Abstract
Differentiation in response to environmental cues is integral to the success of many intracellular pathogens. By characterizing a Legionella pneumophila mutant defective for differentiation in broth and replication in macrophages, we identified a subfamily of major facilitator superfamily transporters, here named Pht (phagosomal transporter), that also is conserved in two other vacuolar pathogens, Coxiella burnetii and Francisella tularensis. Biolog phenotype microarray analysis indicated that PhtA transports threonine, an essential amino acid. Either excess threonine or threonine peptides bypass phtA function. In minimal medium, phtA mutants do not replicate; in rich broth, the bacteria prematurely differentiate to the transmissive phase, as judged by the kinetics of flaA-gfp expression, heat resistance, and sodium sensitivity. PhtA is dispensable for transmissive L. pneumophila to establish and persist within a replication vacuole but is essential for their differentiation to the replicative phase, based on phenotypic and RT-PCR analysis. Accordingly, we propose that the Pht transporter family equips transmissive L. pneumophila, C. burnetii, and F. tularensis to assess their phagosomal nutrient supply before committing to reenter the cell cycle.
Keywords: Coxiella burnetii, Francisella tularensis, major facilitator superfamily transporter
Differentiation is a bacterial strategy for adapting to the environment that is practiced by diverse microbes, ranging from the social Myxococcus xanthus to the motile Caulobacter crescentus to the spore-forming Bacillus anthracis (1–3). As a model organism to study differentiation by intracellular pathogens, we exploit Legionella pneumophila, a Gram-negative bacterium that not only parasitizes freshwater amoeba but also infects human alveolar macrophages to cause Legionnaires' Disease (4). Like several other intracellular pathogens, including Chlamydia species and Coxiella burnetii (5, 6), L. pneumophila alternates between at least two cell types. In the transmissive form, the pathogens express a panel of virulence factors that facilitate survival in the environment and infection of host cells. In particular, infectious L. pneumophila exploit a type IV secretion system to immediately avoid the default endocytic pathway. After establishing a unique vacuole derived from endoplasmic reticulum (ER), the bacteria differentiate to the replicative form (7–9).
In broth, L. pneumophila couples differentiation to nutrient availability using the broadly conserved stringent response pathway (10). When nutrients are abundant, bacteria replicate; when amino acids become scarce, a regulatory cascade activates cytotoxicity, stress resistance, motility, and lysosome avoidance, traits likely to promote transmission of progeny to a new host cell (11). The ability to differentiate in response to nutrient availability is integral to L. pneumophila pathogenesis: replicative phase bacteria fail to evade the macrophage endocytic pathway and are killed by lysosomal enzymes (12). Conversely, when locked genetically in the transmissive phase, L. pneumophila infect macrophages efficiently but do not replicate there (13).
Little is known about the nutrients available to L. pneumophila in its replication vacuole, but several observations predict that amino acid transport is integral to its fitness. In minimal medium, L. pneumophila obtains carbon and energy from amino acids, not sugars (14). Furthermore, the macrophage amino acid transporter SLC1A5 is required to support replication of intracellular L. pneumophila (15). The stringent response model also predicts that amino acids are the primary signal for intracellular L. pneumophila to gauge its environment and govern differentiation (16). Here, we extend knowledge of the L. pneumophila life cycle by identifying a threonine transporter that is essential for both differentiation and virulence in a mouse macrophage model of pathogenesis. This mechanism to link nutrient acquisition to microbial differentiation may be practiced by other intracellular pathogens, because phagosomal transporter A (PhtA) is a member of a unique family of transporters also common to C. burnetii and Francisella tularensis, two other vacuolar pathogens that thrive in both macrophages and amoebae (17).
Materials and Methods
Bacterial Strains, Plasmids, and Cell Culture. L. pneumophila strain Lp02, a virulent thymidine auxotroph, and isogenic strains phtA and dotA (8) were cultured on charcoal yeast extract (CYE) plates supplemented with 100 μg/ml thymidine for 4 days at 37°C as described (18, 19). Single colonies from CYE plates were inoculated into N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered yeast extract (AYE) media supplemented with 100 μg/ml thymidine and incubated at 37°C overnight to the exponential phase. These cultures were subsequently diluted and incubated for an additional 12–18 h to obtain early exponential (EE; OD600 of 1.5), midexponential (ME; OD600 of 2.5), or postexponential (PE; OD600 of 3.5–4.0) cells as described (9).
The phtA mutant was constructed by in vitro transposon mutagenesis of the letE-phtA locus with the mariner 3 transposon (gift of V. DiRita, University of Michigan, Ann Arbor) as described (20). The transposon insertion is ≈300 bp 3′ of the start codon and in the same orientation as the phtA gene. For complementation tests, the phtA locus was amplified by using forward primer GCCGGAATTCTTAATACAGCATGTGGTGGTTATA and reverse primer GCCCAAGCTTTATCATGCAATGCTCAATGT and cloned into pGEMTeasy. The fragment containing phtA, obtained by digesting this plasmid with EcoRI and HindIII, was then cloned into the EcoRI and HindIII sites of plasmid pMMB206 3′ to the isopropyl β-d-thiogalactoside (IP TG)-inducible tac-lac promoter, generating pMMB206::phtA.
