Abstract
Geranyl diphosphate synthase (GPPS) catalyzes the condensation of dimethylallyl diphosphate and isopentenyl diphosphate to form geranyl diphosphate. Geranyl diphosphate is the precursor of monoterpenes, a large family of natural occurring C10 compounds predominately found in plants. Similar to plants but unique to animals, some bark beetle genera (Coleoptera: Scolytidae) produce monoterpenes that function in intraspecific chemical communication as aggregation and dispersion pheromones. The release of monoterpene aggregation pheromone mediates host colonization and mating. It has been debated whether these monoterpene pheromone components are derived de novo through the mevalonate pathway or result from simple modifications of dietary precursors. The data reported here provide conclusive evidence for de novo biosynthesis of monoterpene pheromone components from bark beetles. We describe GPPS in the midgut tissue of pheromone-producing male Ips pini. GPPS expression levels are regulated by juvenile hormone III, similar to other mevalonate pathway genes involved in pheromone biosynthesis. In addition, GPPS transcript is almost exclusively expressed in the anterior midgut of male I. pini, the site of aggregation pheromone biosynthesis. The recombinant enzyme was functionally expressed and produced geranyl diphosphate as its major product. The three-dimensional model structure of GPPS shows that the insect enzyme has the sequence structural motifs common to E-isoprenyl diphosphate synthases.
Keywords: isoprenyl diphosphate synthase, monoterpene biosynthesis, ipsdienol
Short-chain E-isoprenyl diphosphate synthases (E-IPPSs) are a class of prenyltransferases that are central to isoprenoid metabolism. This group includes geranyl diphosphate synthase (GPPS; EC 2.5.1.1), farnesyl diphosphate synthase (FPPS; EC 2.5.1.10) and geranylgeranyl diphosphate synthase (GGPPS; EC 2.5.1.30) which synthesize geranyl diphosphate (GPP), farnesyl diphosphate, and geranylgeranyl diphosphate, respectively. These phosphorylated aliphatic products are the backbone components of >30,000 biologically essential isoprenoid-derived metabolites (e.g., terpenes, sterols, ether-linked lipids, dolichols, ubiquinones, retinoids, chlorophylls, and prenylated proteins) (1, 2). All E-IPPSs catalyze an ordered sequential prenyl transfer involving a 1′-4 condensation reaction between an allylic diphosphate and isopentenyl diphosphate (IPP) (3). The sequential addition of IPP to the growing hydrocarbon chain of the allylic substrate generates polyprenyl homologues of increasing chain length. The reaction proceeds and terminates precisely at a specific carbon chain length according to the enzyme's product chain length specificity (4, 5).
The deduced amino acid sequences of all E-IPPS have high similarity, and sequence alignments reveal five conserved sequence domains (I, II, III, IV, and V) (6). Domain II and IV contain the substrate-binding motifs, the two aspartate-rich regions (DDX2–4D; X represents any amino acid), which are flanked by other conserved residues. These two regions are defined as the first aspartate-rich motif (FARM) and the second aspartate-rich motif. Domain II, which includes the FARM, is designated as the chain length determination region because of the presence of amino acids that are recognized as determinants of product chain length specificity (7, 8).
FPPS and GGPPS are essentially ubiquitous in nature, whereas GPPS has more limited distribution in plants. GPPS catalyzes a single condensation of dimethylallyl diphosphate (DMAPP) with IPP to produce GPP, the C10 backbone component of monoterpenes (9). Monoterpene biosynthesis is primarily localized to plastids of plant cells (10–12), found in the essential oil and resin of plants they function in plant defenses, in allelopathic responses, and as attractants of pollination (1). Recently, the isolation of cDNAs encoding GPPS from Mentha piperita (13), Arabidopsis thaliana (14), Abies grandis (15), Antirrhimum majus, and Clarkia breweri (11) has allowed for its functional characterization.
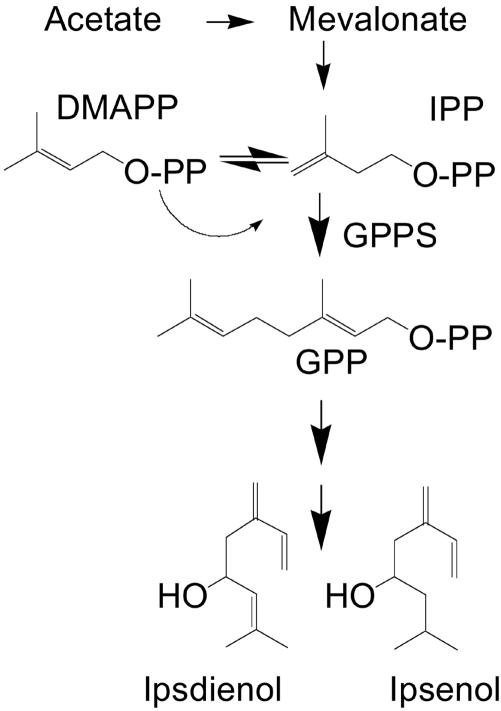
Monoterpene synthesis is usually not associated with animals. Subsequently in animals, GPP serves as an intermediate substrate for FPPS, which synthesizes FPP (C15), an essential precursor of cholesterol in vertebrates (1). In contrast, several bark beetle species produce monoterpene-derived aggregation pheromone to coordinate colonization and mating. Until the last decade, it was widely accepted that bark beetles, in contrast to most other insects (16), derived their monoterpene aggregation pheromones by simple modification of host tree dietary precursors (17, 18). However, it is becoming increasingly clear that most pheromone components are synthesized de novo in the anterior midgut tissue (19–21) of pheromone-producing beetles through the mevalonate pathway (22, 23). The allylic C10 alcohol, ipsdienol, is the major monoterpene-derived aggregation pheromone of male Ips pini (Fig. 1) (24). Many mevalonate pathway genes involved in pheromone biosynthesis of I. pini have been identified, and the regulation of transcript expression has been characterized (25, 26).
