Abstract
An important step toward improving the annotation of the human genome is to identify cis-acting regulatory elements from primary DNA sequence. One approach is to compare sequences from multiple, divergent species. This approach distinguishes multispecies conserved sequences (MCS) in noncoding regions from more rapidly evolving neutral DNA. Here, we have analyzed a region of ≈238kb containing the human α globin cluster that was sequenced and/or annotated across the syntenic region in 22 species spanning 500 million years of evolution. Using a variety of bioinformatic approaches and correlating the results with many aspects of chromosome structure and function in this region, we were able to identify and evaluate the importance of 24 individual MCSs. This approach sensitively and accurately identified previously characterized regulatory elements but also discovered unidentified promoters, exons, splicing, and transcriptional regulatory elements. Together, these studies demonstrate an integrated approach by which to identify, subclassify, and predict the potential importance of MCSs.
Keywords: conserved noncoding elements, gene regulation, globin gene, comparative genomics
With the development of multiple computational algorithms and appropriate biological data, it is now possible to detect coding sequences relatively efficiently and accurately from primary DNA sequence (1). By contrast, it is still difficult to identify and assign function to noncoding sequences that include critical cisacting regulatory elements such as promoters, enhancers, silencers, locus control elements, nuclear matrix attachment sites, and origins of replication. A promising approach is to search for conserved orthologous noncoding sequences from multiple, evolutionarily diverse species (2–8), so-called multispecies conserved sequences (MCSs). However, at present, it is not established how best to design and carry out such searches. Furthermore, even when complete, it is not clear how to evaluate and prioritize MCSs for functional analysis.
An important issue is to know which species to include in searches for MCSs. When comparing the sequences of distantly related species [e.g., birds and mammals, which diverged 310 million years (my) ago], cis elements may have diverged too much to be easily identified. By contrast, when comparing more closely related species (e.g., rodents and primates, which diverged 64–74 my ago), it may be difficult to distinguish between sequence homology resulting from a slow rate of evolution and the conservation of functionally important cis elements. Nevertheless, it is becoming clear that analyzing multiple species in different combinations adds significant power to the identification of functionally important cis-acting sequences (2, 3).
Even when MCS can be unequivocally identified, there is often no way to judge how sensitive the searches have been. Do these routines overcall or undercall regulatory elements? Furthermore, the true functional significance of MCSs often remains untested and/or unknown because there is usually very limited experimental data to link genome structure to function at the locus in question. Therefore, to establish accurate and sensitive routines for evaluating cis-regulatory sequences, it will be necessary to analyze well characterized regions of the human genome so that such sequences can be recognized and their potential importance can be assessed and prioritized for functional studies.
Here, we have sequenced and/or annotated and compared extensive regions (47–238 kb) of conserved synteny containing the α-globin cluster, its known regulatory elements and flanking genes in 22 species representing widely divergent groups, spanning 500 my of evolution. Multiple alignments were analyzed with a variety of bioinformatic approaches, and the results were compared with algorithms for the detection of MCSs described in ref. 9 and a newly developed analysis tool, gumby (S. Prabhakar, unpublished data). The approach taken here allowed us to identify 24 MCSs within a 238-kb region containing and surrounding the human α-globin cluster. Previous studies of this locus have localized all DNase1 hypersensitive sites (DHSs; refs. 10 and 11), characterized its nuclear localization (12), nuclear matrix attachment (13), pattern of replication (14), chromatin structure (10), chromatin modification (15, 16) DNA methylation (17), and pattern of gene expression in normal loci and those with natural deletions (18), allowing us to evaluate in full the functional significance of the MCS elements identified here.
Materials and Methods
Clone Isolation and Sequencing. Bacterial artificial chromosomes (BACs) used in this study were isolated and separated by using standard techniques (19); all details concerning multiple alignments of the discussed elements are available as additional material from the authors on request. All BACs used in this study are available from the BACPAC Resources Centre (http://bacpac.chori.org). The sequences of the chimpanzee, cow, and cat were obtained from the ENCODE project (http://genome.gov/ENCODE), region ENm008.
Sequence Assembly and Finishing. Sequence reads were base called by using phred 0.020425 and assembled by using phrap (www.phrap.org) into a GAP4 database. Standard finishing methods were used.
Sequence Annotation and Multiple Alignment. Sequences were annotated against EST data when available for that species (http://genome.ucsc.edu and www.ensembl.org) or by using cross-species comparison of available ESTs (macvector 7.2, Accelrys, Inc., San Diego). The presence of nonsyntenic features were evaluated by using a modification of HPREP and stored in AceDB (www.acedb.org). Repeat annotation was generated by using repeatmasker (www.repeatmasker.org). Multiple alignments were generated and/or visualized by using vista (www-gsd.lbl.gov/vista/index.shtml); multipipmaker (http://pipmaker.bx.psu.edu/pipmaker), and multilagan (http://lagan.stanford.edu/lagan_web/index.shtml). Multiple alignments were analyzed as described in Results and were also compared with data obtained by using webmcs (http://research.nhgri.nih.gov/MCS/submit.shtml). In addition, multilagan alignments were analyzed for noncoding conservation by using the algorithm gumby, a tool for detecting statistically significant conserved regions in pairwise or multiple alignments of DNA sequences at any alignable evolutionary distance. Individual MCSs were aligned by using clustalw (macvector 7.2, Accelrys); bestfit (wisconsin package 10.3, GCG) and dialign (www.genomatix.de/cgi-bin/dialign/dialign.pl) and manually optimized.