Primary murine bone marrow-derived macrophages (BMM) were isolated from the femur exudate of female A/J mice and cultured in RPMI medium 1640 with 10% heat-inactivated FBS as described (19).
Expression of Transmissive Traits. To assay flaA promoter activity, phtA mutants and WT bacteria were transformed with pflaG, a reporter of the transmissive phase; then GFP fluorescence was quantified as described (11). Experiments were performed twice, and representative data are shown. L. pneumophila heat resistance was determined by measuring viability after incubating cultures at 57°C for 20 min as described (11). Bacterial resistance to 100 mM NaCl was determined as described (9). Infectivity, the ability of L. pneumophila to attach to, invade, and survive inside BMM, and cytotoxicity, the acute killing of BMM, were determined as described (9). Heat resistance, NaCl resistance, and infectivity and cytotoxicity assays were performed in triplicate, and averages and standard deviations are presented.
Intracellular Growth. Replication of L. pneumophila in macrophages was assayed as described (19). For complementation assays, β-d-thiogalactoside (IPTG) was added to the tissue culture medium to 1 mM at the time indicated and maintained for the duration of the experiment. For phtA suppression experiments, amino acids or peptides were added to 3 mM at the time of infection and maintained throughout the infection. Experiments were performed three times; averages and standard deviations are presented.
Immunofluorescence Microscopy. We incubated 2 × 105 BMM per 12-mm glass coverslip in 24-well plates overnight before infection. After infections, as described above for intracellular growth assays, cells were fixed, blocked, and stained as described (21). Anti-lysosome-associated membrane protein 1 (LAMP-1) (1D4B) rat antibody (1:1,000) was acquired from Santa Cruz Biotechnology, anti-calnexin rabbit polyclonal antibody (1:200) was purchased from Stressgen Biotechnologies (Victoria, Canada), anti-L. pneumophila rabbit polyclonal antibody (1:2,000) was a generous gift from R. Isberg (Tufts University School of Medicine, Boston), and anti-L. pneumophila mouse monoclonal hybridoma cell line CRL-1765 (1:5) was acquired from American Type Culture Collection (ATCC). Oregon green conjugated to anti-rat or anti-rabbit IgG antibody and Texas red conjugated to anti-rabbit or anti-mouse IgG antibody (all 1:2,000) were purchased from Molecular Probes.
NaCl Sensitivity and RT-PCR Analysis of Intracellular Bacteria. NaCl sensitivity of intracellular bacteria 1, 2, and 10 h after infection was quantified as described after lysing infected macrophages with 1% saponin in PBS (9).
For RNA purification, ≈5 × 107 macrophages per plate (150 × 25 mm, Fisher Scientific) were infected at a multiplicity of infection of 2 to maximize internalization while minimizing cytotoxicity. After 1 h, the inoculum was removed by three washes with RPMI medium 1640 and replaced with 20 ml of fresh media. At the times indicated, 20 ml of cold 1% saponin in PBS was added, and the cultures were incubated for 10 min at room temperature. Cells were then harvested by scraping, and the 40 ml of lysate was subjected to vortexing for 2 min. To remove host cell nuclei and debris, the lysates were cleared by centrifugation at 400 × g for 10 min at 4°C. To collect the bacteria, supernatants were centrifuged at 5,000 × g for 10 min at 4°C, and RNA was obtained from the resulting bacterial pellet by using the Qiagen (Valencia, CA) RNeasy Mini kit with an on-column DNase digestion. The RNA preparations were reverse-transcribed by using Invitrogen M-MLV reverse transcriptase, then amplified by PCR by using primers specific to the flaA and rpoB genes and 16S rRNA. To compare transcript quantities, PCRs were continued for 20–30 cycles and analyzed by gel electrophoresis. To control for genomic DNA contamination, non-reverse-transcribed RNA was analyzed in parallel. The growth phase-specific gene expression of flaA and rpoB and the utility of 16S rRNA as a loading control were confirmed by analyzing RNA purified from WT EE and PE broth cultures.
Growth in Defined Media Supplemented with Casamino Acid or Tryptone Peptone. Defined medium, modified Ristroph media (MRM), was prepared according to the protocol of Ristroph et al. (22) except that cystine, choline, and rhamnose were excluded. Where indicated, 5 mg of Difco Bacto casamino acids or Difco Tryptone was added per ml of MRM; single amino acids (Sigma) or peptides (Bachem) were added to 3 mM.
