Abstract
Down-regulation of the androgen receptor (AR) is being evaluated as an effective therapy for the advanced stages of prostate cancer. We report that Ebp1, a protein identified by its interactions with the ErbB3 receptor, down-regulates expression of AR and AR-regulated genes in the LNCaP prostate cancer cell line. Using microarray analysis, we identified six endogenous AR target genes, including the AR itself, that are down-regulated by ebp1 overexpression. Chromatin immunoprecipitation assays revealed that Ebp1 was recruited to the prostate-specific antigen gene promoter in response to the androgen antagonist bicalutamide, suggesting that Ebp1 directly affected the expression of AR-regulated genes in response to androgen antagonists. Ebp1 expression was reduced in cells that had become androgen-independent. Androgens failed to stimulate either the growth of ebp1 transfectants or transcription of AR-regulated reporter genes in these cells. The agonist activity of the antiandrogen cyproterone acetate was abolished in ebp1 transfectants. In severe combined immunodeficient mice, Ebp1 overexpression resulted in a reduced incidence of LNCaP tumors and slower tumor growth. These findings suggest that Ebp1 is a previously unrecognized therapeutic target for treatment of hormone refractory prostate cancer.
Keywords: ErbB receptors, transcriptional corepressors, androgen independence
Prostate cancer is currently the most prevalent cancer among men in the United States and ranks second to lung cancer in terms of annual mortality (1). Prostate cancer begins as an androgen-dependent tumor that undergoes clinical regression in response to pharmacological and surgical strategies that reduce testosterone concentration. Despite this treatment, a majority of patients develop lethal androgen-independent tumors. The androgen receptor (AR) is central to the initiation and growth of prostate cancer and to the therapeutic response to hormones. AR continues to be expressed even in androgen-independent tumors, and aberrant AR signaling is postulated to be an important mechanism of progression to androgen independence (2). Increases in AR protein levels are associated with hormone refractory disease clinically (3, 4) and in xenograft models of prostate cancer (5). Inhibition of AR mRNA expression (6, 7), destabilization of AR protein (8), and pharmacological inhibition of AR protein synthesis or function (9, 10) have been used in preclinical models for prostate cancer treatment. However, the manipulation of endogenous AR corepressors to down-regulate AR levels or function has not yet been reported.
Recent studies indicate that the ErbB2/ErbB3 pathway is a critical target in hormone refractory prostate cancer; activation of this pathway stabilizes AR level and enhances AR binding to promoter elements of AR responsive genes (11). Our laboratory has demonstrated that Ebp1, a member of the PA2G4 family of proliferation-regulated proteins (12, 13) that interacts with the ErbB3 receptor (14), is also an AR corepressor (15). Activity of both exogenous and endogenous AR-regulated promoters was inhibited by ectopic expression of ebp1 independent of prostate cell type (16). Ebp1 interacted with histone deacetylase (HDAC) 2 and also inhibited transcription of cell cycle regulators such as E2F1, cyclin D, and c-myc (17). Ectopic expression of ebp1 resulted in growth inhibition of both breast cancer and prostate cancer cell lines (15).
To further understand Ebp1-mediated inhibition of AR signaling and cell growth, we evaluated the effects of ebp1 overexpression on AR-regulated genes or those related to prostate cancer by using microarray analysis. Ectopic expression of Ebp1 in LNCaP cells resulted in a down-regulation of several AR-regulated genes, including AR itself. We demonstrate both in vitro and in vivo that overexpression of ebp1 in LNCaP cells results in a less transformed phenotype. Ebp1 expression was reduced in two models of androgen-independent prostate cancer. These studies suggest that Ebp1 may be a target for the development of therapies in prostate cancer.
Methods
Microarray Analysis. RNA was prepared by using TRIzol reagent as described in ref. 15. Microarray processing and data analysis were performed at Genome Explorations (Nashville, TN) as described in ref. 18. U133A oligonucleotide arrays (Affymetrix, Santa Clara, CA) containing ≈33,000 full-length annotated genes together with additional probe sets designed to represent EST sequences were used for the analysis. Only genes with a minimum expression level of 500 were included in this analysis. Genes whose expression varied >3-fold with P < 0.05 were considered to be significantly different between the two cell lines.