Fig. 1.
Abbreviated pathway of de novo biosynthesis defining the reaction catalyzed by GPPS in production of monoterpene pheromone components of male I. pini.
In this study, we report the isolation of a GPPS of an animal and present evidence that it functions in bark beetle pheromone biosynthesis. A full-length GPPS cDNA from I. pini was cloned and functionally expressed. GPPS is a single-copy gene, and transcript levels are induced specifically in the anterior midgut of pheromone-producing male beetles. The three-dimensional model structure of GPPS, built on the crystal structure of the avian FPPS (27, 28), folded as a single all-α-helical domain with variable loop regions and showed the conserved structural features that are essential for E-IPPS activity.
Materials and Methods
Substrates, Reagents, and cDNA Library. [1-14C]IPP was purchased from American Radiochemicals (St. Louis). Unlabeled IPP, DMAPP, GPP, FPP, and GGPP were purchased from Sigma. Oligonucleotide primers were obtained from GIBCO/BRL or Integrated DNA Technologies (Coralville, IA).
Sources of Beetles and Treatments. Immature I. pini were collected from infested Jeffrey pine (Pinus jeffreyi) from the University of Nevada Whittell Forest, NV, and Lassen National Forest, CA. Infested log bolts were cut into ≈1.5-m lengths and placed in rearing boxes in a greenhouse as described by Browne (29). Beetles were reared to adults before collection. Approximately 50 adult male beetles were topically treated with 10 μg of JH III dissolved in 0.5 μl of acetone or 0.5 μl of acetone (control) to the abdominal venter by using a 10-μl syringe with a 33-guage blunt-tipped needle (Hamilton, Reno, NV) and incubated for 16 h at 25°C in the dark. Midguts were dissected and tissue was flash frozen in liquid nitrogen and immediately homogenized on ice in 250–500 μl of 25 mM Hepes pH 7.2/2 mM MgCl2/5 mM KF/1 mM DTT/10% glycerol. Homogenized samples were centrifuged initially at 8,000 × g for 5 min, followed by a second spin at 16,000 × g for 20 min. The crude soluble tissue extract was then stored at –80°C or assayed directly for prenyltransferase activity.
Isolation and Cloning of GPPS. A standard PCR cloning strategy was used to isolate the cDNA sequence of GPPS from male I. pini. Template DNA from an I. pini cDNA library in λZAP II (19) was amplified by using degenerate primers (5′-GTNGARATGYTICAYACN-3′; 5′-RTCRTCYTGIACYTGRAA-3′) designed to recognize E-IPPS cDNAs (30). Preliminary amplified products included a fragment corresponding to FPPS. A 532-bp cDNA fragment corresponding to GPPS was generated by using a reverse primer [5′-GGCAGTCAGACCGATACCTGG-3′ (a sequence specific primer previously designed for amplifying FPPS)] and a R20 vector primer. The remaining downstream 716-bp fragment was amplified by PCR walking techniques with two forward sequence-specific internal primers (5′-CTTAGTGGAGTCCTA-3′; 5′-TTGGAGTTGACCAGTGCC-3′) and a T7 vector primer. cDNA amplification was carried out in a 50-μl reaction volume containing 50 pmol of each primer, 2.5 mM MgCl2, and 2 μl of cDNA library as template. The cycling parameters were as follows: 95°C for 60 s; followed by 35 cycles at 94°C for 60 s, 55°C for 60 s, and 72°C for 40 s. Amplified fragments were directionally ligated into pST-Blue-1 Acceptor vector (Novagen) and transformed in E. coli Novablue or Novagen Singles competent cells (Novagen). Sequence analysis of the 1.2-kb cDNA clone (IpiGPPS) was done by a dRhodamine dye-terminator kit (Applied Biosystems) and an Applied Biosystems Prism 3730 DNA analyzer (Nevada Genomics Center, University of Nevada, Reno). IpiGPPS was identified as an E-IPPS through a blast search. Protein sequence alignments were performed by using clustalw. The nearest neighbor-joining method was applied to create phylogenetic trees for short-chain E-IPPSs (Vector nti advance 9.0 sequence analysis software, Invitrogen).