Biolog Phenotype Array Analysis. Peptide array plates were purchased from BioLog Chemical (San Diego). Each well of the 96-well Biolog Phenotype Microarray (PM) plates PM6, PM7, and PM8 contains a different di- or tripeptide combination. Starting with an initial OD600 of ≈0.03 in 100 μl of MRM, growth in each well during a 96-h incubation was determined by measuring the OD600 at 24-h intervals. Bypass of the mutant growth defect was defined as a >50-fold increase in OD600 relative to a control well that contained no peptide supplement.
Results
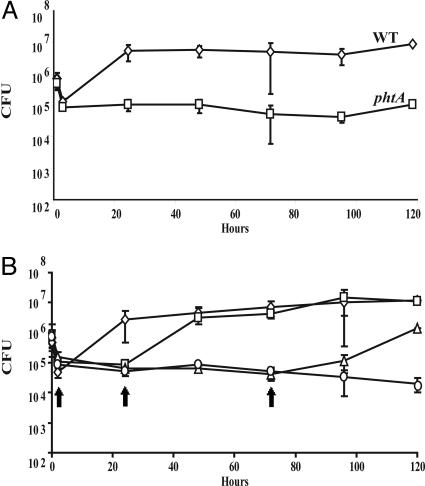
In Macrophages, phtA Mutants Exhibit a Strong but Reversible Growth Defect. While conducting a saturation mutagenesis of the letE locus, we discovered that L. pneumophila require the adjacent phtA gene (23) for virulence. When A/J mouse BMM were incubated with phtA mutants, the bacteria infected as efficiently as WT yet did not grow during the subsequent 120 h (Fig. 1A). The mutants were rescued by expression of the phtA locus from an inducible plasmid before infection. Furthermore, unlike dotA type IV secretion mutants, which cannot be rescued once internalized (24), intracellular phtA mutants resumed replication when phtA expression was induced at 0, 24, or even 72 h post infection (Fig. 1B). Therefore, L. pneumophila absolutely require PhtA for growth but not survival within BMM.
Fig. 1.
phtA mutants are defective for intracellular replication but can be rescued by complementation in trans any time after infection. (A) WT Legionella (diamonds) infect macrophages and replicate efficiently, whereas phtA mutants (squares) infect macrophages like WT, but their yield does not increase. (B) β-d-Thiogalactoside (IPTG) induction of the phtA locus on pMMB206::phtA (arrows) after 0 h (diamonds), 24 h (squares), or even 72 h (triangles) restores replication, whereas the yield of uninduced mutants (circles) remains constant, as judged by enumerating colony-forming units (CFU). Each experiment was performed in triplicate, and means and standard deviations are shown.
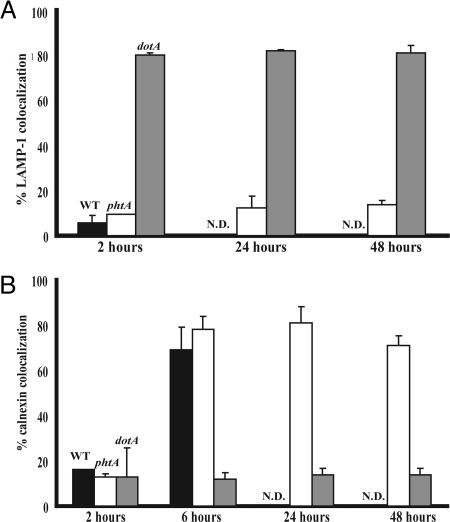
phtA Mutants Properly Avoid LAMP-1 Compartments and Traffic to Calnexin-Labeled Compartments. Because aberrant trafficking in macrophages confers an intracellular replication defect (24), we investigated whether phtA is required to establish a protected vacuole in macrophages. Like WT Legionella, phtA mutants avoided the endosomal pathway 2 h postinfection, because <10% of the bacteria colocalized with LAMP-1. The phtA mutants maintained their ability to avoid LAMP-1 for at least 48 h, whereas >80% dotA mutants immediately associated with LAMP-1-labeled compartments (Fig. 2A). By 6 h, >75% of WT and phtA mutant vacuoles were marked by calnexin, an ER-resident protein (Fig. 2B). phtA mutants remained associated with the ER for at least 48 h, whereas dotA mutants rarely did so (Fig. 2B). Therefore, L. pneumophila do not require PhtA to establish a protected ER-derived vacuole but do require its activity to initiate intracellular replication, because single phtA mutant cells persisted in each of the >300 vacuoles observed.
Fig. 2.
phtA mutants traffic in macrophages like WT Legionella. (A) WT bacteria (black bars) and phtA mutants (white bars) initially avoid LAMP-1-labeled compartments whereas dotA mutants (gray bars) do not. phtA mutants continue to avoid LAMP-1 endosomes throughout the infection. (B) WT bacteria (black bars) and phtA mutants (white bars) associate with calnexin by 6 h after infection, unlike dotA mutant vacuoles (gray bars). phtA mutant vacuoles remain associated with ER and isolated from the endosomal pathway for the duration of the infection. Fifty vacuoles per condition were scored for each time point, and experiments were performed in triplicate (A) or duplicate (B). Means and standard deviations are presented.