Real-Time Quantitative RT-PCR. The method of Nakanishi et al. (19) was used as previously described. Real-time quantitative RT-PCR was performed on a LightCycler (Roche Diagnostics) platform. The following forward and reverse primers were selected by using primer express software and were synthesized by the Core Laboratory of University of Maryland School of Medicine. Ebp1, sense 5′-GCACGCCAATAGAAGG-3′ and antisense 5′-GTAAACGGCATGGCATC-3′; AR, sense 5′-AAGGCTATGAATGTCAGCCCA-3′ and antisense 5′CATTGAGGCTAGAGAGCAAGGC-3′; kallikrein 2, sense 5′-CATCCAGTCTCGGATTG-3′ and antisense 5′-CTCATATTGTAGAGCGGGT-3′; POV-1, sense 5′-AGTGCTGTGTTCGCCTTG-3′ and antisense 5′-CACCTCAGAGCCGCTAAG-3′; β-actin, sense 5′ GCT ATC CAG GCT GTG CTA TC-3′ and antisense TGT CAC GCA CGA TTT CC-3′. A SYBR green PCR kit (Applied Biosystems) was used per the manufacturer's instructions, and the analyses were performed in duplicate or triplicate. Target mRNA values were normalized by using β-actin mRNA as an internal control. The relative quantitation of gene expression was performed by using the comparative ΔΔCt (threshold method) with β-actin as an internal control (20).
Western Blot Analysis. Western blot analysis was performed as described in ref. 21. The AR antibody was from Santa Cruz Biotechnology, the Ebp1 antibody was from Upstate Biotechnology (Lake Placid, NY), the polyclonal antibody to actin was from Sigma, and the POV-1 antibody was a gift from Rodrigo Chugai (National Cancer Institute).
Chromatin Immunoprecipitation Assays. Briefly, LNCaP ebp1 transfectants were grown in RPMI medium 1640 supplemented with 5% charcoal-stripped FBS (Sigma). After 3 days of culture, cells were treated with 5 μM bicalutamide for 1 h. Chromatin was prepared, and chromatin immunoprecipitation assays were performed as described in ref. 22. Nested PCR amplification of a 210-bp prostate-specific antigen (PSA) promoter fragment (–250 to –39) was carried out by using a 5′ primer (5′-TCTGCCTTTGTCCCCTAGAT-3′) and a 3′ primer (5′-AACCTTCATTCCCCAGGACT-3′). The PCR products were resolved on 2.5% agarose gels and visualized with ethidium bromide.
Cell Growth Assays. Cell growth measurement in complete media was performed as described in ref. 23 by using a hemocytometer. For soft agar growth assays, increasing concentrations of cells (as indicated) were plated in 35-mm Petri dishes in 0.3% agar in complete media, and colonies were counted after 10 days of incubation (23). To assess the effect of dihydrotestosterone (DHT) on cell growth, cells (2 × 104) were plated in 12-well plates in complete medium. After 24 h, the medium was replaced with steroid-free medium [phenol red-free RPMI medium 1640/5% charcoal-stripped FBS (Sigma)] for 48 h. After 48 h of steroid depletion, cells were refed with fresh steroid-reduced medium with or without the indicated concentrations of DHT and bicalutamide (10 μM), and total cell numbers were assessed 7 days later.
Luciferase Reporter Assays. Vector control or ebp1 transfected LNCaP cells (5 × 104) were plated in 12-well plates in complete media. When cells reached 50–60% confluence, they were transfected with 0.5 μg of the androgen responsive mouse mammary tumor virus (MMTV)-luciferase reporter construct by using Fu-GENE 6 reagent (Roche Applied Science, Indianapolis). Cells were also transfected with the RL-TK vector as an internal control. Complete medium was replaced after 24 h with phenol red and serum-free DMEM-F12 or RPMI medium 1640 with or without 10 nM R1881 (NEN) or cyproterone acetate (CA) (Sigma). Luciferase activity was determined as described in ref. 16 by using a dualluciferase kit (Promega).
In Vivo Studies in Severe Combined Immunodeficient (SCID) Mice. Male SCID mice, 4–6 weeks of age (National Cancer Institute), were housed in a pathogen-free environment under controlled conditions of light and humidity and received food and water ad libitum. LNCaP cells grown in complete medium and 800 μg/ml G418 were suspended in Matrigel (10 mg/ml, Collaborative Research) at 1 × 107 cells per ml. Each mouse received s.c. injections at one site on each flank with 100 μl of cell suspension. Tumors were measured three times a week with calipers, and tumor volumes were calculated by the formula 0.5236 × r12 × r2 (r1 < r2) (24). The animal protocols were approved by the Institutional Animal Care and Use Committee at University of Maryland. After the mice were killed, tumors were excised and fixed in 10% buffered neutral formalin. Sections of formalin-fixed, paraffin-embedded tissues were cut to 5 μm and stained with an Ebp1 antibody (Upstate Biotechnology) diluted 1:100 by using the standard avidin-biotin method (Vector Laboratories).