Sequence Analysis. Potential transcription factor binding sites (TFBs) were identified by using transfac 6.0 (www.generegulation.com); tres (http://bioportal.bic.nus.edu.sg/tres), and regulatory vista (http://gsd.lbl.gov/vista/rvista/submit.shtml). Plots of Observed/Expected for the CpG dinucleotide were generated by using window and statplot 10.3 (GCG) comparing the local frequency to the global expected value over a 1,000-bp window.
Results
A Region of Conserved Synteny Maintained Throughout 500 my of Evolution. The structure of the α-globin cluster was established by DNA sequence analysis (6- to 8-fold coverage) in 22 divergent species representing well defined time points throughout 500 my of evolution (Fig. 1). In most species, there is an embryonic α-like gene(s) (ζ), adult gene(s) (α), and theta gene(s) (θ). In addition, we found that, in most species, there is a functional homologue of the previously characterized avian αD gene (20), lying between the ζ and α genes; in man, this gene was previously annotated as a pseudogene (ψα2; ref. 21). The structure and evolution of the α-like genes are summarized in Figs. 1, 2 and 5 (see also Table 2, which is published as supporting information on the PNAS web site).
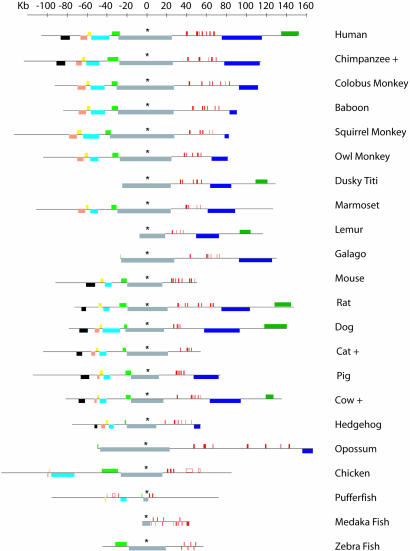
Fig. 1.
The arrangement (to scale) of globin genes (red boxes) and nonglobin genes (colored boxes) flanking the α-globin cluster. The extent of each black horizontal line indicates the amount of DNA sequenced in each species. Further details of these genes are shown in Fig. 2 (see also Fig. 5, which is published as supporting information on the PNAS web site). Sequences of species denoted + were obtained from encode, the rat sequence is from (http://rgd.mcw.edu). The loci are aligned on the highly conserved sixth exon of the C16orf35 gene, which is indicated by an asterisk.
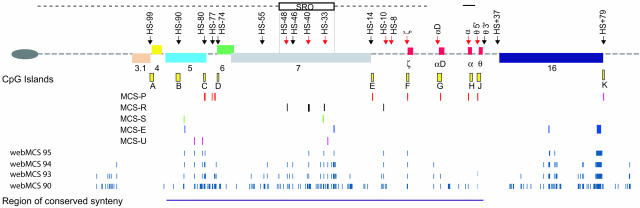
Fig. 2.
The prototypical α-globin cluster derived from multispecies comparisons. The telomere represented as an oval. The annotated globin genes (red boxes) and flanking genes (colored boxes) are shown. These genes are annotated as described in ref. 22: 3.1, POLR3K;4, C16orf33;5, C16orf8;6, MPG;7, C16orf35; and 16, LUC7L. Above the line are DNase1 hypersensitive sites described in the text (black arrows for constitutive and red for erythroid-specific sites). The extentof previously described deletions (black dashed horizontal line) is shown, with a small line over the α gene representing many common deletions (23) that remove these gene(s). The shortest region of overlap (SRO) of all upstream deletions is shown. Below are CpG islands (yellow boxes), conserved promoter elements (MCS-P, red lines), conserved regulatory elements (MCS-R, black lines), conserved splicing intronic regulatory elements (MCS-S, green lines), conserved alternative exons (MCS-E, blue lines), and conserved elements of unknown function (MCS-U, purple lines). In each line, the elements represent MCS-1, MCS-2, MCS-3, MCS-n from left to right. Aligned below the MCSs are the noncoding conservation scores for webmcs analyzed at the 95th, 94th, 93rd, and 90th percentiles. The region of conserved synteny is shown at the bottom as a dark purple line.