The phtA Locus Encodes a Major Facilitator Superfamily (MFS) Transporter with Homologues in C. burnetii and F. tularensis. Because phtA is not involved in intracellular trafficking, we pursued functions predicted by its sequence. Protein structure programs (http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html) indicate that the PhtA protein spans the inner membrane 12 times. L. pneumophila strain Philadelphia 1 encodes 10 additional phtA homologues (e values e–64 to e–28), the Lens strain carries 11 homologues (e–64 to e–28), and the Paris strain has nine (e–64 to e–27). Furthermore, C. burnetii encodes nine close homologues (e–51 to e–39) and F. tularensis contains four (e–35 to e–28), whereas three other species contain one each (Fig. 8, which is published as supporting information on the PNAS web site). All of the phtA homologues (proteins with an e value of <–28, which corresponds to the most distant L. pneumophila family member) are members of the MFS of transporters (25) but have no assigned function. The closest known homologue is the UhpC sugar transporter of Escherichia coli (≈e–14). Because MFS proteins contribute to diverse processes, including amino acid transport, we postulated that PhtA transports nutrients that are essential for intracellular L. pneumophila to replicate.
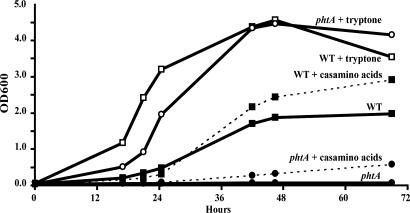
The phtA Mutant Minimal Medium Growth Defect Is Bypassed by Supplemental Tryptone. To test the hypothesis that PhtA equips L. pneumophila to acquire nutrients, we applied an in vitro system to model the phtA mutant intracellular growth defect. In a defined minimal media containing only amino acids and salts (22), MRM, WT L. pneumophila replicated, but phtA mutants did not (Fig. 3). Supplementation of MRM with casamino acids (5 g/liter) had little effect on the growth of either strain, but addition of tryptone (5 g/liter) fully restored phtA mutant growth to that of WT (Fig. 3). The amino acids of tryptone are predominantly in peptide form (62.4% of amino acid content), whereas casamino acids contain free amino acids (only 2.67% of amino acid content in peptide form; Difco). These data suggested that PhtA is an amino acid transporter whose function can be bypassed by peptide permeases.
Fig. 3.
phtA mutants, which do not grow in MRM, can be fully rescued by the addition of tryptone but only partially rescued with casamino acids. WT bacteria (solid lines, filled squares) replicate slowly and to low densities in MRM, but phtA mutants (solid line, filled circles) do not. When MRM is supplemented with 5 g/liter casamino acids, WT (dotted line, filled squares) and phtA mutants (dotted line, filled circles) are only partially aided, but supplementation with 5 g/liter tryptone restores WT (solid lines, open squares) and phtA mutant (solid line, open circles) growth alike. A representative of three independent experiments performed in triplicate is shown.
Biolog Phenotype Microarrays Indicate That Threonine Dipeptides Can Suppress the MRM Growth Defect. We reasoned that the substrate of PhtA could be deduced by identifying the composition of peptides that rescue growth of phtA mutants. For this purpose, we applied a Biolog phenotype microarray to screen a library of di- and tripeptide combinations that span all 20 amino acids. Of the 282 peptide combinations tested, only 15 supported phtA growth in MRM, and each of these peptides contained threonine (Fig. 9, which is published as supporting information on the PNAS web site). Therefore, PhtA likely equips L. pneumophila to acquire threonine.
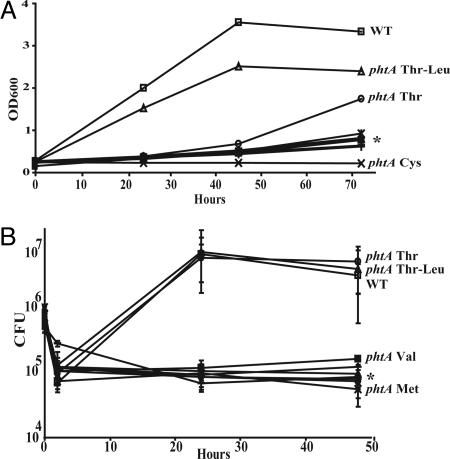
To confirm that threonine-containing dipeptides bypass phtA function, we quantified replication by phtA mutants in MRM supplemented with Thr-Leu or Ala-Leu dipeptides (3 mM). In addition, we tested whether the phtA mutant growth defect was bypassed by any of the six amino acids that are essential to L. pneumophila, namely 3 mM Arg, Cys, Met, Ser, Thr, or Val (22). Threonine-containing peptides and free threonine supported growth of the mutant, but Ala-Leu peptides or any other free amino acid did not (Fig. 4A). The growth-defect suppression by free threonine was dose-dependent (data not shown); it also was sensitive to competition, because a mixture of all six essential amino acids did not bypass PhtA activity (data not shown). Therefore, phtA is predicted to encode a high-affinity threonine transporter whose function can be bypassed by less efficient peptide and amino acid transporters, possibly serine transporters (26, 27).