Measurement of PSA Levels. PSA levels were determined by using a PSA ELISA kit from DSL (Webster, TX) as described in ref. 24.
Statistical Analysis. Results of growth and luciferase assays were analyzed by using Student's t test. Significance was established at P < 0.05. The linear mixed-effects models approach was used to estimate groups' average tumor volume and growth rate. The proportion of developed tumors in control and ebp1 transfectants were compared by using Fisher's exact test. All hypothesis tests were two-sided. The different groups were compared at the 0.05 level of significance.
Results
Ectopic Expression of Ebp1 Down-Regulates Androgen-Regulated Genes. Gene expression profiling of LNCaP cells stably transfected with ebp1 was used to determine the range of androgen-dependent genes affected by Ebp1 overexpression. Of 8,000 genes that were evaluable, the expression of 167 genes was found to be activated 3-fold or repressed to 1/3 (P < 0.05; 500 minimum expression units). Forty-one genes were induced by ebp1 overexpression, and 126 were repressed. Seventy-two of these genes with Human Genome Organization-approved names are displayed (see Fig. 7, which is published as supporting information on the PNAS web site). We noted that six androgen responsive genes were significantly changed in ebp1-overexpressing cells compared with controls (Table 1). These genes include the AR, PSA (kallikrein 3) as previously reported (15), Kallikrein 2, POV-1, TMPRSS2, and prostate-derived factor. The kallikrein 2 gene is regulated by AR and being evaluated as a marker for prostate cancer progression (25). The POV-1 gene encodes a transcript for an L amino acid transporter (26) that is up-regulated in aggressive prostate carcinoma (27). The TMPRSS2 gene is androgen-regulated and also highly expressed in the prostate and prostate cancer (28). Prostate-derived factor, a member of the bone morphogenetic protein family, is androgen-regulated and expressed at high levels in the prostate (29).
Table 1. Androgen-regulated genes decreased in Ebp1-transfected LNCaP cells.
| Accession no. | Gene name | Fold decrease | P value |
|---|---|---|---|
| NM_003527 | Prostate cancer overexpressed POV | 5.0 | 0.001 |
| U17040 | PSA | 3.7 | 0.0002 |
| AF1627 | AR | 3.7 | 0.001 |
| BC005196 | Prostatic kallikrein 2 | 3.2 | 0.001 |
| AF2700487 | Androgen-regulated serine protease TMPRSS2 | 3.2 | 0.001 |
| AF003934 | Prostate differentiation factor mRNA | 3.2 | 0.0002 |
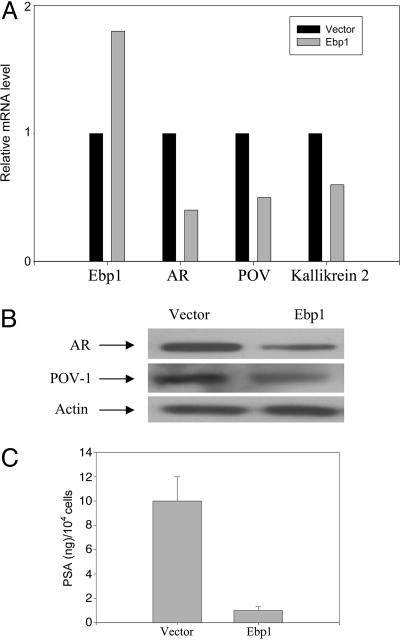
Real-time RT-PCR methods indicated that the level of AR mRNA was halved in ebp1 transfectants (Fig. 1A). Western blot analysis indicated that AR protein was decreased 80% in LNCaP ebp1 transfectants (Fig. 1B). The expression of POV-1 mRNA (Fig. 1A) and protein (Fig. 1B) each was decreased by ≈50%. PSA secretion in the conditioned media of ebp1 and vector control cells was down-regulated 90% in ebp1 transfectants compared with controls (Fig. 1C). Finally, kallikrein 2 mRNA expression was decreased ≈40% (Fig. 1A).
Fig. 1.
