR | ACACAACTGTGTTCACTAGCAAC | |
| β-Reverse for RT-PCR | TGCAGCTTGTCACAGTGCAGCTCACT | 460 |
| γ-Forward for RT-PCR | CACTCGCTTCTGGAACGTCTGAGA | |
| γ-Reverse for RT-PCR | GGACTCACCTTGAAGTTCTCAGGA | 497 |
| HS5 forward | GACCTATATCTGGCAGGAC | |
| HS5 reverse | GTGATGTCTTACTAACTAGC | 295 |
| HS4 forward | TGGCATCTAGCGCAATGACTT | |
| HS4 reverse | GGGCAAGCCATCTCATAGCTG | 194 |
| HS3 forward | CAGGAGTCTCTAAGGACTTGG | |
| HS3 reverse | CATAGGAGTCAAGGCACTTGC | 374 |
| HS2 forward | CTGTGTGTCTCCATTAGTGACCTCCC | |
| HS2 reverse | TGATGCCGTTTGAGGTGGAGTTTTA | 303 |
| HS1 forward | CTGAGAAGGCAATAGCAGGAGC | |
| HS1 reverse | GACACTAGTGTCACCAGTCTCC | 291 |
| ε-Promoter forward | GAGCCTCAGGATCCAGCACAC | |
| ε-Promoter reverse | GATGCCAGGCCTGAGAGCTTGC | 243 |
| γ-Promoter forward | ATTAAGCAGCAGTATCCTC | |
| γ-Promoter reverse | GGCGTCTGGACTAGGAGC | 282 |
| β-Promoter forward | CAATTTGTACTGATGGTATGG | |
| β-Promoter reverse | GGTGTCTGTTTGAGGTTGC | 280 |
Whole Chromosome Painting. The biotin-labeled human chromosome 11 probe was from ALTech Biomedical (Rockville, MD), and the biotin-labeled mouse chromosome 7 probe was from Opensystem Biotechnology (Huntsville, AL). Cultured A181γ cells were incubated with colchicines (Invitrogen) for 40 min, the cells were resuspended in 75 mM KCl hypotonic buffer for 15 min at 37°C, several drops of fixative (3:1, methanol/acetic acid) were added to the cell suspension, and the cells were washed three more times with the fixative. Twenty-five microliters of cell suspension in fixative was dropped on each slide and spread evenly by moving the pipette tip gently parallel to the slide surface. When the surface of the slides turned grainy, the slides were placed face down in the steam of a 75°C water bath for 1–3 sec and turned face up on a hot metal platform placed over a water bath until the slide was dry. After chemically aging in 99% ethanol for 2 min, the slides were put into the prewarmed pepsin working solution (99 ml H2O/1 ml 1 M HCl/60 μl of 10% pepsin) for 30 sec, rinsed in PBS, dehydrated in 70% and 99% ethanol for 5 min each, and air dried. Sixty microliters of probe in hybridization buffer (15 μl of 20 × SSC/45 μl of 30% dextran sulfate) was added and the slides covered with coverslips. The chromatin and probes were denatured at 75°C for 2 min on a PCR machine, gradually cooled down to room temperature, and incubated at 37°C in a humid chamber for 12–16 h. The slides were washed, and avidin conjugated with FITC was applied. After 20 min incubation at room temperature, the slides were washed. FITC-conjugated antiavidin antibodies were applied to amplify the signals, and the slides were mounted with antifade solution containing DAPI. Photographs were taken with a fluorescence microscope (Zeiss Axiophot) and overlapped by adobe photoshop 5.5 software (zcomAdobe Systems, San Jose, CA).