Fig. 4.
Dipeptides that contain threonine or excess free threonine bypass phtA function in broth and macrophages. (A) Defined media were supplemented with 3 mM of the indicated amino acid or peptide; then phtA mutant growth was assessed by measuring culture density (OD600). The asterisk represents the following independent samples: phtA Met, phtA, phtA Val, phtA Ala-Leu, phtA Ser, and phtA Arg from top to bottom, respectively, none of which replicated. This experiment was performed in triplicate, and a representative of three independent experiments is shown. (B) Macrophage culture media were supplemented with 3 mM peptide or amino acid as indicated. The asterisk represents the following independent samples: phtA Arg, phtA Ala-Leu, phtA Cys, phtA Ser, and phtA, from top to bottom, respectively; none of these replicated. This experiment was performed in triplicate; means and standard deviations are presented.
Threonine Dipeptides or Excess Free Threonine Suppresses the Growth Defect of phtA Mutants in Macrophages. To determine whether L. pneumophila also rely on PhtA to acquire threonine in macrophages, we tested the same series of peptides and free amino acids for their ability to rescue the intracellular replication defect of phtA mutants. As predicted by the in vitro model, both threonine-containing peptides and excess free threonine (3 mM) suppressed the phtA mutant growth defect in macrophages (Fig. 4B). To a first approximation, the suppression seemed complete; however, intracellular growth assays are not sufficiently sensitive to detect modest differences in replication rates. The suppression of the phtA phenotype was threonine-specific, because other amino acids and peptides did not rescue growth (Fig. 4B), and additional threonine in any form did not alter replication by WT bacteria (data not shown).
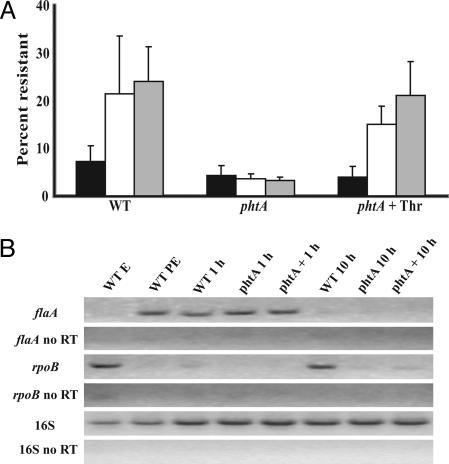
In Macrophages, phtA Mutants Fail to Differentiate to the Replicative Form. Because L. pneumophila that lack the PhtA transporter traffic properly but do not initiate intracellular replication, we determined whether phtA is required for transmissive bacteria to differentiate to the replicative form. To test whether the mutants persisted in macrophages in the transmissive or the replicative phase, we first measured their sensitivity to sodium, a trait that distinguishes the two forms of WT L. pneumophila. After a 1-h infection, both WT and phtA mutants were sodium-sensitive, a feature characteristic of the transmissive phase (Fig. 5A). In contrast, by 2 and 10 h after infection, WT bacteria were sodium-resistant, a hallmark of the replicative phase, but phtA mutants remained sodium-sensitive, indicating they had failed to differentiate (Fig. 5A). The addition of 3 mM free threonine to macrophage cultures triggered differentiation of intracellular phtA mutants to the replicative form, as judged by their sodium resistance at 2 and 10 h postinfection (Fig. 5A)
Fig. 5.
Intracellular phtA mutants fail to differentiate. (A) WT, phtA mutants, and phtA mutants supplemented with 3 mM threonine obtained from macrophages after 1 h (black bars) are sensitive to 100 mM NaCl. By 2 h (white bars) or 10 h (gray bars) after infection, WT and threonine-supplemented phtA mutants are NaCl-resistant, but without supplementation, phtA mutants remain sodium-sensitive. Experiments were performed in triplicate, and means and standard deviations are presented. (B) WT, phtA mutants, and phtA mutants supplemented with 3 mM threonine were lysed from host cells 1 h or 10 h postinfection; then their expression of the replicative phase rpoB gene and the transmissive phase flaA gene was analyzed by RT-PCR. As a reference for the replicative (WT EE) and transmissive (WT PE) differentiation state, RNA purified from broth cultures was analyzed; to control for genomic DNA contamination, non-reverse-transcribed RNA samples also were analyzed (no RT). 16s rRNA was analyzed as a loading control.