Validation of differential gene expression in vector and ebp1 transfectants. (A) Real-time quantitative RT-PCR analysis of AR, POV-1, Kallikrein 2, and Ebp1 mRNA. The relative levels of all test mRNAs were normalized to β-actin. Results are representative of three experiments using different sets of cells. (B) Expression of AR and POV-1 protein in vector and ebp1 LNCaP transfectants. Lysates of vector control or Ebp1 transfected cells were resolved by SDS/PAGE and analyzed by Western blotting with the indicated antibodies. (C) Secreted PSA as measured by ELISA. PSA levels in conditioned media from Ebp1 transfected and vector control LNCaP were assessed by ELISA and adjusted to total cell number.
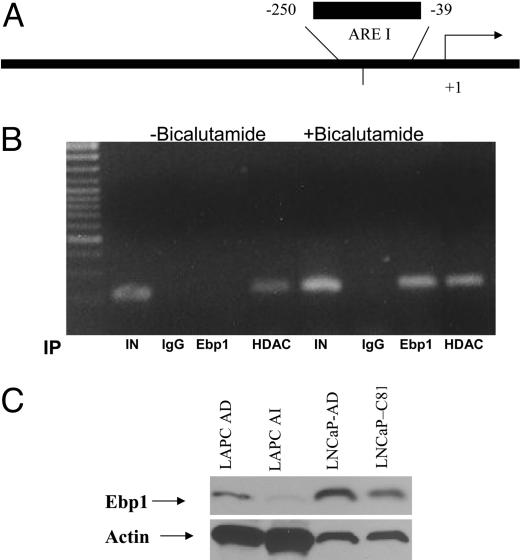
Ebp1 Is Involved in the Response to Antiandrogens and Is Decreased in Androgen-Independent Tumors. To determine whether Ebp1 directly affects the transcription of AR-regulated genes, we conducted chromatin immunoprecipitation experiments. Ebp1 transfectants were serum-starved and then treated with the androgen antagonist bicalutamide for 1 h. HDAC2 was recruited to the PSA promoter after exposure to bicalutamide as reported in ref. 30. Ebp1 was not associated with the promoter in the absence of bicalutamide but was recruited to the PSA promoter after bicalutamide exposure (Fig. 2 A and B).
Fig. 2.
Ebp1 is involved in the response to antiandrogens and is decreased in androgen-independent tumors. (A and B) Ebp1 is recruited to the PSA promoter by the androgen antagonist bicalutamide. (A) The positions of the PCR primers on the PSA promoter relative to the transcriptional start site are indicated. The primers amplify the first androgen response element (ARE I) within the PSA promoter. The arrow represents the transcriptional start site. (B) LNCaP ebp1 transfectants, growing in steroid-depleted media, were treated with 5 μM bicalutamide for 1 h. Soluble chromatin was prepared as described in Methods and immunoprecipitated (IP) with preimmune IgG or antibodies to Ebp1 or HDAC2. Immunoprecipitated DNA was amplified by using primers specific for the human PSA promoter. PCR products were visualized by ethidium bromide staining. “IN” is input, which represents 1% of the total amount of chromatin added to each immunoprecipitation reaction. (C) Ebp1 expression is decreased in androgen-independent tumors. Western blot analysis of lysates from LAPC tumors that were growing in intact mice (LAPC AD), castrated mice (LAPC AI), androgen-dependent LNCaP cells (LNCaP-AD), or androgen-independent LNCaP cells (LNCaP-C81) was performed with the indicated antibodies.
The recruitment of Ebp1 to the PSA promoter in the presence of bicalutamide suggests that Ebp1 is involved in the response to antiandrogens and may play a role in the development of the androgen-independent phenotype. We tested for the presence of Ebp1 in two models of androgen independence. The C-81 LNCaP subline has been made androgen-independent by continuous long-term passage in complete media (31). The LAPC-4 xenograft has wild-type AR, grows as an androgen-dependent cancer in male SCID mice, and regresses in response to androgen ablation but eventually regrows as an androgen-independent tumor that overexpresses ErbB2 (32). LAPC xenografts that were grown in either intact or castrated mice were examined. The results indicated that expression of Ebp1 protein was decreased in the LAPC androgen-independent xenografts and in the C81 androgen-independent cell line grown in vitro (Fig. 2C).