Antibodies. Polyclonal antibodies to Sp1 (07-124), Sp3 (07-107), acetylated histone 3 (06-598), histone 4 (06-599), and histone deacetylase 1 (HDAC1) (05-614) were obtained from Upstate Biotechnology (Lake Placid, NY); antibodies to p300/cAMP-response element-binding protein-associated factor (PCAF) were from Santa Cruz Biotechnology (sc-8999); phosphothreonine antibodies were from Cell Signaling Technology (Beverly, MA); and FKLF2 antibodies were from Orbigen (San Diego, CA)
ChIP. ChIP analysis was performed as described (13). Briefly, 12 × 107 cells were crosslinked by 0.4% formaldehyde in the original medium for 10 min at room temperature. The formaldehyde was neutralized by glycine at a final concentration of 125 mM. After washing in PBS, the cells were resuspended in 2.5 volume of cell lysis buffer (10 mM Tris, pH 8.0/10 mM NaCl/0.2% Nonidet P-40/10 mM sodium butyrate/proteinase inhibitors mixture from Roche Applied Science), and incubated on ice for 10 min. The nuclei were isolated by centrifugation at 1,000 × g for 10 min, resuspended in 1 ml of nucleus lysis buffer (50 mM Tris, pH 8.1/10 mM EDTA/1% SDS/10 mM sodium butyrate/proteinase inhibitors) and incubated on ice for 10 min. One milliliter of immunoprecipitation (IP) dilution buffer (20 mM Tris, pH 8.1/150 mM NaCl/2 mM EDTA/0.01% SDS/10 mM sodium butyrate/proteinase inhibitors) was added and the chromatin sheared to ≈500 bp by sonication. After centrifugation at 10,000 × g for 10 min at 4°C, 3 ml of IP dilution buffer was added to dilute SDS concentration to 0.2%. To remove nonspecific protein binding, 100 μg of rabbit IgG and 100-μl bed volume of protein-A Sepharose were added and the mixture rotated on a rotating wheel overnight at 4°C, spun at 1,500 × g for 10 min, and the supernatant recovered. After saving 180 μl to indicate the DNA input, 900 μl of the supernatant was transferred to a 1.5-ml tube, and 7 μg of specific antibodies was added and incubated at 4°C for 3 h. Thirty-microliter bed volume of protein-A Sepharose was added and mixed on a rotating wheel at 4°C for 2 h. The Sepharose beads were spun down and washed extensively. The protein–DNA complexes were then eluted with 300 μl of elution buffer (100 mM NaHCO3/1% SDS). Crosslinking was reversed by heating at 65°C for 5 h followed by 100 μg/ml proteinase K treatment overnight at 45°C. DNA were extracted with phenol/chloroform, precipitated with ethanol, and suspended in 30 μlof distilled water. As a control, input DNAs were also extracted and dissolved in 64 μl of distilled water. Immunoprecipitated and input DNAs were analyzed by PCR with primers that generate 200- to 350-bp products. The input DNA was serially diluted twice and amplified with the immunoprecipitated DNA simultaneously for similar cycles to ensure the PCR amplifications were in exponential range. The PCR products were resolved on 2% agarose gel stained with ethidium bromide and quantified by an Alpha Image system (San Leandro, CA). Primers used to amplify the HS1, HS2, HS3, HS4, and HS5 regions and the ε, γ, and β promoters are shown in Table 1.
Western Blot Analysis. Nuclear extracts from induced and uninduced A181γ cells were resolved by electrophoresis on an SDS/7% polyacrylamide gel. Proteins were electrotransferred onto poly(vinylidene difluoride) membrane (Hybond-P, Amersham Pharmacia) and probed with rabbit anti-Sp1 antibodies. Horseradish peroxidase-conjugated donkey anti-rabbit antibodies were applied to detect the primary antibodies signals by luminol reagent (Santa Cruz Biotechnology). The membrane was submerged in a buffer containing 100 mM 2-mercaptoethanol, 2% SDS, and 62.5 mM Tris·HCl, pH 6.7, for 30 min at 60°C to strip off the anti-Sp1 antibodies. After washing in PBS containing 0.05% Tween-20, the membrane was incubated with luminol reagent to ensure removal of antibodies, and rabbit antiphosphothreonine antibodies were applied to detect the phosphorylation of Sp1.
Results
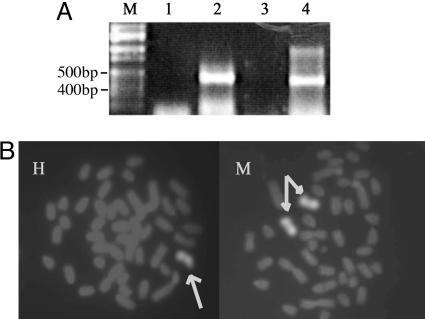
A181γ cells are hybrid cells from human fetal liver cells and mouse erythroleukemia cells. Because continuous culture of these cells could result in switching from γ- to β-globin gene expression as well as to loss of human chromosomes (12), we assessed the status of the cells we were using by RT-PCR and whole chromosome painting. Fig. 1A shows that both human γ- and β-globin genes were transcribed in the cells after induction, indicating that the cells were undergoing a γ-to-β switch. One human chromosome 11 and two mouse chromosomes 7 that contained the β-globin locus were found in this cell line (Fig. 1B).