We have previously shown that the human α cluster lies in a GC-rich, repeat-rich, gene dense region of the genome close to the telomere of the short arm of chromosome 16, associated with internal, direct repeats of telomeric DNA (TTAGGG)n characteristic of such regions (22). The α-globin cluster in the rabbit (24) and horse (25) are also known to lie close to telomeric regions. Here, we found telomeric repeats upstream of the α cluster in 8 of 16 species analyzed, suggesting that, in many species, it is located or originated close to a telomere; a clear exception occurs in the mouse and rat clusters, which contain no telomeric repeats and lie at interstitial chromosomal locations (26, 27).
Previous comparisons of the mouse, chicken, and fugu (spheroides nephelus) α-globin clusters with a 376-kb segment of DNA containing the human α-globin cluster (28) allowed us to define accurately the limits of a ≈135-kb region of conserved synteny. This syntenic segment of DNA, although variable in size, mainly due to differences in the numbers of repeats and duplications/deletions of the α-like globin genes, contains the same set of genes arranged in the same order in all 22 species studied here, even in fish, which diverged from primates >500my ago. Together, these observations define a region of conserved synteny, which should contain all of the α-like globin genes and the cis-acting elements required for their fully regulated expression (Fig. 2).
The Evolution of CpG Islands and Their Role as Cis Elements. Within this region, CpG islands are among the simplest of cis elements to identify. They are short, unmethylated, GC-rich, CpG-dense regions of DNA usually located at the 5′ ends of genes and, potentially, they mark the position of a large proportion of promoters in mammalian genomes. Their role, if any, in regulating normal gene expression is not clear (29). We have previously characterized the CpG islands associated with the tissue-specific human α-globin genes and the widely expressed genes that flank them (10) (A–K, Fig. 6, which is published as supporting information on the PNAS web site).
At present, there are no clear definitions that easily allow the identification of CpG islands in widely diverse species. Parameters devised for one species may be inappropriate for another and most algorithms overcall CpG islands (30). However, by analyzing the CpG observed (in a 1,000-bp window)/CpG expected (estimated from the entire sequence of the α-globin cluster being analyzed), in most species, we could identify sequences corresponding to the known CpG islands previously identified in the human genome (Fig. 6).
Prominent CpG islands were detected in all primates, although we noted that islands associated with the α genes were less prominent in some species (e.g., Galago) than others. Similar patterns were seen in carnivores, cows, and pigs, although CpG islands were somewhat difficult to identify in pigs because there is a general lack of CpG suppression in the α cluster of this species (Fig. 5). A major difference was observed in rodents in which, as noted in ref. 31, some islands (including α globin) were substantially eroded or absent, whereas others (e.g., C and E) remained readily detectable. Whatever the mechanism by which they are lost, it appears that CpG islands are certainly not essential for gene regulation and, although often a valuable marker, may not provide a consistent guide to the presence of regulatory elements in all species.
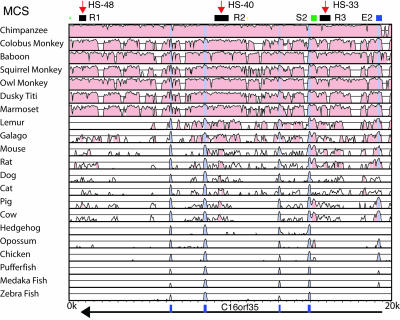
The Identification of Multispecies Conserved Sequences. To identify MCSs, we initially used a global alignment (avid visualized with vista, see Fig. 3), identifying and noting all conserved, noncoding regions that showed a minimum of 50% conservation in multiple mammalian species. vista highlights, in pink, regions with >75% conservation in 100 bp (Fig. 3). We next identified MCSs by direct visual inspection of local blastz alignments generated by pipmaker by using the sequences of the entire set of 22 species. All previously known coding regions were excluded from these analyses, and we focused on those conserved noncoding sequences, which aligned in >75% of the mammalian species available for study. All conserved sequences identified by the global alignment were also found in the local blastz alignment, but the latter identified additional candidate regions. To focus on sequences containing conserved motifs, the regions identified by vista and blastz were extracted and aligned by using clustalw, dialign, and bestfit and manually optimized. Sequences in which we found any 6 bp perfectly conserved in a 10-bp window, in >75% of mammalian species analyzed, defined the subset of MCSs described here. Using this approach, we found a total of 24 MCSs across a region of 238 kb. These MCSs were identified entirely independently of any prior knowledge of previously characterized regulatory elements in this area. For each MCS, we scored the length of conservation, noted all of the species in which conservation could be detected, and calculated the degree of conservation (Table 1).
Fig. 3.
Example of vista output for the 22 species between coordinates 94,273 to 114,273 from the telomere of the human α globin locus. Minimum conservation is 50% in a 100-bp window. Conservation >75% is colored pink, and annotated exons are shaded gray. At the top, shown are three classes of MCS: MCS-R1–3, MCS-S2, and MCS-E2. The position of the erythroid DHSs, HS-48, HS-40 and HS-33, are indicated by red arrows above. The direction of transcription (black arrow) and positions of exons (blue boxes) of the C16orf35 gene are shown below the vista plot.