Tissue Distribution. The tissue localization of GPPS mRNA of I. pini was determined by semiquantitative RT-PCR. Male adult beetles were dissected 8 h after treatment with 10 μg of JH III or acetone. This time point was chosen because GPPS transcript levels are already induced at 8 h (data not shown). Each sample contained tissues pooled from five insects that were separated into head, anterior midgut, posterior midgut, hindgut, fat body, appendages (legs and antennae), and carcass. The tissues were kept at –by 80°C until total RNA was prepared by using the MasterPure RNA Purification kit (Epicentre, Madison, WI). First strand cDNA template was synthesized by using Superscript III reverse transcriptase (Invitrogen) and an oligo(dT) primer (Promega) according to the manufacturer's instructions. To normalize the template amounts and determine the range of linear amplification, PCR was performed with I. pini cytosolic actin primers (5′-CATGTGTGACGAAGAAGTAGC-3′; 5′-GGCATAACCTTCATAGATGG-3′) at several different cycle numbers. Once the template amounts were equalized, the same amounts were used with the GPPS primer set (5′-GTACCTCAGTACGAGGAAAT-3′; 5′-CTCTTCAGTGAAATCGTTGA-3′) at the previously established number of cycles. All PCR products were separated on 1.5% agarose gels.
Functional Expression of Recombinant GPPS. A truncated version of IpiGPPS was generated for functional expression analysis. A cDNA coding region corresponding to amino acids P51-E416 was amplified by PCR with a Pfu Turbo Taq polymerase (Stratagene) and subcloned with the pET Trx Fusion System 32 (Novagen) into the pET-32c vector that had been previously digested with EcoRI and HindIII. The N-terminal Hisx6 tagged truncated clone was generated by using an adaptor oligonucleotide (5′-GCGAATTCCAAGTTCTCTGTCC-3′) as the upstream primer designed to create an EcoRI restriction site built into the start site and a downstream adaptor primer (5′-GCAAGCT TATTCAGGCCTCCTCTT-3′) generating a HindIII restriction site at the terminal stop codon. The pET-32c construct was transformed into Escherichia coli BL21-Star(DE3)pLysS cells (Invitrogen). The cell culture was grown to an OD600 of 0.5 and then induced in 1 mM isopropyl β-d-thiogalactoside for 3 h. Levels of protein expression were verified by SDS/PAGE (31).
Prenyltransferase Activity and Product Distribution Analysis. Prenyltransferase activity was measured by a standard acid lability assay that determined [1-14C]IPP incorporation into polyisoprenols (32). Samples were run in triplicate. Purified enzyme suspension (1–10 μg), clarified crude E. coli extract, or soluble beetle tissue extract (100–500 μg total protein) was incubated with 50 μM DMAPP and 10 μM [1-14C]IPP (55 μCi/μmol; 1 Ci = 37 GBq) for 1 h at 32°C in a 200 μl reaction volume containing 20 mM Hepes (pH 7.2), 2.5 mM MgCl2, 5 mM KF, 1 mM DTT, 10% glycerol, and protease inhibitor mixture. Protein concentration was determined by the Bradford method (33) or estimated by SDS/PAGE. The identification of isoprenyl diphosphate product formation was performed by reverse phase-HPLC. To terminate the reaction before loading the sample onto the column, samples were boiled for 5 min and spun at (16,000 × g for 5 min). Clear homogenates (injection volume 200 μl: 100 μl of reaction sample mixed with 75 μl of buffer and 25 μl of authentic mixed standards [(DMAPP, GPP, FPP, GGPP; 1:1:0.5:0.5)] were directly loaded onto a Discovery 250 × 4.6 C18, 5 μm, silica column (Supelco) by using a HP 1050 series instrument. Phosphorylated short-chain isoprenyls were cleanly separated by using a solvent system of 25 mM NH4HCO3 pH 7.0: acetonitrile described by Zhang and Poulter (34). Samples were eluted at a flow rate 1 ml/min by using a linear gradient program: 100% 25 mM NH4HCO3 for 5 min and 0–100% acetonitrile for 30 min. Eluent was monitored by UV (214 nm). One milliliter fractions were collected by using a Amersham Pharmacia LKB-FRAC-100 Fraction Collector (Amersham Pharmacia Biotech), and fractions corresponding to IPP/DMAPP, GPP, FPP, and GGPP peaks (retention times 4, 16, 20, and 25 min, respectively) were assayed for radioactivity by using a TRI-CARB 2900 TR liquid scintillation analyzer (Packard).