Fig. 1.
Human γ- and β-globin gene expression and the number of chromosome-containing β-loci in the A181γ cells. (A) RT-PCR analysis of γ- and β-globin gene RNA. Lane M, Hi-Lo DNA ladder marker (Bionexus, Oakland, CA); lanes 1 and 2, human γ-globin cDNA before and after induction; lanes 3 and 4, human β-globin cDNA before and after induction. (B) Chromosome painting. Probe for human (H) chromosome 11 and for mouse (M) chromosome 7. The arrows point to one copy of human chromosome 11 and two copies of mouse chromosome 7 in this cell line.
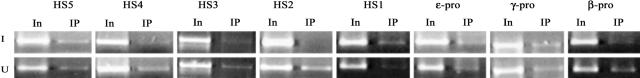
In uninduced A181γ cells, Sp1 binding was seen on HS1 to five ε-, γ-, and β-promoters, all of the known cis-elements within the human β-locus. When the cells were induced with hexamethylenebisacetamide, no Sp1 binding was observed at HS2, HS3, HS4, and the β-globin promoter, whereas the binding of Sp1 at HS1, HS5, and the ε and γ promoters could still be observed (Fig. 2). This suggests that Sp1 represses the expression of human β-like globin genes by binding at HSs and the globin promoters, and that the release of Sp1 binding at HS2–4 accompanied erythroid terminal differentiation.
Fig. 2.
Binding of Sp1 to hypersensitive sites and globin gene promoters in induced and uninduced A181γ cells. Input DNA or DNA immunoprecipitated by Sp1 antibodies was amplified for 28 cycles in 25 μl of reaction using pairs of the primers indicated in Table 1. I, induced; U, uninduced; In, input DNA; IP, immunoprecipitated DNA.
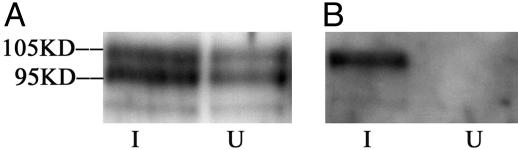
Sp1 is subjected to posttranslational modifications that can influence its activity. The three types of posttranslational modification thought to be involved in transcription regulation are glycosylation, phosphorylation, and acetylation (7, 14). Phosphorylation at Thr-579 by casein kinase II has been reported to inhibit Sp1 DNA-binding activity in the liver (15). The phosphorylation of serine residues at C-terminal that is regulated by cell cycle-related kinase can also reduce the DNA-binding activity of Sp1 (16). We performed Western blot analysis by using antiSp1 antibodies and reprobed the membrane with antiphosphothreonine antibodies. Phosphorylation of the 105-kD isoform of Sp1 was found after induction (Fig. 3). Hence, phosphorylation of Sp1 accompanies its release from DNA binding.
Fig. 3.
Sp1 expression and phosphorylation before and after induction. I, induced A181γ cell; U, uninduced A181γ cells. The membrane was first probed with anti-Sp1 antibodies (A) and then stripped and reprobed with antiphosphorylated threonine antibodies (B).