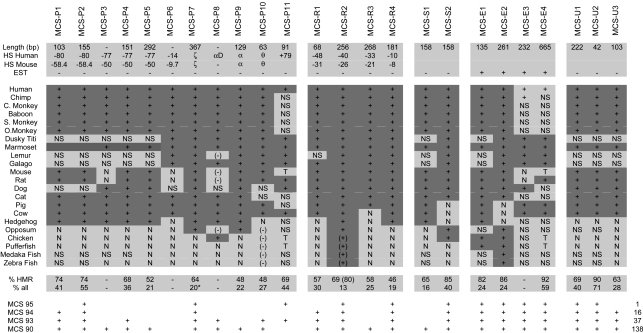
Table 1. Conservation of MCSs in 22 species.
The conservation of the described MCSs are shown for each species. MCSs are separated into five subclasses (see text). The length, names of the DHS or promoters with which the MCS are associated in human and mouse, and whether it is transcribed are shown in rows 2–5, respectively. For each species, MCSs are labelled as follows: +, present; N, not detectable; (-), gene not orthologous; (+), similar TFBs but not homologous; NS, no sequence. Values for % conservation in human, mouse, and rat (HMR) and proportion of bases perfectly conserved in all species are given. The asterisk in MCS-P7 indicates this figure was derived from an alignment without hedgehog and opossum due to evolutionary shuffling of the conserved motifs in these species. + indicates in which percentile webMCS detected the described MCS and, at right, the number of additional MCSs scored. Coordinates for start and stop of each MCS are given relative to the telomere of human chromosome 16.
To compare this approach with current algorithms, we searched for MCSs by using webmcs (9) set at the default value (95%), which assumes that 5% of the genome is under purifying selection. When all known exons were excluded, webmcs identified 13 individual MCSs, including 12 of the 24 MCSs described here. When set at 90%, webmcs found 138 elements, including 23 of the 24 MCS elements identified here. To search for MCSs in an another independent manner, we also ran gumby on multilagan alignments of closely related eutherians in the region of conserved synteny and the adjacent region containing Luc7L. At the default P value threshold of 0.5, gumby identified 20 of the 24 MCSs defined here and 19 additional regions. At a stricter threshold of 0.1, gumby identified 16 of the 24 MCS and 10 other predictions. These findings are discussed in detail elsewhere (S. Prabhakar, unpublished data).
Classification of MCSs. Together with previously described mapping of CpG islands (10, 22), localization of DHSs (10, 11), identification of ESTs, and annotation of genes (22), the 24 MCSs identified here could be subclassified into five groups. These groups are promoters (MCS-Ps), other transcriptional regulatory elements (MCS-Rs), splicing elements (MCS-Ss) previously unidentified exons (MCS-Es) and unknown elements (MCS-Us) (Fig. 2 and Table 1).
We found 11 MCSs associated with known promoters. Four (MCS-P1, MCS-P2, MCS-P6, and MCS-P11; Fig. 2) were found within CpG islands associated with the constitutively expressed DHSs of widely expressed genes. MCS-P3–MCS-P5 were found associated with a DHS marking an alternative promoter of the gene encoding methyladeneine DNA glycosylase (gene 6 in Fig. 2). Conserved elements MCS-P7–MCS-P10 were found in the promoters and CpG islands (when present) of the ζ, αD, α, and θ globin genes, respectively. Each of these genes is associated with erythroid-specific (ζ, αD, and α) or constitutive (θ) DHSs. Of the globin gene promoters, the embryonic (ζ) globin gene was the most highly conserved (64% conservation over 367 bp between man and mouse and detectable in all mammals).
A second group of MCSs (MCS-R1–MCS-R4 in Fig. 2 and Table 1) corresponded to known and presumed cis-acting regulatory elements associated with erythroid-specific DHSs. MCS-R1 identified a previously uncharacterized erythroid-specific DHS (ref. 16 and M.D.G., E.A., and J.R.H., unpublished data). Most of these MCSs were conserved, to variable extents, in all mammals (68–100my, Table 1) although MCS-R3 was not detectable in cows. MCS-R2 was the most highly conserved regulatory element, based on its 69% conservation between mouse and human over a region of 256 bp, and the fact that it could be detected in all mammals. This finding is consistent with its previously established role as the human α-globin major regulatory element (αMRE). None of the elements was readily detectable in birds (310 my) or fish (415–485 my), although the same repertoire of TFBs found in MCS-R2 were also found in the orthologous region in these vertebrate groups (28).