Three-Dimensional Structural Modeling of GPPS. A three-dimensional model structure of GPPS from I. pini was constructed through knowledge-based methods by using sybyl 6.91–7.0 (Tripos Associates, St. Louis) on Silicon Graphics (Mountain View, CA) Indigo2 workstations. The solution conformation was based on energy and statistical criteria by using a biased-threading algorithm [genefold (35), as implemented in sybyl]. The crystallographic structure of avian mutant FPPS (Protein Data Bank ID code 1UBY) was used as a template to build the I. pini GPPS model structure. Gaps in the structurally conserved regions were closed by a loop search utility. Selection of loops from solved structures (Protein Data Bank ID codes 1UBY and 1FPS) was based on sequence homology, end-to-end distance and absence of severe steric clashes with the previously built sections of the insect target structure. The conformation of loops was refined by molecular dynamics and minimizations by using an amber7 force field. Pseudoenergy functions (matchmaker) were used to evaluate energetically favorable conformers. The average positional energies were calculated, and the model was refined through energy minimizations (Powell, steepest descent) or molecular dynamics to obtain a local and global minimum, respectively. Pairwise alignments were made between the I. pini GPPS model structure and avian crystal structures (1FPS and 1UBY) to establish positions and nearby contacts of analogous catalytic residues located at or nearby the substrate binding site.
Results
Molecular Cloning and Sequence Analyses. A full-length cDNA clone encoding GPPS was isolated from a male I. pini cDNA library. The ORF is 1,248 bp, encoding a predicted 416-aa protein with a calculated molecular mass of 48.5 kDa and a pI of 8.33 (SwissProt/TrEMBL). The predicted translated product has the conserved sequence domains present in all E-IPPSs (6), including the FARM (DDIMD, D157–D161) and second aspartate-rich motif (NDFKD, N299–D303) (Fig. 2). Within the first 20 amino acids, residues from the start codon is a consensus N-terminal peroxisomal targeting sequence (S11–L19).
Fig. 2.
The cDNA coding region of GPPS is 416 amino acids. The conserved sequence domains (I–V) are underlined. The aspartate-rich motifs, common to all-E-isoprenyl diphosphate synthases, are bold and underlined. A peroxisomal targeting signaling sequence (italicized and underlined) is present at the N terminus.
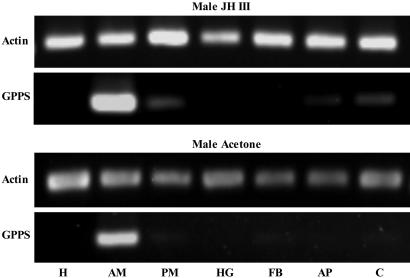
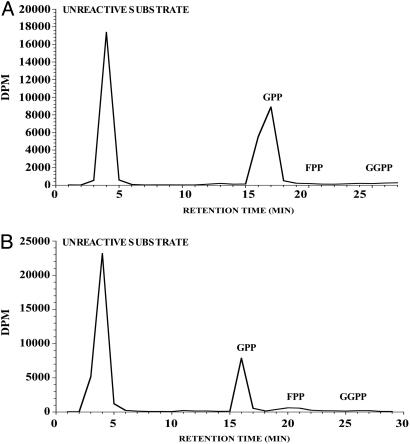
Localization of GPPS to the Midgut of Male I. pini. Northern analysis showed that GPPS is up-regulated by JH III in a dose- and time-dependent manner in male thoraces, similar to the transcriptional pattern described for other mevalonate pathway genes involved in pheromone biosynthesis (data not shown) (19, 36, 37). Semiquantitative RT-PCR demonstrated that the expression of GPPS was predominately localized to the anterior midgut tissue of pheromone-producing male I. pini (Fig. 3). Notably, in the midgut of JH III-treated male beetles, GPP was the only short-chain isoprenyl diphosphate product detected (Fig. 4A). Southern blot analysis showed a banding pattern consistent with a single-copy gene for GPPS of I. pini (data not shown).
Fig. 3.
Semiquantitative RT-PCR shows the tissue distribution of GPPS transcript from JH III- and acetone-treated male I. pini (head, H; anterior midgut, AM; posterior midgut, PM; hindgut, HG; fat body, FB; appendages, AP; carcass, C).
Fig. 4.
Radio-HPLC analyses of short-chain isoprenyl diphosphate products. Products are from midgut homogenates of JH III-treated male I. pini (A) and the recombinant-truncated GPPS (B).
Functional Expression and Product Formation. Induction of the transformed expression host with isopropyl β-d-thiogalactoside resulted in a highly expressed 60-kDa protein. Although the majority of protein was expressed in inclusion bodies, there was a sufficient level of active protein in the soluble fraction for functional analysis. The recombinant GPPS yielded GPP (C10)as the major product (Fig. 4B). FPP (C15) was also produced but at a much lower level (<3% of total radioactivity). GGPP (C20) was not detected under the assay conditions.