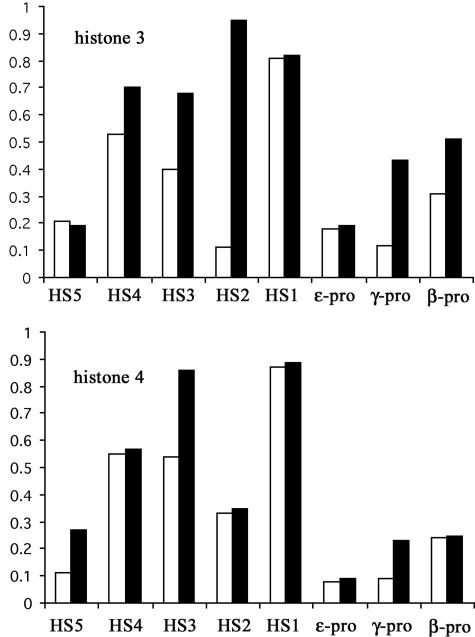
Sp1 has been reported to activate the expression of some genes, as well as repress that of other genes, by recruiting HDAC1 to deacetylate histones (17). To see whether Sp1 represses β-like globin gene expression in a similar manner, we assayed histone acetylation and HDAC1 binding at the human β-LCR in A181γ cells. Fig. 4 shows the changes in histone acetylation within the human β-locus. Histone 3 acetylation at HS-4 and the γ and β globin promoters was increased, but no significant differences were observed in HS1, HS5, or the ε-globin promoter. Histone 4 acetylation at HS3, HS5, and the γ-globin promoter was increased, and no significant changes were observed over HS1, HS2, HS4, and the ε and β globin promoters. These results indicate there is an overall increase in histone acetylation at the human β-globin locus after induction. Because the A181γ cells we used transcribed both human γ- and β-globin genes, the acetylation changes found in the β- and γ-promoters are compatible with the expression of both γ- and β-globin in the cell line we are using. The absence of changes in histone acetylation at the human ε-globin promoter on induction is in agreement with the silencing of ε-globin gene expression in this cell line.
Fig. 4.
Acetylation of histones 3 and 4 on the human β-globin locus in A181γ cells. Chromosome DNA was precipitated with antibodies against acetylated histones 3 and 4. The acetylation of histones 3 and 4 before and after induction was represented by the ratio of band intensity of PCR products from 25 cycles on agarose gel stained with ethidium bromide amplified from immunoprecipitated chromosome DNA to that of input DNA. ▪, induced; □, uninduced.
Because the HS2–4 regions lost Sp1 binding after induction, and because the binding of transcription factors at the γ- and β-globin promoters has been studied extensively (10, 11, 18), we planned to study the three HS regions in additional experiments. However, the HS4 fragment could not be amplified even after 35 cycles, when we used all of the available transcription factor antibodies, a phenomenon also observed by Horak et al. (19) in their studies. Hence we focused our studies on only the HS2 and HS3 regions in our subsequent experiments.
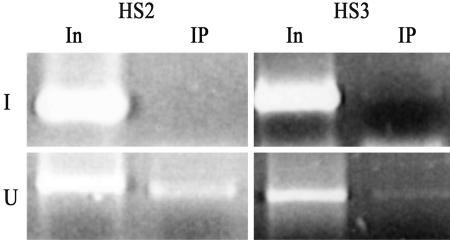
We studied the role of HDAC1 on the changes of the histone acetylation pattern. Fig. 5 shows the recruitment of HDAC1 at HS2 and HS3 before induction. After induction, HDAC1 binding disappeared at HS2 and HS3, along with the loss of Sp1.
Fig. 5.
Recruitment of HDAC1 to HS2 and HS3 in induced and uninduced A181γ cells. Input DNA or DNA immunoprecipitated by HDAC1 antibodies was amplified for 33 cycles in 25 μl of reaction by using pairs of the primers in Table 1.
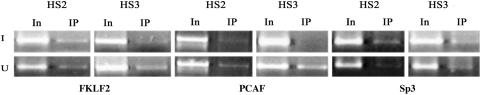
FKLF2, EKLF, and Sp1 belong to the same Sp1/XKLF family. Because Sp1 appears to inhibit, whereas FKLF2 and EKLF activate, globin gene expression (20, 21), we investigated the relation between Sp1 and FKLF2. ChIP results showed that FKLF2 binding could be seen at HS2 and HS3 in both induced and uninduced A181γ cells. PCAF, one of the histone acetyl-transferases that has been shown to interact with FKLF2, was found to bind HS2 and HS3 in induced but not in uninduced cells (Fig. 6). This suggests that FKLF2 binds to HS2 and HS3 in the uninduced state, but its function is inhibited by Sp1 binding. After induction, with the release of Sp1 binding, PCAF is recruited to FKLF2 to initiate γ-globin gene transcription.
Fig. 6.