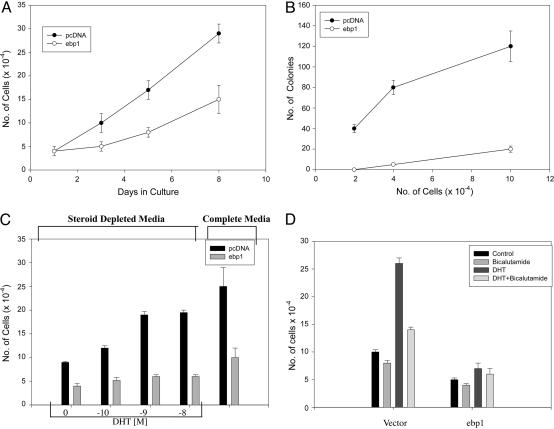
Growth Characteristics of LNCaP ebp1 Transfectants. We next determined whether the changes observed in AR-regulated gene expression affected cell growth or the transformed properties of LNCaP cells in vitro. The growth rate of the ebp1 transfectants in complete media was significantly decreased (P < 0.05) compared with that of the vector control. The doubling time for ebp1 transfectants increased to 72 h from 48 h for the vector control line (Fig. 3A). Ectopic expression of ebp1 also decreased colony growth in soft agar ≈90% at the highest cell concentration tested (Fig. 3B).
Fig. 3.
Growth of Ebp1 and vector transfected LNCaP cells. (A) Growth in monolayer culture. Equal numbers (5 × 104) of pcDNA or ebp1 stably transfected cells were plated in complete media, and viable cell numbers were determined at the indicated times. Each point represents mean ± SE of three wells. Data are representative of three experiments. (B) Colony growth in soft agar. LNCaP cells stably transfected with either pcDNA or ebp1 were plated in soft agar at the cell densities indicated, and colony numbers were assessed at day 10. Each point represents mean ± SE of three dishes. Data are representative of three experiments. (C) Growth response to DHT. Ebp1 and vector control transfected cells were plated in complete media at 2 × 104 cells per well for 1 day. Cells were then switched to steroid-reduced medium for 2 days, and DHT was added at the indicated concentrations. Total cell numbers were determined 7 days later. Each point represents mean ± SE of three wells. Data are representative of two experiments. (D) Growth response of cells to bicalutamide. Cells were plated as described in Methods. After 2 days in steroid-reduced medium, cells were fed with fresh steroid-reduced medium with or without bicalutamide in the presence or absence of 10 nM DHT. Total cell numbers were assessed 7 days later. The data shown are mean ± SE of triplicate wells and are representative of two experiments.
We examined the sensitivity of ebp1 transfectants to the androgen metabolite DHT. Both LNCaP vector controls and ebp1 transfectants were placed in serum-free media and then stimulated with DHT. The growth of the vector controls was inhibited ≈70% by the withdrawal of androgens as reported in ref. 31 (Fig. 3C). The growth of ebp1 transfectants was inhibited 40% (P < 0.05) in the absence of androgens. Thus, ebp1 transfectants had not become androgen-independent. As previously reported (31), DHT at increasing concentrations stimulated the growth of LNCaP vector controls with a maximal 250% stimulation at 10–9 M DHT as compared with no DHT. In contrast, DHT at 10–9 M only slightly (35%) stimulated the growth of Ebp1 transfectants (Fig. 3C). To determine whether the growth of both cell lines in the presence of DHT was mediated by means of the AR, we examined the effect of the antiandrogen bicalutamide on DHT-stimulated growth. The DHT-stimulated growth of LNCaP vector controls and ebp1 transfected cells was partially suppressed by 10 μM bicalutamide (Fig. 3D).
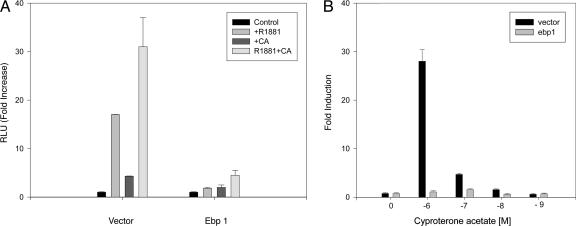
We then analyzed whether the stable overexpression of ebp1 affected the transactivation of the MMTV-luciferase reporter gene by either the synthetic androgen agonist R1881 or the partial agonist CA. Addition of R1881 (10–8 M) led to a 2,000% increase in luciferase activity of LNCaP vector controls. CA (10–7 M) activated AR by 500% as previously reported (33) and enhanced the transcriptional response in the presence of R1881. In contrast, R1881, CA, or the combination of the two did not stimulate AR activation in ebp1 transfectants (Fig. 4A). Although CA induced activation of AR at concentrations of 10–6–10–8 in vector control cells, it failed to induce activation of the AR in the ebp1 transfectants at any concentration tested (Fig. 4B).
Fig. 4.