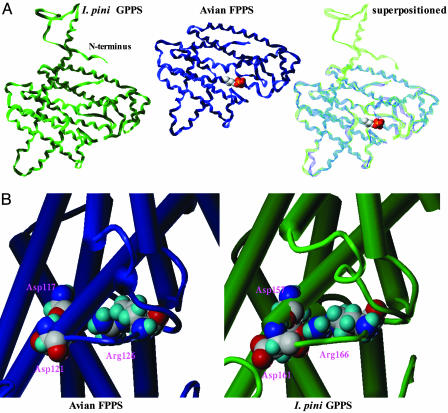
Model Structure of I. pini GPPS. A knowledge-based approach was used to propose a three-dimensional model structure of the I. pini GPPS. Pure sequence homology (composer, Tripos Associates, St. Louis) could not provide a protein structure for the GPPS enzyme because of its low sequence identity (<30%) with available structures deposited in the Protein Data Bank. Therefore, genefold was used to search for energetically favorable secondary motifs by using the mutant avian FPPS crystal structure 1UBY as a template. The best structure obtained for GPPS had an average positional energy of –0.13 kT compared with –0.20 kT of the avian crystal structure. The monomeric structure folds as a single all-α-helical domain with antiparallel α-helices connected by loop regions (Fig. 5A). The backbone atoms of the modeled GPPS superpositioned onto the crystal structure of the avian FPPS (1FPS or 1UBY) has a respective rms deviation of 2.2 Å, indicating the structural conformation of the insect enzyme is very similar to the avian crystal structure. The major structural feature is a large central core surrounded by 10 α-helices. The allylic and homoallyic substrate binding sites, the FARM and second aspartate-rich motif, respectively, are located in the largest of several solvent accessible channels.
Fig. 5.
Three-dimensional I. pini GPPS model structure and the avian FPPS crystal structure (1UBY). (A) GPPS model and avian crystal structures are superpositioned with DMAPP bound at the FARM region in the avian enzyme. The GPPS-extended N-terminal tail is not resolved in the avian crystal structure. (B) Catalytic Asp and Arg residues (avian FPPS, D117,D121,R126; I. pini GPPS, D157,D161,R166) are shown at the allylic binding site of the FARM region in the avian FPPS crystal structure and the I. pini GPPS model structure. The alpha helices are represented as barrels and the loop regions as tubes.
To establish conserved function and catalytic competence of the proposed GPPS model, we identified and compared the location and positional orientation of reactive residues with the solved avian FPPS structure (27, 28). The conserved catalytic residues (38, 39) at the FARM and second aspartate-rich motif binding motifs in the avian template (1FPS and 1UBY) were aligned with analogous residues in the I. pini GPPS model structure. Importantly, the conserved aspartate residues (avian FPPS, D117, D121; I. pini GPPS, D157, D161), which are involved in substrate binding at the FARM region were in isosteric positions within the active site (Fig. 5B).
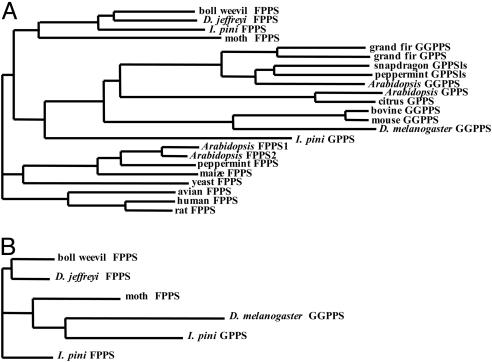
Phylogenetic Analyses. To date, FPPS cDNAs have been cloned from three coleopterans. A multiple sequence alignment of I. pini FPPS (AY953507), Dendroctonus jeffreyi FPPS (AY966009) and Anthonomus grandis (boll weevil) FPPS (AY966008) has a relatively high amino acid sequence identity of 60%. When I. pini GPPS (AY953508) is included in the alignment, the sequence identity lowers to 21%. Phylogenetic analysis based on the degree of sequence similarity between short-chain E-IPPSs shows that the I. pini GPPS shares a more recent ancestral origin with GGPPS (plant/animal) and GPPS (plant) rather than with FPPS (plant/animal) (Fig. 6A). Moreover, when comparing short-chain E-IPPSs from several insects, the I. pini GPPS clusters with GGPPS from Drosophila melanogaster (Fig. 6B).
Fig. 6.
Neighbor-joining trees based on sequence similarity of short-chain E-IPPSs. Sequence comparisons of short-chain enzymes from plants and animals (A) and insects (B).
Discussion
Until now, the distribution of GPPS has been restricted to plants, which produce an abundant array of monoterpenes. Here, we report cDNA clone-encoding GPPS from an animal. GPPS of I. pini functions at a late stage in the mevalonate pathway. It is required to generate GPP, the C10 precursor to ipsdienol, the major monoterpene-derived aggregation pheromone of male I. pini.
JH III regulates pheromone biosynthesis by coordinately regulating gene expression of mevalonate pathway genes in pheromone-producing midgut tissue (40). Likewise, GPPS mRNA is up-regulated in midgut tissue of JH III-treated male I. pini. Notably, there is an ≈41-fold higher basal level of GPPS in the midgut of males compared with female beetles, suggesting that GPPS may be modulated by sex-specific, developmental factors rather than directly by JH III (26). In turn, GPP was the only detectable short-chain isoprenyl diphosphate product produced in the midgut tissue of male beetles. Martin et al. (41) demonstrated the conversion of GPP to the acyclic monoterpene myrcene in male I. pini. Moreover, monoterpene synthase activity was shown to be up-regulated by JH III. These data suggest that GPPS plays a regulatory role in controlling the flux of carbon into monoterpenoids.