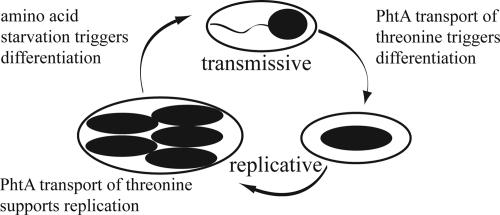
To examine differentiation of intracellular bacteria by an independent approach, we analyzed expression of genes that are specific to either the replicative or the transmissive phase. Consistent with their sodium sensitivity, both WT and phtA mutants contained transcripts of the transmissive phase gene flaA 1 h after infection (Fig. 5B). By 10 h after infection, mRNA of the replicative phase gene rpoB was detectable in WT bacteria but not in phtA mutants, indicating that WT bacteria had differentiated to the replicative phase but phtA mutants had not (Fig. 5B). Minimal phtA mutant expression of rpoB 10 h after infection was observed in cultures supplemented with excess threonine (Fig. 5B). These findings suggest that L. pneumophila exploit PhtA to identify nutrient rich environments as a prerequisite for differentiation to the replicative phase (Fig. 7).
Fig. 7.
PhtA couples nutrient acquisition to microbial differentiation at two stages of the L. pneumophila life cycle. PhtA threonine transport triggers intracellular transmissive L. pneumophila to differentiate to the replicative form (Fig. 5). Threonine acquisition by means of PhtA also is essential for L. pneumophila to replicate in macrophages (Figs. 1 and 4). When threonine acquisition by replicating bacteria is inefficient, L. pneumophila prematurely express transmission traits (Fig. 6), perhaps due to an early accumulation of the alarmone ppGpp.
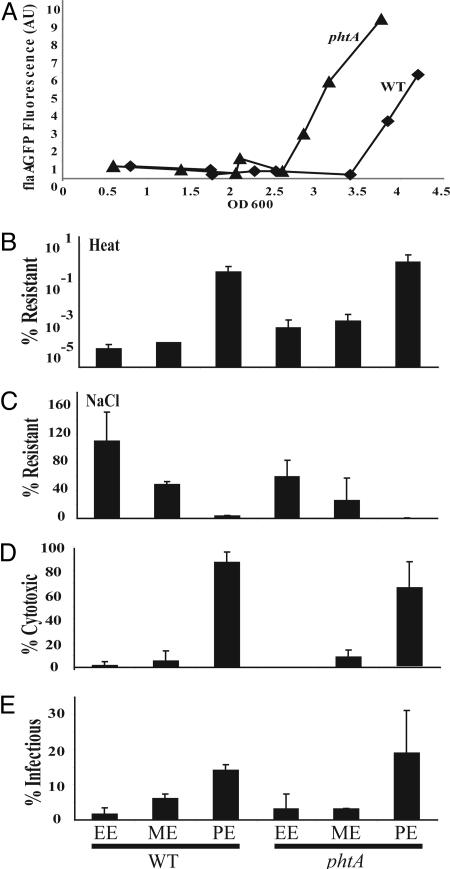
In Broth, phtA Mutants Prematurely Express Certain Transmission Traits. In broth, replicating L. pneumophila differentiate to the transmissive form in response to (p)ppGpp, a second messenger of the stringent response pathway that accumulates when amino acids become scarce (11). To determine whether the efficiency of threonine transport affects L. pneumophila induction of transmission traits, the phenotype of phtA mutants was assayed during growth in broth. Replication of the mutant in rich ACES-buffered yeast extract (AYE) media was comparable with WT, as judged by both their growth rates and maximal OD600 (data not shown); however, phtA mutants became motile prematurely. To analyze the kinetics of motility expression, we quantified GFP fluorescence of bacteria transformed with pflaG, a plasmid in which the flaA promoter is fused to a gfp gene to report transcription of the flagellin gene (11). Flagellin promoter expression was activated prematurely by phtA mutants at culture densities of OD600 ≈2.5 compared with WT bacterial cultures, which became fluorescent at an OD600 of ≈3.5 (Fig. 6A). Of the five transmission phenotypes examined, motility was the most drastically affected; however heat resistance and NaCl sensitivity were somewhat premature, whereas cytotoxicity and infectivity were indistinguishable from WT (Fig. 6 B–E). Consistent with the stringent response paradigm, in the absence of PhtA, replicating bacteria likely experience threonine starvation prematurely; consequently, the mutants induce expression of certain transmissive traits at lower cell densities (Fig. 7).
Fig. 6.
Premature activation of certain transmission traits by phtA mutants replicating in broth. (A) Fluorescence (AU, arbitrary units) from cultures carrying the pflaG reporter indicates that phtA mutant (triangles) express flagellin earlier than WT cells do (squares). The experiment was performed in duplicate, and a representative experiment is shown. (B–E) phtA mutants prematurely express heat resistance (B) and NaCl resistance (C), but not cytotoxicity (D) or infectivity (E), as judged by comparing phtA mutants and WT bacteria obtained from ME cultures (OD600 2.5). Replicative phase (OD600 1.5; EE) and transmissive phase (OD600 3.5–4.0; PE) WT and phtA mutant cultures serve as references. Each assay was conducted in triplicate, and the means and standard deviations are shown.