A third group includes MCS-S1 and MCS-S2. Initially, the role of these elements was not clear because they were not represented as ESTs and did not associate with any known DHSs (Table 1). However, we noted that MCS-S2 lies close to an alternative coding exon (MCS-E2, see below) within the gene C16orf35 (gene 7 in Figs. 2 and 3) but does not itself contain an ORF and has no EST database matches (UCSC/Ensembl genome browsers and ncbi blast). These data, combined with the relatively high degree of conservation in this element (MCS-S2 is 85% conserved over 158 bp between human and mouse and is detectable in most mammals, opossum, and chickens; Table 1) is reminiscent of conserved elements thought to be associated with RNA processing, although it does not fulfil the strict criteria for an ultraconserved element (32). Support for such a role in RNA splicing came from multispecies analysis. In cows, pigs and hedgehogs, the only evolutionary lines in which the neighboring coding exon MCS-E2 is not conserved (Table 1), MCS-S2 has also undergone rapid sequence divergence, suggesting that conservation of MCS-S2 is required to act as a signal for the alternative splicing of MCS-E2. In support of this hypothesis, a phylogenetic footprint of MCS-S2 revealed two highly conserved (100% from human to chicken) sequences (AGCATG), previously shown to be functional components of intronic control regions (ICRs), which regulate alternative splicing (33, 34). Similar conserved ICRs were also found in MCS-S1, which lies close to another known alternatively spliced exon in the rhomboid-like gene (gene 5, Fig. 2).
Four conserved elements (MCS-E1–MCS-E4) were present within spliced ESTs and are thought to represent exons. MCS-E1 is an alternative coding exon of the rhomboid-like gene (gene 5 in Fig. 2), and MCS-E2 is an alternative coding exon of C16orf35 (gene 7 in Fig. 2). MCS-E3 is an alternative coding exon of the LUC7L gene (gene 16 in Fig. 2), which encodes a component of the U1Snrp complex (35). MCS-E4 is a large, highly conserved element (1,600 bp) lying in the first intron of LUC7L. Part of MCS-E4 is incorporated as a noncoding exon, although its inclusion disrupts the normal reading frame of LUC7L. A 204-bp segment of MCS-E4 fulfils the criteria for an ultraconserved element (32) with only one base pair difference between the human, mouse, and rat.
We could not assign any known function to the remaining three elements MCS-U1–MCS-U3 (Fig. 2) even though they appear to be relatively highly conserved (Table 1).
Erythroid MCSs Contain Clusters of Conserved TFBs. Because expression of the α-globin cluster has been particularly well characterized, we analyzed the association of MCSs with erythroid-specific DHSs in depth. In particular, we optimized alignments of each MCS corresponding to the erythroid-specific promoters (MCS-P7–MCS-P10) and regulatory elements (MCS-R1–MCS-R4) and analyzed them by using the transfac database (version 6.0, both directly and utilizing tres and rvista), looking for proteins that might bind these conserved elements. Many of the phylogenetically conserved motifs corresponded to known sites of functional importance that have been previously shown to bind transcription factors in vitro and in vivo (16, 36–38). In particular, we found that six (MCS-R1–MCS-R4 and MCS-P7 and MCS-P8) of the eight MCSs associated with erythroid-specific DHSs contain at least one very highly conserved GATA binding site (WGATAR). Such sites are characteristically found in elements regulating genes that are switched on or off during erythropoiesis (39). MCS-R2 (αMRE) also contained two highly conserved Maf regulatory elements (40). MCS-P9 (α-globin promoter) contained conserved CCAAT and ATA motifs but no conserved GATA binding sites. Some elements (MCS-P7, MCS-P8, and MCS-P10) also contained highly conserved GC-rich elements, and some MCSs also included highly conserved elements that do not obviously correspond to the binding sites for any known transcription factors.
In many regulatory MCSs, individual TFBs have been completely conserved across all species (e.g., GATA binding site in MCS-R2, Fig. 4a). Others are fully conserved in most species but have mutated one or more critical residues that would alter binding in one or two species (e.g., GATA binding site in MCS-R2, Fig. 5b). In some MCSs, we noted that the disappearance of a conserved GATA site in one position was associated with the appearance of a new GATA site elsewhere in the element (e.g., in the dog in MCS-R4). Although the function of these new sites has not yet been tested, these observations are consistent with the concept of transcription factor “turnover” and coevolution (41), in which a regulatory element conserves overall function, albeit by using different sequence motifs.
Fig. 4.
Examples of conserved TFBs. The example is from MCS-R2. (a) A fully conserved GATA binding site followed by two conserved Maf recognition elements (MAREs) that form the core of this MCS. (b) A conserved GATA binding site in MCS-R2 that is lost in rodents and hedgehog.