We expressed the IpiGPPS clone in E. coli to characterize for functional activity. The recombinant enzyme expressed as a single subunit is shown to be active and is not suggested to be a heterodimer. The recombinant enzyme yielded GPP as its major product. Although FPP was also detected in trace amounts, GGPP and longer-chain products were not present under the assay conditions. Short-chain E-IPPSs effectively use several allylic substrates to synthesize products with carbon chains longer than their native product specificities (8, 15, 42). Whether FPP production is due to the intrinsic activity of the recombinant GPPS or the in vitro assay conditions is unclear.
Southern blot analysis performed on genomic DNA of I. pini revealed GPPS as a single-copy gene. Nevertheless, it cannot be ruled out that several isoforms of GPPS translated from a single gene may exist for I. pini. Bouvier et al. (14) proposed two isoforms of GPPS translate from the same gene in A. thaliana. Depending on the transcriptional start site, a plastid- or a cytosolic-targeted enzyme may be generated.
The deduced amino acid sequence of GPPS from I. pini has the conserved sequence domains required by all E-IPPSs for substrate binding and catalytic activity (6), included is a peroxisomal targeting sequence [SVX5QL (X being any amino acid); S11-L19] at the amino-terminal end. The same PTS-2-like signaling peptide (SVX5QL; S94-L102) has been identified in 3-hydroxy-3-methylglutaryl-CoA synthase (HMG-S) in D. jeffreyi (43). Olivier et al. (44) showed this version of PTS-2 (SVX5QL) targeted cytosolic HMG-S and mevalonate diphosphate decarboxylase to the peroxisomes in Chinese hamster ovary cells. The absence of a peroxisomal signaling peptide for I. pini FPPS indicates that the two short-chain E-IPPSs are possibly targeted to different subcellular locations for functional activity. Subcellular compartmentalization may function in the regulation of GPPS activity and pheromone production in bark beetles.
The I. pini GPPS model structure has consensus with the avian FPPS crystal structure (27, 28). Reasonable assumptions were drawn from the deduced hypothetical structure based on the known mechanism of reaction and product chain length specificity of E-IPPSs (3, 8, 45). Importantly, analogous substrate binding and catalytic residues are similarly located and orientated in the I. pini GPPS model and avian FPPS crystal structure.
A three-dimensional model structure of FPPS from I. pini was also constructed through knowledge-based methods. Currently, ligand–protein interactions are being simulated at the binding site of both the I. pini GPPS and I. pini FPPS model structures. The substrate binding pocket of I. pini GPPS model structure is smaller in size than that of both the avian FPPS crystal and I. pini FPPS model structures, which reflects their known product chain-length specificities (2, 28). Preliminary analysis of steric interactions within the substrate binding pocket of the GPPS model structure shows that GPP is well fitted within the binding pocket, the distal end of the isoprene tail extends and is located at the pocket floor, whereas FPP is not well accommodated and tightly kinks within the binding pocket. Importantly, these comparative structural studies will help define determinants of product chain-length specificity between C10- and C15-producing enzymes within the same species.
Phylogenetic analyses support that IPPSs have evolved from a common ancestor into three distinct clades. The earliest divergence was a separation of the long-chain and short-chain E-IPPSs (6). It has been suggested that an archaebacterial GGPPS-type enzyme is the origin of short-chain biosynthesis. GPPS of plants are more evolutionary related to GGPPS than to FPPS. A phylogenetic tree based on the degree of sequence similarity between selected eukaryotic short-chain E-IPPSs show that FPPS (insect), GGPPS (plant/animal) and GPPS (plant/I. pini) all share a common origin. The next branching event within this cluster separates insect FPPS from GGPPS and GPPS. This event signifies that GPPS of I. pini evolved along the same lineage as GGPPS and GPPS of other eukaryotes. It should be noted that the I. pini GPPS is the least derived member of this group, implying it is more related to the insect FPPS enzymes than the other members of the cluster. GPPS of I. pini clusters with GGPPS of D. melanogaster, providing a clustering pattern for the insect GPPS that is similar to what is found for GPPS of plants. Both define GPPS activity as more related to GGPPS than to FPPS on an evolutionary scale. To date, the GGPPS of D. melanogaster is the only C20 producing enzyme isolated from insects (46).
The isolation of GPPS from I. pini provides conclusive evidence that bark beetles derive their monoterpene aggregation pheromone components de novo through the mevalonate pathway. In turn, this previously uncharacterized animal enzyme is an excellent candidate to selectively target and disrupt pheromone-mediated behavior of bark beetles. Moreover, the isolation of a GPPS from an animal also provides new insight into the regulatory mechanism of product chain-length specificity used by an important class of prenyltransferases.