Inhibition of AR transcriptional activation in Ebp1 transfectants. (A) AR activation in response to R1881 or CA. Ebp1 or vector control LNCaP transfectants were transfected with an MMTV-luciferase reporter plasmid. Twenty-four hours after transfection, cells were switched to phenol red-free RPMI medium 1640 with 5% charcoal-stripped serum (CSS) containing R1881 (10–8 M), CA (10–7 M), the two in combination, or vehicle control. Sixteen hours later, luciferase activity was measured. Each point represents mean ± SE of triplicate wells. Data are representative of three experiments. (B) Ebp1 expression inhibits the agonist effects of CA. LNCaP vector or ebp1 transfected cells were transfected with the MMTV-luciferase reporter. Twenty-four hours after transfection, cells were switched to phenol red-free RPMI medium 1640 with 5% CSS containing the indicated concentrations of CA or vehicle control. Sixteen hours later, luciferase activity was measured. Each point represents mean ± SE of triplicate wells. Results are representative of three experiments.
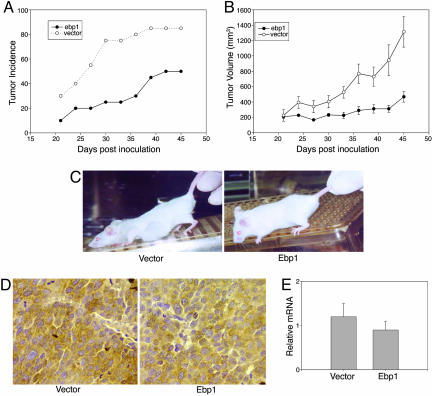
Ebp1 Expression Suppresses Growth of Prostate Cancer Xenografts. To test whether the in vitro effects of Ebp1 could be observed in vivo, we examined the effect of ectopic expression of ebp1 on the tumorigenicity of LNCaP cells. Ebp1 and vector transfected cells were injected s.c. into SCID mice, and tumor growth was monitored. Tumor growth was first noted in both groups at day 20 (Fig. 5A). However, on day 20, tumors were observed at only 10% of the ebp1 inoculation sites, as opposed to 35% of the sites for vector controls. At the end of the study, tumors had developed at >85% of the sites injected with the vector control cells, whereas <50% of sites inoculated with ebp1 transfectants developed tumors. This difference was significant (Fisher's exact test, two-sided P = 0.04). The average tumor volume for the ebp1 transfectants observed at the end of the study was 268 ± 70 mm3, compared with 1,214 ± 168 mm3 for the vector controls (P = 0 .0001). The growth rate of ebp1 transfectants was reduced compared with the vector controls at all time points measured (P = 0.0001) (Fig. 5 B and C). Immunohistochemical staining of tissue sections of the tumors harvested at the conclusion of the experiment (day 45) indicated that Ebp1 expression was equivalent in both groups (Fig. 5D). Real-time quantitative RT-PCR of ebp1 mRNA extracted from the tumors showed no change between the two groups (Fig. 5E). Thus, the ebp1 transfected cells that grew to form tumors had lost overexpression of the transgene.
Fig. 5.
Ebp1 overexpression decreases growth of LNCaP cells in SCID mice. LNCaP vector or ebp1 (1 × 106) transfected cells were injected into SCID mice s.c. Tumor growth was assessed every 3 days. (A) Tumor incidence. Tumor development with time is shown (n = 10 × 2). (B) Tumor volumes and growth rate. Tumor volumes were calculated as described in Methods. Each point represents the mean ± SD of the tumor volume of a representative experiment (of two) with 10 mice injected at 20 sites. Results shown are the average tumor size for tumors actively growing at the indicated days. (C) Growth of Ebp1 and vector transfected LNCaP cells in SCID mice. Mice are depicted 35 days after inoculation with vector or epb1 transfected cells. The ebp1 mouse was one in which tumors failed to develop. (D and E) Ebp1 protein and RNA are not increased in LNCaP ebp1 xenografts. (D) Photomicrographs of sections of ebp1 or vector transfected LNCaP xenografts stained with an antibody to Ebp1. (E) RNA was isolated from xenografts of mice bearing tumors derived from Ebp1 or vector transfected cells. Realtime quantitative RT-PCR was performed as described in Methods. Data represent mean ± SE of three tumors.
Discussion
Our laboratory has demonstrated that Ebp1, an ErbB3-binding protein, is a potent repressor of AR signaling. Gene expression profiling identified a cohort of AR target genes, including AR itself, that are potentially involved in androgen-independent growth of prostate cancer cells and that are down-regulated in cells with only moderate Ebp1 expression compared with the vector transfected cells. To our knowledge, this demonstration is the first that shows that an AR corepressor can down-regulate levels of AR protein.