FKLF2, PCAF, and Sp3 binding at HS2 and HS3. Chromosome DNA was precipitated by antibodies against FKLF2, PCAF, and Sp3. Input DNA or DNA immunoprecipitated by FKLF2, PCAF, and Sp3 antibodies was amplified for 33 cycles in 25 μl of reaction by using pairs of the primers in Table 1.
Because Sp3 has been reported to be a competitor of Sp1 and can repress the function of Sp1 (22), we performed ChIP using Sp3 antibodies (Fig. 6). Like Sp1, the binding of Sp3 on HS2 and HS3 sites disappeared also after induction. Because both Sp1 and Sp3 bind HS2 and HS3 in the uninduced state, it appears that in the HSs, they collaborate with each other to repress the expression of human β-globin genes. A similar cooperative result was found on the human telomerase reverse transcriptase promoter (23).
Discussion
More than 20 members of the Sp1/XKLF family of transcription factors have been described. They all have a conserved Cys2His2 zinc-finger DNA-binding domain within the 81-aa residues at the C terminus (24). They recognize the same GC and GT boxes, which are important cis-acting elements for the expression of many different housekeeping, tissue-specific, and viral genes. Some transcriptional factors, such as Sp1, Sp3, and UKLF (25), are ubiquitously expressed, whereas others such as basic Krüppel-like factor (BKLF) and LKLF (26, 27) are tissue-specifically expressed. Thus, it is likely they control gene transcription by competing for or cooperating within the same binding sites. The most impressive example came from an experiment on BKLF and EKLF (26), both of which are erythroid-specific proteins: EKLF activates β-globin transcription, whereas BKLF, a repressor of ε-, γ-, and β-globin gene transcription, maintains the proper expression level of β-like globin genes. Our results suggest that in erythroid progenitor cells that do not express globin genes, the GC and GT boxes at the β-LCR are occupied by Sp1, which recruits HDAC1 to deacetylate histones and keeps the chromatin structure at a closed status, and, at the same time, represses the function of FKLF2 by preventing it from recruiting histone acetyltransferases (PCAF). During hematopoiesis, Sp1 is phosphorylated. The phosphorylation of Sp1 in the N terminus has been shown to augment transcription, whereas phosphorylation at threonine 579 correlates with the inactivation of Sp1. Although the antibody we used could not distinguish the different threonine phosphorylation sites of Sp1, our results are inconsistent with the phosphorylation of threonine 579. The phosphorylated Sp1 is then displaced from the motif, thus allowing the FKLF2 situated at this motif to recruit PCAF to acetylate histones to activate γ-globin gene transcription. The interaction among FKLF2, NF-E2, and GATA-1 may also occur at this stage, to assemble the preinitiation complex (28–30). A similar mechanism may occur during adult β-globin gene expression. The loss of Sp1 on LCR and the β-globin promoter would allow EKLF to recruit the holocomplex consisting of all other known transcription factors and initiate β-globin gene transcription. Unfortunately, we cannot show this directly, because anti-mouse EKLF antibodies suitable for ChIP studies are not available.
We noticed that the acetylation of the histones is different in several regions after induction. For example, at the HS2 region, only histone 3 acetylation was increase, but at the HS3 region, both histone 3 and 4 acetylation could be observed. These results suggest that histones at different regions are acetylated at different times and recruit different transcriptional factors to assemble sequentially the preinitiation complex.
Sp1 binding also maintains the silencing of β-like globin genes. The expression of the ε-globin gene has already turned off in the cell line, and the binding of Sp1 at the ε-globin promoter could be seen both before and after induction. Because the A181γ cells under study are undergoing a γ-to-β switch, the expression of γ-globin is subjected to silencing soon. Hence, the binding of Sp1 at the γ-globin promoter could still be seen but was totally lost at the β-globin promoter after induction. Sp1 and EKLF binding at the β-LCR can repress the spreading of heterochromatin in erythroid cells (31). HS5 has been reported as an insulator in human β-globin locus (32), and the binding of Sp1 at HS5 after induction is in agreement with these findings. With the induction of the A181γ cells to erythroid terminal differentiation, the binding of Sp1 at HS5 prevents the spreading of the heterochromatin structure outside the locus and maintains the open chromatin conformation at HS2, HS3, and HS4 so they can interact with the β-like globin gene promoters through the transcription factors.