Acknowledgments
We thank the Whittell Forest management board for allowing us to collect beetle-infested trees. This work is supported in part by National Science Foundation Grants IBN 9906530 and IBN 0316370 and the Nevada Agricultural Experiment Station publication no. 03055518.
Author contributions: A.B.G., C.T., W.H.W., and G.J.B. designed research; A.B.G. and J.C.B. performed research; A.B.G., J.C.B., C.T., and W.H.W. analyzed data; and A.B.G. and G.J.B. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DMAPP, dimethylallyl diphosphate; E-IPPSs, E-isoprenyl diphosphate synthases; FARM, first aspartate-rich motif; FPPS, farnesyl diphosphate synthase; GGPPS, geranylgeranyl diphosphate synthase; GPP, geranyl diphosphate; GPPS, geranyl diphosphate synthase; IPP, isopentenyl diphosphate.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AY953508 (Ips pini GPPS), AY953507 (Ips pini FPPS), AY966009 (Dendroctonus jeffreyi FPPS), and AY966008 (Anthonomus grandis FPPS)].
References
- 1.Poulter, C. D. & Rilling, H. C. (1981) in Biochemistry of Isoprenoid Compounds, eds. Porter, J. W. & Spurgeon, S. L. (Wiley, New York), Vol. 1, pp. 161–224. [Google Scholar]
- 2.Wang, K. C. & Ohnuma, S.-I. (2000) Biochim. Biophys. Acta 1529, 33–48. [DOI] [PubMed] [Google Scholar]
- 3.Poulter, C. D. & Rilling, H. C. (1978) Acc. Chem. Res. 307–313.
- 4.Kellogg, B. A. & Poulter, C. D. (1997) Curr. Opin. Chem. Biol. 1, 570–578. [DOI] [PubMed] [Google Scholar]
- 5.Ogura, K. & Koyama, T. (1998) Chem. Rev. (Washington, D.C.) 98, 1263–1275. [DOI] [PubMed] [Google Scholar]
- 6.Chen, A., Kroon, P. A. & Poulter, C. D. (1994) Protein Sci. 3, 600–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohnuma, S.-I., Hirooka, K., Ohto, C. & Nishino, T. (1997) J. Biol. Chem. 272, 5192–5198. [DOI] [PubMed] [Google Scholar]
- 8.Liang, P.-H., Ko, T.-P. & Wang, A. H.-J. (2002) Eur. J. Biochem. 269, 3339–3354. [DOI] [PubMed] [Google Scholar]
- 9.Gershenzon, J. & Croteau, R. (1993) in Lipid Metabolism in Plants, ed. Moore, T. C. J. (CRC, Boca Raton, FL), pp. 339–388.
- 10.Soler, F., Feron, G., Clastre, M., Dargent, R., Gleizes, M. & Ambid, C. (1992) Planta 187, 171–175. [DOI] [PubMed] [Google Scholar]
- 11.Tholl, D., Kish, C. M., Orlova, I., Sherman, D., Gershenzon, J., Pichersky, E. & Dudareva, N. (2004) Plant Cell 16, 977–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner, G. W. & Croteau, R. (2004) Plant Physiol. 136, 4215–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burke, C. C., Wildung, M. R. & Croteau, R. (1999) Proc. Natl. Acad. Sci. USA 96, 13062–13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouvier, F., Suire, C., d'Harlingue, A., Backhaus, R. A. & Camara, B. (2000) Plant J. 24, 241–252. [DOI] [PubMed] [Google Scholar]
- 15.Burke, C. & Croteau, R. (2002) Arch. Biochem. Biophys. 405, 130–136. [DOI] [PubMed] [Google Scholar]
- 16.Blomquist, G. J., Jurenka, R., Schal, C. & Tittiger, C. (2005) in Comprehensive Molecular Insect Science, eds. Gilbert, L. I., Iatrou, K. & Gill, S. (Elsevier Academic, San Francisco), Vol. 3, pp. 705–751. [Google Scholar]
- 17.Borden, J. H. (1985) in Comprehensive Insect Physiology, Biochemistry, and Pharmacology, eds. Kerkut, G. A. & Gilbert, L. I. (Pergamon, Oxford), pp. 257–285.