Discussion
The ability of microbes to recognize and react to their surroundings is a hallmark of their evolutionary success both in the environment and during infection. Cycling between a transmissive form and a replicative phase in response to the intracellular cues is integral not only to L. pneumophila virulence, but also to pathogens such as C. burnetii, Chlamydia species, and B. anthracis and even the eukaryotic Leishmania species (3, 5, 6, 28). Here, we show that acquisition of an essential amino acid by PhtA, a member of a subclass of MFS transporters, is a prerequisite for transmissive L. pneumophila to differentiate to a replicative form in host cells (Figs. 1 and 5).
The L. pneumophila Pht subfamily of MFS transporters (25) consists of 11 members in strains Philadelphia 1 and Lens, and nine loci in strain Paris (29). Analysis of the phylogeny and chromosomal position of the Philadelphia 1 pht family reveals likely gene duplications (Fig. 8). For example, phtJ (milA) and phtG are 41% identical and are the closest relatives of one another. Gene duplication may provide functional redundancy: phtJ mutants replicate efficiently in MRM (data not shown) and have only a modest growth defect in macrophages (30), whereas cells that lack the nonpartnered phtA locus have severe growth defects (Figs. 1 and 3). Based on sequence alignments, the Philadelphia 1 Pht family can be classified into seven functional groups. Additional experiments can determine whether each group is dedicated to one of the six amino acids that are essential for L. pneumophila [arginine, cysteine, methionine, serine, threonine, and valine (22)], or whether the Pht proteins have distinct functions, some of which are essential for growth in macrophages. We propose that, before reentering the cell cycle, the pathogen utilizes its Pht transporter family to gauge whether each of the amino acids it requires for growth is available. A similar strategy may be used by two other pathogens that replicate in vacuoles of amoebae and macrophages: C. burnetii encodes nine pht genes, and F. tularensis carries four (7, 17, 31, 32). Thus, Pht transporters may have evolved specific attributes that are critical for pathogens to exploit vacuoles of professional phagocytes.
Very little is known about the nutrient composition of the vacuoles inhabited by L. pneumophila or by other pathogens. Immediately after internalization, L. pneumophila is sequestered for hours from the endocytic pathway, a virulence strategy that trades access to nutrients for protection from lysosomal enzymes. The macrophage amino acid transporter SLC1A5 promotes replication by WT L. pneumophila (25). Furthermore, PhtA function can be bypassed by either exogenous peptides or excess threonine (Fig. 4), indicating that L. pneumophila also is equipped with less efficient mechanisms to acquire this essential amino acid. Nevertheless, intracellular phtA mutants exhibit a severe differentiation and growth defect (Figs. 1 and 5). Therefore, either phagosomes do not contain sufficient peptides or threonine to bypass the phtA defect, or other amino acids effectively compete with their transport, as observed in broth cultures (data not shown). Accordingly, to replicate in nutrient poor phagosomes, L. pneumophila require an efficient threonine transporter, PhtA.
The role that the Pht family transporters play in the life cycle of the Gram-negative bacterium L. pneumophila is conceptually similar to the function of the ger operons of the Gram-positive spore-forming Bacillus species. In both L. pneumophila and B. anthracis, membrane proteins recognize specific molecules in the environment, including amino acids, then trigger resilient infectious particles to differentiate to initiate replication (3). Although Ger proteins have been shown not to be transporters (33), each ger operon responds to either a distinct molecule or a combination of germinants, and a single ger operon is sufficient to trigger germination (34). For L. pneumophila, it is logical that each of the Pht transporters must recognize its substrate(s) before the cells differentiate, because loss of phtA alone confers a complete replication defect. Transport of essential nutrients to the cytosol by Pht proteins would then trigger the bacterium to reenter the cell cycle, perhaps by releasing the stringent response. The impact of Pht proteins on both the transition to the replicative phase (Fig. 5) and its duration (Figs. 6 and 7) makes this class of transporters an attractive target for the rational design of molecules to manage not only spread of L. pneumophila but perhaps also the potential bioterrorist agents F. tularensis and C. burnetii.
Supplementary Material
Acknowledgments
We thank Drs. Victor DiRita and Mary O'Riordan for their critical review of the manuscript. We also thank K. T. Young for performing in vitro mariner transposon saturation mutagenesis of the letE-phtA locus and Dr. Yousef Abu Kwaik (University of Louisville, Louisville, KY) for the phtJ mutant strain.
Author contributions: J.-D.S., M.A.B., and M.S.S. designed research; J.-D.S. and M.A.B. performed research; J.-D.S., M.A.B., and M.S.S. analyzed data; and J.-D.S. and M.S.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ER, endoplasmic reticulum; EE, early exponential; ME, midexponential; PE, postexponential; BMM, bone marrow-derived macrophage; LAMP, lysosome-associated membrane protein; MRM, modified Ristroph media; MFS, major facilitator superfamily.