Spacing Between Conserved TFBs in MCSs. From these and other data (16, 42), it seems likely that MCS-R1–MCS-R4 and MCS-P7 and MCS-P8 act as binding sites for multiple interacting proteins regulating gene expression and/or chromosome function. If such proteins bind and/or interact cooperatively, the spacing between binding sites may also have been conserved. The distances between the most consistently conserved binding sites (Fig. 7a, which is published as supporting information on the PNAS web site) in each MCS are plotted in Fig. 7b. In most elements, the distances between such sites are variable. However, in MCS-R2 (αMRE), the spacing and sequences between the fully conserved GATA binding site and two Maf recognition elements (Fig. 4 and intervals IIb and IIc in Fig. 7a), which form the core of this MCS, are highly conserved in 18 species, whereas spacing of the flanking GATA sites (intervals IIa and IId in Fig. 7a) are not (Fig. 7b). This result suggests that not only the sequence, but also the spacing and orientation of TFBs on the DNA helix, are highly conserved in some MCSs, which correspond to key regulatory elements.
Discussion
This study demonstrates that a combination of CpG analysis, mapping of DHSs and evaluation of MCSs, as defined here, can identify and subclassify most, if not all, critical, noncoding, cis-acting sequences. The additional question we have asked here is to what extent MCS analysis alone could achieve this result.
The multispecies analysis described here was highly sensitive and accurate for independently identifying previously characterized cis-acting elements within and surrounding the human α-globin cluster. Using the MCS data alone, we would have identified seven of eight promoters. The major promoter of the methyladenine DNA glycosylase gene (gene 6 in Fig. 2) is associated with a CpG island (D) but not an MCS. All of these promoters are associated with DHSs (Fig. 2). MCS analysis also identified an unknown alternative promoter of the MPG gene associated with a previously mapped DHS (HS-77). In general, these promoter elements are well conserved in mammals but not in birds and fish (Table 1).
In addition to the globin gene promoters, MCS analysis identified all previously known erythroid regulatory elements and one new element (HS-48), all of which are associated with erythroid-specific DHSs. As for promoters, these regulatory elements are well conserved in mammals but not in birds and fish (Table 1). The most conserved of these sequences (the promoters of the globin genes and the α globin major regulatory element, αMRE or HS-40) correspond to the most functionally important regulatory elements (11, 43, 44). In the αMRE, both the sequences and spacing of TFBs were conserved, and this finding may provide an important general criterion for subclassifying and prioritizing elements for functional analysis.
Two MCSs identified previously unknown elements containing very highly conserved IREs that were found to be conserved between mammals and birds. These MCSs, most likely, provide signals for mRNA splicing. Neither of these elements was associated with DHSs or ESTs. This analysis also identified four previously unrecognized exons, which were present in mammals, birds, and fish. One of these exons (MCS-E3) contains an ultraconserved element (200 bp 100% identical in human mouse and rat, ref. 32) whose function remains unknown.
The remaining question is whether the MCS analysis based on the criteria defined here may have missed additional cis-acting elements because we have taken a simple but stringent approach to identify such sequences. Independent of any previous knowledge of the region, we ultimately focused on 24 elements, which are conserved in a wide range (>75%) of distantly related mammals. These elements included 12 of 20 fully mapped constitutive and erythroid-specific DHSs. Further close inspection of the sequences underlying the remaining eight DHS showed that although three are associated with CpG islands, none contains an MCS. It is of interest that whereas most DHS can be detected in humans and mice (16, 45) four of the seven DHS analyzed were not found in mice. Furthermore, in contrast to the 17 DHS associated with MCSs and/or CpG islands, there is no observational or experimental data to show that the remaining 3 DHSs (+37, –8, and –55) are of functional importance.
It is of interest that by using the standard parameters and same data set, webmcs identified 13 conserved elements, including 12 of the 24 MCSs described here and one additional element that failed our criteria. Several of the 24 MCS elements were missed, including some of known functional significance. Most elements were found by using a less stringent webmcs search (Table 1), but this search also highlighted six times as many additional candidate MCSs. gumby identified 20 of the 24 MCSs and 19 additional candidates. Therefore, it appears that judicious use of current algorithms for identifying MCSs can detect many cis elements with good sensitivity and without an overwhelming number of false positives.
Independently of webmcs and gumby, the simple but stringent bioinformatic approach described here identified most of the key cis elements in a previously well characterized segment of the genome. Whether these criteria would identify such elements with similar efficiency in other areas of the genome remains to be tested. Classification of MCSs clearly requires integration with other bioinformatic analyses and experimental data, particularly identification of ESTs and DHSs as described here.
Acknowledgments
Sequences of the chimpanzee, cow, and cat were from encode. We thank The Oxford Computational Biology Research Group for support and Prof. W. Wood, Dr. R. Gibbons, Prof. R. Patient, and Dr. M. de Bruijn for helpful comments. This work was funded by the Medical Research Council (U.K.) and partly supported by National Heart, Lung, and Blood Institute Programs for Genomic Applications Grant HL66728 (to E.R. and J.-F.C.).