- 18.Vanderwel, D. (1994) Arch. Insect Biochem. Physiol. 25, 347–362. [Google Scholar]
- 19.Hall, G. M., Tittiger, C., Andrews, G. L., Mastick, G. S., Kuenzli, M., Luo, X., Seybold, S. J. & Blomquist, G. J. (2002) Naturwissenschaften 89, 79–83. [DOI] [PubMed] [Google Scholar]
- 20.Hall, G. M., Tittiger, C., Blomquist, G. J., Andrews, G., Mastick, G., Barkawi, L. S., Bengoa, C. & Seybold, S. J. (2002) Insect Biochem. Mol. Biol. 32, 1525–1532. [DOI] [PubMed] [Google Scholar]
- 21.Nardi, J. B., Gilg-Young, A., Ujhelyi, E., Tittiger, C., Lehane, M. J. & Blomquist, G. J. (2002) Tissue Cell 34, 221–231. [DOI] [PubMed] [Google Scholar]
- 22.Seybold, S. J., Quilici, D. R., Tillman, J. A., Vanderwel, D., Wood, D. L. & Blomquist, G. J. (1995) Proc. Natl. Acad. Sci. USA 92, 8393–8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tillman, J. A., Holbrook, G. L., Dallara, P. L., Schal, C., Wood, D. L., Blomquist, G. J. & Seybold, S. J. (1998) Insect Biochem. Mol. Biol. 28, 705–715. [Google Scholar]
- 24.Wood, D. L., Browne, L. E. & Silverstein, R. M. (1966) J. Insect Physiol. 12, 523–536. [DOI] [PubMed] [Google Scholar]
- 25.Eigenheer, A. L., Keeling, C. I., Young, S. & Tittiger, C. (2003) Gene 316, 127–136. [DOI] [PubMed] [Google Scholar]
- 26.Keeling, C. I., Blomquist, G. J. & Tittiger, C. (2004) Naturwissenschaften 91, 324–328. [DOI] [PubMed] [Google Scholar]
- 27.Tarshis, L. C., Yan, M., Poulter, C. D. & Sacchettini, J. C. (1994) Biochemistry 33, 10871–10877. [DOI] [PubMed] [Google Scholar]
- 28.Tarshis, L. C., Proteau, P. J., Kellogg, B. A., Sacchettini, J. C. & Poulter, C. D. (1996) Proc. Natl. Acad. Sci. USA 93, 15018–15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Browne, L. E. (1972) J. Econ. Entom. 65, 1499–1501. [Google Scholar]
- 30.Gilg-Young, A. (2004) Ph.D. thesis (University of Nevada, Reno, NV).
- 31.Laemmli, U. K. (1970) Nature 277, 680–695. [DOI] [PubMed] [Google Scholar]
- 32.Fujii, H., Koyama, T. & Ogura, K. (1982) Biochim. Biophys. Acta 712, 716–718. [PubMed] [Google Scholar]
- 33.Bradford, M. M. (1976) Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, D. & Poulter, C. D. (1993) Anal. Biochem. 213, 356–361. [DOI] [PubMed] [Google Scholar]
- 35.Jaroszewski, L., Rychlewski, L., Zhang, B. & Godzik, A. (1998) Protein Sci. 7, 1431–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivarsson, P., Tittiger, C., Blomquist, C., Borgeson, C. E., Seybold, S. J., Blomquist, G. J. & Hogberg, H.-E. (1998) Naturwissenschaften 85, 507–511. [Google Scholar]
- 37.Tillman, J. A., Lu, F., Goddard, L. M., Donaldson, Z. R., Dwinell, S. C., Tittiger, C., Hall, G. M., Storer, A. J., Blomquist, G. J. & Seybold, S. J. (2004) J. Chem. Ecol. 30, 2459–2494. [DOI] [PubMed] [Google Scholar]
- 38.Joly, A. & Edwards, P. A. (1993) J. Biol. Chem. 268, 26983–26989. [PubMed] [Google Scholar]
- 39.Song, L. & Poulter, C. D. (1994) Proc. Natl. Acad. Sci. USA 91, 3044–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tittiger, C. (2003) in Insect Pheromone Biochemistry and Molecular Biology: The Biosynthesis and Detection of Pheromones and Plant Volatiles, eds. Blomquist, G. J. & Vogt, R. G. (Elsevier Academic, San Francisco), pp. 209–215.
- 41.Martin, D., Bohlmann, J., Gershenzon, J., Francke, W. & Seybold, S. J. (2003) Naturwissenschaften 90, 173–179. [DOI] [PubMed] [Google Scholar]
- 42.Ohnuma, S.-I., Koyama, T. & Ogura, K. (1992) J. Biochem. 112, 743–749. [DOI] [PubMed] [Google Scholar]
- 43.Tittiger, C., O'Keeffe, C., Bengoa, C. S., Barkawi, L. S., Seybold, S. J. & Blomquist, G. J. (2000) Insect Biochem. Biol. 30, 1203–1211. [DOI] [PubMed] [Google Scholar]
- 44.Olivier, L. M., Kovacs, W., Masuda, K., Keller, G.-A. & Krisans, S. K. (2000) J. Lipid Res. 41, 1929–1935. [PubMed] [Google Scholar]
- 45.Ohnuma, S.-I., Hirooka, K., Tsuruoka, N., Yano, M., Ohto, C., Nakane, H. & Nishino, T. (1998) J. Biol. Chem. 273, 26705–26713. [DOI] [PubMed] [Google Scholar]
- 46.Lai, C., McMahon, R., Young, C., Mackay, T. & Langley, C. H. (1998) Genetics 149, 1051–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]