References
- 1.Ohta, N. & Newton, A. (1996) Trends Microbiol. 4, 326–332. [DOI] [PubMed] [Google Scholar]
- 2.Kaiser, D. (1999) Trends Genet. 15, 273–277. [DOI] [PubMed] [Google Scholar]
- 3.Setlow, P. (2003) Curr. Opin. Microbiol. 6, 550–556. [DOI] [PubMed] [Google Scholar]
- 4.Glavin, F. L., Winn, W. C., Jr., & Craighead, J. E. (1979) Ann. Intern. Med. 90, 555–559. [DOI] [PubMed] [Google Scholar]
- 5.Moulder, J. W. (1991) Microbiol. Rev. 55, 143–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman, S. A., Fischer, E. R., Howe, D., Mead, D. J. & Heinzen, R. A. (2004) J. Bacteriol. 186, 7344–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horwitz, M. A. (1983) J. Exp. Med. 158, 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger, K. H., Merriam, J. J. & Isberg, R. R. (1994) Mol. Microbiol. 14, 809–822. [DOI] [PubMed] [Google Scholar]
- 9.Byrne, B. & Swanson, M. S. (1998) Infect. Immun. 66, 3029–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterji, D. & Ojha, A. K. (2001) Curr. Opin. Microbiol. 4, 160–165. [DOI] [PubMed] [Google Scholar]
- 11.Hammer, B. K. & Swanson, M. S. (1999) Mol. Microbiol. 33, 721–731. [DOI] [PubMed] [Google Scholar]
- 12.Hammer, B. K., Tateda, E. S. & Swanson, M. S. (2002) Mol. Microbiol. 44, 107–118. [DOI] [PubMed] [Google Scholar]
- 13.Molofsky, A. B. & Swanson, M. S. (2003) Mol. Microbiol. 50, 445–461. [DOI] [PubMed] [Google Scholar]
- 14.Tesh, M. J. & Miller, R. D. (1981) J. Clin. Microbiol. 13, 865–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wieland, H., Ullrich, S., Lang, F. & Neumeister, B. (2005) Mol. Microbiol. 55, 1528–1537. [DOI] [PubMed] [Google Scholar]
- 16.Molofsky, A. B. & Swanson, M. S. (2004) Mol. Microbiol. 53, 29–40. [DOI] [PubMed] [Google Scholar]
- 17.Oyston, P. C., Sjostedt, A. & Titball, R. W. (2004) Nat. Rev. Microbiol. 2, 967–978. [DOI] [PubMed] [Google Scholar]
- 18.Feeley, J. C., Gibson, R. J., Gorman, G. W., Langford, N. C., Rasheed, J. K., Mackel, D. C. & Baine, W. B. (1979) J. Clin. Microbiol. 10, 437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swanson, M. S. & Isberg, R. R. (1995) Infect. Immun. 63, 3609–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendrixson, D. R., Akerley, B. J. & DiRita, V. J. (2001) Mol. Microbiol. 40, 214–224. [DOI] [PubMed] [Google Scholar]
- 21.Swanson, M. S. & Isberg, R. R. (1996) Ann. N.Y. Acad. Sci. 797, 8–18. [DOI] [PubMed] [Google Scholar]
- 22.Ristroph, J. D., Hedlund, K. W. & Gowda, S. (1981) J. Clin. Microbiol. 13, 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachman, M. A. & Swanson, M. S. (2004) Infect. Immun. 72, 3284–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy, C. R., Berger, K. H. & Isberg, R. R. (1998) Mol. Microbiol. 28, 663–674. [DOI] [PubMed] [Google Scholar]
- 25.Pao, S. S., Paulsen, I. T. & Saier, M. H., Jr. (1998) Microbiol. Mol. Biol. Rev. 62, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Templeton, B. A. & Savageau, M. A. (1974) J. Bacteriol. 120, 114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sumantran, V. N., Schweizer, H. P. & Datta, P. (1990) J. Bacteriol. 172, 4288–4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burchmore, R. J. & Barrett, M. P. (2001) Int. J. Parasitol. 31, 1311–1320. [DOI] [PubMed] [Google Scholar]
- 29.Cazalet, C., Rusniok, C., Bruggemann, H., Zidane, N., Magnier, A., Ma, L., Tichit, M., Jarraud, S., Bouchier, C., Vandenesch, F., et al. (2004) Nat. Genet. 36, 1165–1173. [DOI] [PubMed] [Google Scholar]
- 30.Harb, O. S. & Abu Kwaik, Y. (2000) Infect. Immun. 68, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anthony, L. D., Burke, R. D. & Nano, F. E. (1991) Infect. Immun. 59, 3291–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baca, O. G., Akporiaye, E. T., Aragon, A. S., Martinez, I. L., Robles, M. V. & Warner, N. L. (1981) Infect. Immun. 33, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott, I. R. & Ellar, D. J. (1978) Biochem. J. 174, 635–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paidhungat, M. & Setlow, P. (2000) J. Bacteriol. 182, 2513–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.