Author contributions: J.R.H., E.R., and D.R.H. designed research; J.R.H., J.-F.C., and K.C. performed research; S.P. and P.d.J. contributed new reagents/analytic tools; J.R.H., J.-F.C., N.V., S.P., E.A., M.D.G., and D.R.H. analyzed data; and J.R.H., S.P., and D.R.H. wrote the paper.
Abbreviations: αMRE, human α-globin major regulatory element; DHS, DNase1 hypersensitive site; MCS, multispecies conserved sequences; MCS-E, MCS-previously unidentified exon; MCS-P, MCS promoters; MCS-R, MCS-other transcriptional regulatory element; MCS-S, MCS splicing element; my, million years; TFB, transcription factor binding site.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [(species, clone names, and accession nos.): baboon, RP41-98L24, AC145461; colobus, CH272–179O16, AC148220; dusky titi, LB5-13F19, AC145465; marmoset, CH259-50K11, AC146591, and CH259-177O2, AC145483; owl monkey, CH258-305M22, AC146782; squirrel monkey, CH254-200G16, AC146463, and CH254-167L1, AC151883; lemur, LB2-99K6, AC145463; pig, RP44-210L18, AC120214, and RP44-293L23, AC120215; dog, RP81-84F12, AC120212, and RP81-397M1, AC120213; hedgehog, LB4-358K2, AC150435; opossum, LB3-33C20, AC120504, and LB3-206D4, AC139599, and LB3-171C2, AC148752].
References
- 1.Kanehisa, M. & Bork, P. (2003) Nat. Genet. 33 Suppl., 305–310. [DOI] [PubMed] [Google Scholar]
- 2.Thomas, J. W., Touchman, J. W., Blakesley, R. W., Bouffard, G. G., Beckstrom-Sternberg, S. M., Margulies, E. H., Blanchette, M., Siepel, A. C., Thomas, P. J., McDowell, J. C., et al. (2003) Nature 424 788–793. [DOI] [PubMed] [Google Scholar]
- 3.Boffelli, D., Nobrega, M. A. & Rubin, E. M. (2004) Nat. Rev. Genet. 5 456–465. [DOI] [PubMed] [Google Scholar]
- 4.Sidow, A. (2002) Cell 111 13–16. [DOI] [PubMed] [Google Scholar]
- 5.Hardison, R. C. (2000) Trends Genet. 16 369–372. [DOI] [PubMed] [Google Scholar]
- 6.Loots, G. G., Locksley, R. M., Blankespoor, C. M., Wang, Z. E., Miller, W., Rubin, E. M. & Frazer, K. A. (2000) Science 288 136–140. [DOI] [PubMed] [Google Scholar]
- 7.Thomas, J. W. & Touchman, J. W. (2002) Trends Genet. 18 104–108. [DOI] [PubMed] [Google Scholar]
- 8.Nardone, J., Lee, D. U., Ansel, K. M. & Rao, A. (2004) Nat. Immunol. 5 768–774. [DOI] [PubMed] [Google Scholar]
- 9.Margulies, E. H., Blanchette, M., Haussler, D. & Green, E. D. (2003) Genome Res. 13 2507–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vyas, P., Vickers, M. A., Simmons, D. L., Ayyub, H., Craddock, C. F. & Higgs, D. R. (1992) Cell 69 781–793. [DOI] [PubMed] [Google Scholar]
- 11.Higgs, D. R., Wood, W. G., Jarman, A. P., Sharpe, J., Lida, J., Pretorius, I. M. & Ayyub, H. (1990) Genes Dev. 4 1588–1601. [DOI] [PubMed] [Google Scholar]
- 12.Brown, K. E., Amoils, S., Horn, J. M., Buckle, V. J., Higgs, D. R., Merkenschlager, M. & Fisher, A. G. (2001) Nat. Cell Biol. 3 602–606. [DOI] [PubMed] [Google Scholar]
- 13.Jarman, A. P. & Higgs, D. R. (1988) EMBO J. 7 3337–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith, Z. E. & Higgs, D. R. (1999) Hum. Mol. Genet. 8 1373–1386. [DOI] [PubMed] [Google Scholar]
- 15.Anguita, E., Johnson, C. A., Wood, W. G., Turner, B. M. & Higgs, D. R. (2001) Proc. Natl. Acad. Sci. USA 98 12114–12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anguita, E., Hughes, J., Heyworth, C., Blobel, G. A., Wood, W. G. & Higgs, D. R. (2004) EMBO J. 23 2841–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bird, A. P., Taggart, M. H., Nicholls, R. D. & Higgs, D. R. (1987) EMBO J. 6 999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgs, D. R. (2001) in Disorders of Hemoglobin, eds. Steinberg, M. H., Forget, B. G., Higgs, D. R. & Nagel, R. L. (Cambridge Univ. Press, Cambridge, U.K.), pp. 405–430.