Because changes in AR protein were greater than those observed for AR mRNA, we postulate that posttranscriptional mechanisms may affect steady-state AR protein levels. A pool of Ebp1 localized in the nucleolus is part of ribonucleoprotein complexes and may affect protein translation (34). Another possible mechanism by which changes in AR protein are greater than those of AR mRNA may be due to Ebp1 destabilization of AR protein. Ebp1 physically associates with the N-terminal domain of AR (15) and might disrupt interactions between the N- and C-terminal domains of the AR that are important for AR stability (35, 36). Finally, interactions of Ebp1 with the ErbB3 receptor may play a role in maintaining AR protein levels by an unknown mechanism, because the ErbB2/ErbB3 pathway stabilizes AR protein by means of effects on ubiquitin-mediated degradation (11).
The molecular basis for the global effects of Ebp1 on the activity of AR target genes is incompletely understood. First, reduction in AR protein by Ebp1 could result in decreased transcription of its target genes by a negative feedback mechanism. In addition, Ebp1, through binding the N-terminal domain of the AR, may decrease transcription by displacing coactivators that also bind this domain (37). Ebp1 may also inhibit AR-mediated transcription by directly binding to androgen response elements within AR-regulated promoters and recruiting HDACs to repress transcription (17). The association of Ebp1 with the PSA promoter supports our hypothesis that Ebp1 is directly involved in AR-mediated transcription and suggests that Ebp1 may be a mediator of bicalutamide's ability to inhibit AR-regulated transcription.
The importance of AR corepressors in mediating the inhibitory responses of antiandrogens was first demonstrated by Shang et al. (30), who showed that bicalutamide recruits the corepressors NCoR and SMRT along with HDACs to the PSA promoter. Recruitment of NCoR binding to AR-regulated promoters by RU486 also results in decreased transcriptional activation of target genes (38). Conversely, increases in AR protein levels that result in an androgen-independent phenotype lead to a decrease in corepressor recruitment to AR-regulated promoters after bicalutamide treatment (5). The fact that Ebp1 expression is decreased in two models of androgen-independent prostate cancer suggests that loss of Ebp1 expression may be one reason that patients fail to respond to antiandrogen therapy in advanced stages of prostate cancer. In fact, inhibition of endogenous Ebp1 by small interfering RNA results in the activation of the MMTV reporter plasmid in the absence of androgen (16). Therefore, defects in Ebp1 protein or in factors controlling Ebp1 function may confer increased sensitivity to low levels of androgens, leading to an androgen-independent phenotype. Heregulin enhances the ability of Ebp1 to physically associate with AR and suppress AR transcriptional activity (16). In the presence of low concentrations of heregulin, such as that observed in clinical prostate cancer tissues (39), the activity of Ebp1 may be suboptimal. Alternatively, high levels of IL-6, associated with the etiology of prostate cancer (40), may silence Ebp1 expression, as was recently demonstrated in myeloma cells (41).
The decreases of AR-regulated genes induced by ebp1 overexpression correlated with changes in the growth rate of the transfected cells and a loss of sensitivity to DHT stimulation. Furthermore, the transcriptional response to R1881 was greatly attenuated in the ebp1 transfectants, possibly in part due to the reduced expression of AR. The AR antagonist CA, which is an agonist for LNCaP cells expressing the mutant T877A AR (42), did not stimulate activity of an AR reporter construct in ebp1 transfectants. The ability of Ebp1 to inhibit the transactivation of a mutant AR by androgen antagonists would be of potential clinical interest, because mutation of AR leading to promiscuous activation has been postulated to be one mechanism for developing antiandrogen resistance (43).
Whether the slower growth rate and attenuation of the transformed properties of ebp1 transfectants was solely attributable to changes in AR expression is not clear at this time. Ebp1 also affects transcription of E2F1-regulated cell cycle genes (17, 44), and this repression may also contribute to the overall biological effects of ebp1 overexpression. However, it is unlikely that slower growth alone accounts for the changes in AR expression. For example, Igawa et al. (31) have found that LNCaP cells growing at different rates do not have changes in AR expression per cell. In addition, AR expression is unchanged in LNCaP cells in which growth is slowed by p53 (45).