The effects of interactions between Sp1 and Sp3 apparently vary in different genes. Sp1 can act as an activator of house-keeping gene expression that is repressed by Sp3 (22). On the other hand, in normal human somatic cells, Sp1 and Sp3 cooperate at the promoter to repress transcription of the human telomerase reverse transcriptase gene (23), similar to our finding in the human β-like globin gene locus.
In terminally differentiated tissues such as brain, lung, and liver, Sp1 has been shown to be highly phosphorylated to release the inhibition of tissue-specific gene transcription. In contrast, Sp1 is underphosphorylated in cancer cells and regenerating liver cells, so its binding would inhibit the expression of tissue-specific genes and prevent the terminal differentiation of these cells (33). In Sp1 knockout mice, most genes containing Sp1-binding sites are transcribed normally, but the embryos die at an early stage of embryonic development (34). Our results suggest that without the repression of Sp1, tissue-specific transacting factors such as FKLF2 and EKLF, as well as other tissue-specific transcriptional factors, might prematurely activate tissue-specific gene expression. Therefore, the embryos would undergo premature differentiation, and the inappropriate early expression of tissue-specific genes causes the embryos to die at an early stage of development.
Acknowledgments
We thank Dr. Tohru Ikuta (Medical College of Georgia, Augusta, GA) for providing the A181γ cell line originally obtained from Drs. George Syamatoyannopoulos and Thalia Papayannopoulou (University of Washington, Seattle). We also thank Mei-Chi Cheung (Howard Hughes Medical Institute, University of California, San Francisco) for the preparation of the FISH images.
Author contributions: D.F. and Y.W.K. designed research; D.F. performed research; D.F. and Y.W.K. analyzed data; and D.F. and Y.W.K. wrote the paper.
Abbreviations: LCR, locus control region; HS, DNase I hypersensitive site; EKLF, erythroid Krüppel-like factor; FKLF2, fetal Krüppel-like factor 2; PCAF, p300/cAMP-response element binding protein-associated factor; HDAC1, histone deacetylase 1; ChIP, chromatin immunoprecipitation.
References
- 1.Huang, Y., Liu, D. P., Feng, D. X., Wu, M., Shen, W., Tang, Y., Tang, X. B. & Liang, C. C. (2004) Int. J. Biochem. Cell Biol. 36, 1261–1265. [DOI] [PubMed] [Google Scholar]
- 2.Li, Q., Peterson, K. R., Fang, X. & Stamatoyannopoulos, G. (2002) Blood 100, 3077–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orlando, V. & Jones, K. A. (2002) Genes Dev. 16, 2039–2044. [DOI] [PubMed] [Google Scholar]
- 4.Harju, S., McQueen, K. J. & Peterson, K. R. (2002) Exp. Biol. Med. 227, 683–700. [DOI] [PubMed] [Google Scholar]
- 5.Ikuta, T., Papayannopoulou, T., Stamatoyannopoulos, G. & Kan, Y. W. (1996) J. Biol. Chem. 271, 14082–14091. [DOI] [PubMed] [Google Scholar]
- 6.Dynan, W. S. & Tjian, R. (1983) Cell. 32, 669–680. [DOI] [PubMed] [Google Scholar]
- 7.Bouwman, P. & Philipsen, S. (2002) Mol. Cell Endocrinol. 195, 27–38. [DOI] [PubMed] [Google Scholar]
- 8.Huang, Y., Liu, D. P., Wu, L., Li, T. C., Wu, M., Feng, D. X. & Liang, C. C. (2000) Blood Cell Mol. Dis. 26, 598–610. [DOI] [PubMed] [Google Scholar]
- 9.Dubourg, C., Toutain, B., Le Gall, J. Y. & Le Treut, A. (2003) Gene 316, 91–101. [DOI] [PubMed] [Google Scholar]