- 19.Wilson, R., Ainscough, R., Anderson, K., Baynes, C., Berks, M., Bonfield, J., Burton, J., Connell, M., Copsey, T., Cooper, J., et al. (1994) Nature 368 32–38. [DOI] [PubMed] [Google Scholar]
- 20.Engel, J. D. & Dodgson, J. B. (1980) Proc. Natl. Acad. Sci. USA 77 2596–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardison, R. C., Sawada, I., Cheng, J. F., Shen, C. K. & Schmid, C. W. (1986) Nucleic Acids Res. 14 1903–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flint, J., Thomas, K., Micklem, G., Raynham, H., Clark, K., Doggett, N. A., King, A. & Higgs, D. R. (1997) Nat. Genet. 15 252–257. [DOI] [PubMed] [Google Scholar]
- 23.Xu, J. & Hardison, R. C. (1991) Genomics 9 362–365. [DOI] [PubMed] [Google Scholar]
- 24.Oakenfull, E. A., Buckle, V. J. & Clegg, J. B. (1993) Cytogenet. Cell Genet. 62 136–138. [DOI] [PubMed] [Google Scholar]
- 25.Tan, H. & Whitney, J. B., 3rd (1993) Biochem. Genet. 31 473–484. [DOI] [PubMed] [Google Scholar]
- 26.Tufarelli, C., Hardison, R., Miller, W., Hughes, J., Clark, K., Ventress, N., Frischauf, A. M. & Higgs, D. R. (2004) Genome Res. 14 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flint, J., Tufarelli, C., Peden, J., Clark, K., Daniels, R. J., Hardison, R., Miller, W., Philipsen, S., Tan-Un, K. C., McMorrow, T., et al. (2001) Hum. Mol. Genet. 10 371–382. [DOI] [PubMed] [Google Scholar]
- 28.Bird, A. (2002) Genes Dev. 16 6–21. [DOI] [PubMed] [Google Scholar]
- 29.Takai, D. & Jones, P. A. (2002) Proc. Natl. Acad. Sci. USA 99 3740–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuo, K., Clay, O., Takahashi, T., Silke, J. & Schaffner, W. (1993) Somatic Cell Mol. Genet. 19 543–555. [DOI] [PubMed] [Google Scholar]
- 31.Bejerano, G., Pheasant, M., Makunin, I., Stephen, S., Kent, W. J., Mattick, J. S. & Haussler, D. (2004) Science 304 1321–1325. [DOI] [PubMed] [Google Scholar]
- 32.Lim, L. P. & Sharp, P. A. (1998) Mol. Cell Biol. 18 3900–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hedjran, F., Yeakley, J. M., Huh, G. S., Hynes, R. O. & Rosenfeld, M. G. (1997) Proc. Natl. Acad. Sci. USA 94 12343–12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tufarelli, C., Frischauf, A. M., Hardison, R., Flint, J. & Higgs, D. R. (2001) Genomics 71 307–314. [DOI] [PubMed] [Google Scholar]
- 35.Jarman, A. P., Wood, W. G., Sharpe, J. A., Gourdon, G., Ayyub, H. & Higgs, D. R. (1991) Mol. Cell Biol. 11 4679–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pondel, M. D., Murphy, S., Pearson, L., Craddock, C. & Proudfoot, N. J. (1995) Proc. Natl. Acad. Sci. USA 92 7237–7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, Q., Reddy, P. M. S., Yu, C.-Y., Bastiani, C., Higgs, D., Stamatoyannopoulos, G., Papayannopoulou, T. & Shen, C.-K. J. (1993) Mol. Cell Biol. 13 2298–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cantor, A. B. & Orkin, S. H. (2002) Oncogene 21 3368–3376. [DOI] [PubMed] [Google Scholar]
- 39.Motohashi, H., O'Connor, T., Katsuoka, F., Engel, J. D. & Yamamoto, M. (2002) Gene 294 1–12. [DOI] [PubMed] [Google Scholar]
- 40.Ludwig, M. Z. (2002) Curr. Opin. Genet. Dev. 12 634–639. [DOI] [PubMed] [Google Scholar]
- 41.Higgs, D. R., Sharpe, J. A. & Wood, W. G. (1998) Semin. Hematol. 35 93–104. [PubMed] [Google Scholar]
- 42.Yu, C. Y., Chen, J., Lin, L. I., Tam, M. & Shen, C. K. (1990) Mol. Cell Biol. 10 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mellon, P., Parker, V., Gluzman, Y. & Maniatis, T. (1981) Cell 27 279–288. [DOI] [PubMed] [Google Scholar]
- 44.Kielman, M. F., Smits, R., Hof, I. & Bernini, L. F. (1996) Genomics 32 341–351. [DOI] [PubMed] [Google Scholar]
- 45.Horsley, S. W., Daniels, R. J., Anguita, E., Raynham, H. A., Peden, J. F., Villegas, A., Vickers, M. A., Green, S., Waye, J. S., Chui, D. H., et al. (2001) Eur. J. Hum. Genet. 9 217–225. [DOI] [PubMed] [Google Scholar]