- 10.Song, C. Z., Keller, K., Murata, K., Asano, H. & Stamatoyannopoulos, G. (2002) J. Biol. Chem. 277, 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, W. & Bieker, J. J. (1998) Proc. Natl. Acad. Sci. USA 95, 9855–9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papayannopoulou, T., Brice, M. & Stamatoyannopoulos, G. (1986) Cell 46, 469–476. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, K. D. & Bresnick, E. H. (2002) Methods 26, 27–36. [DOI] [PubMed] [Google Scholar]
- 14.Leggett, R. W., Armstrong, S. A., Barry, D. & Mueller, C. R. (1995) J. Biol. Chem. 270, 25879–25884. [DOI] [PubMed] [Google Scholar]
- 15.Armstrong, S. A., Barry, D. A., Leggett, R. W. & Mueller, C. R. (1997) J. Biol. Chem. 272, 13489–13495. [DOI] [PubMed] [Google Scholar]
- 16.Black, A. R., Jensen, D., Lin, S. Y. & Azizkhan, J. C. (1999) J. Biol. Chem. 274, 1207–1215. [DOI] [PubMed] [Google Scholar]
- 17.Doetzlhofer, A., Rotheneder, H., Lagger, G., Koranda, M., Kurtev, V., Brosch, G., Wintersberger, E. & Seiser, C. (1999) Mol. Cell. Biol. 19, 5504–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brand, M., Ranish, J. A., Kummer, N. T., Hamilton, J., Igarashi, K., Francastel, C., Chi, T. H., Crabtree, G. R., Aebersold, R. & Groudine, M. (2004) Nat. Struct. Mol. Biol. 1, 73–80. [DOI] [PubMed] [Google Scholar]
- 19.Horak, C. E., Mahajan, M. C., Luscombe, N. M., Gerstein, M., Weissman, S. M. & Snyder, M. (2002) Proc. Natl. Acad. Sci. USA 99, 2924–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, J. S., Ngo, H., Kim, D. & Chung, J. H. (2000) Proc. Natl. Acad. Sci. USA 97, 2468–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asano, H., Li, X. S. & Stamatoyannopoulos, G. (2000) Blood 95, 3578–3584. [PubMed] [Google Scholar]
- 22.Hagen, G., Muller, S., Beato, M. & Suske, G. (1994) EMBO J. 13, 3843–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Won, J., Yim, J. & Kim, T. K. (2002) J. Biol. Chem. 277, 38230–38238. [DOI] [PubMed] [Google Scholar]
- 24.Philipsen, S. & Suske, G. (1999) Nucleic Acids Res. 27, 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turner, J. & Crossley, M. (1999) Trends Biochem. Sci. 24, 236–240. [DOI] [PubMed] [Google Scholar]
- 26.Crossley, M., Whitelaw, E., Perkins, A., Williams, G., Fujiwara, Y. & Orkin, S. H. (1996) Mol. Cell. Biol. 16, 1695–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Endrizzi, B. T. & Jameson, S. C. (2003) Int. Immunol. 15, 1341–1348. [DOI] [PubMed] [Google Scholar]
- 28.Hung, H. L., Kim, A. Y., Hong, W., Rakowski, C. & Blobel, G. A. (2001) J. Biol. Chem. 276, 10715–10721. [DOI] [PubMed] [Google Scholar]
- 29.Boyes, J., Byfield, P., Nakatani. Y. & Ogryzko, V. (1998) Nature 396, 594–598. [DOI] [PubMed] [Google Scholar]
- 30.Sawado, T., Halow, J., Bender, M. A. & Groudine, M. (2003) Genes Dev. 17, 1009–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMorrow, T., van den Wijngaard, A., Wollenschlaeger, A., van de Corput, M., Monkhorst, K., Trimborn, T., Fraser, P., van Lohuizen, M., Jenuwein, T., Djabali, M., et al. (2000) EMBO J. 19, 4986–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, Q. & Stamatoyannopoulos, G. (1994) Blood 84, 1399–1401. [PubMed] [Google Scholar]
- 33.Zannetti, A., Del Vecchio, S., Carriero, M. V., Fonti, R., Franco, P., Botti, G., D'Aiuto, G., Stoppelli, M. P. & Salvatore, M. (2000) Cancer Res. 60, 1546–1551. [PubMed] [Google Scholar]
- 34.Marin, M., Karis, A., Visser, P., Grosveld, F. & Philipsen, S. (1997) Cell 89, 619–628. [DOI] [PubMed] [Google Scholar]