The growth of ebp1 transfected cells was also suppressed in an LNCaP xenograft model. Both tumor incidence and tumor size were reduced. In our study, the tumors that did grow did not show increased Ebp1 expression levels as determined by both immunohistochemistry and real-time PCR, indicating selection for cells that had escaped Ebp1 overexpression. The slowing of tumor growth may have been due to the loss of the Ebp1-overexpressing cells early in the time course of the experiment, essentially eliminating such cells from the proliferating pool. Thus, with a smaller proliferative fraction, cell growth will be delayed or decreased.
In summary, a reduction in AR activity or expression is a key component of prostate cancer treatment (46). Overexpression of Ebp1, an AR corepressor that interacts with ErbB3, reduces AR protein levels and transcription of AR-regulated genes in LNCAP cells, resulting in a less tumorigenic phenotype. In addition, endogenous Ebp1 expression was decreased in two different models of androgen-independent prostate cancer growth. Our work provides a rationale for the design of innovative therapeutic approaches for advanced prostate cancer based on a knowledge of the biology of Ebp1.
Supplementary Material
Acknowledgments
We thank Dr. Joe Fondell (University of Medicine and Dentistry of New Jersey, Piscataway) for the MMTV reporter gene and Dr. Yun Qiu (University of Maryland) for a critical reading of the manuscript. This work was supported in part by National Institutes of Health Grants R01 CA76047 and R21 088882-01, a grant from the University of Maryland School of Medicine Department of Pathology (to A.W.H.), and a Department of Veterans Affairs Merit Review grant (to D.D.R.).
Author contributions: Y.Z. and A.W.H. designed research; Y.Z., D.J., T.N., M.-h.Y., D.A., and A.W.H. performed research; Y.Z., X.-W.W., D.J., T.N., D.D.R., and A.B. contributed new reagents/analytic tools; Y.Z., X.-W.W., T.N., O.G., and A.W.H. analyzed data; and Y.Z., X.-W.W., and A.W.H. wrote the paper.
Abbreviations: AR, androgen receptor; PSA, prostate-specific antigen; SCID, severe combined immunodeficient; HDAC, histone deacetylase; DHT, dihydrotestosterone; MMTV, mouse mammary tumor virus; CA, cyproterone acetate.
References
- 1.Jemal, A., Tiwari, R. C., Murray, T., Ghafoor, A., Samuels, A., Ward, E., Feuer, E. J. & Thun, M. J. (2004) CA Cancer J. Clin. 54, 8–29. [DOI] [PubMed] [Google Scholar]
- 2.Taplin, M. E. & Balk, S. P. (2004) J. Cell. Biochem. 91, 483–490. [DOI] [PubMed] [Google Scholar]
- 3.Edwards, J., Krishna, N. S., Grigor, K. M. & Bartlett, J. M. (2003) Br. J. Cancer 89, 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taplin, M. E., Bubley, G. J., Shuster, T. D., Frantz, M. E., Spooner, A. E., Ogata, G. K., Keer, H. N. & Balk, S. P. (1995) N. Engl. J. Med. 332, 1393–1398. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C. D., Welsbie, D. S., Tran, C., Baek, S. H., Chen, R., Vessella, R., Rosenfeld, M. G. & Sawyers, C. L. (2004) Nat. Med. 10, 33–39. [DOI] [PubMed] [Google Scholar]
- 6.Eder, I. E., Hoffmann, J., Rogatsch, H., Schafer, G., Zopf, D., Bartsch, G. & Klocker, H. (2002) Cancer Gene Ther. 9, 117–125. [DOI] [PubMed] [Google Scholar]
- 7.Zegarra-Moro, O. L., Schmidt, L. J., Huang, H. & Tindall, D. J. (2002) Cancer Res. 62, 1008–1013. [PubMed] [Google Scholar]
- 8.Solit, D. B., Zheng, F. F., Drobnjak, M., Munster, P. N., Higgins, B., Verbel, D., Heller, G., Tong, W., Cordon-Cardo, C., Agus, D. B., et al. (2002) Clin. Cancer Res. 8, 986–993. [PubMed] [Google Scholar]
- 9.Mitchell, S. H., Zhu, W. & Young, C. Y. (1999) Cancer Res. 59, 5892–5895. [PubMed] [Google Scholar]
- 10.Zhu, W., Zhang, J. S. & Young, C. Y. (2001) Carcinogenesis 22, 1399–1403. [DOI] [PubMed] [Google Scholar]
- 11.Mellinghoff, I. K., Vivanco, I., Kwon, A., Tran, C., Wongvipat, J. & Sawyers, C. L. (2004) Cancer Cell 6, 517–527. [DOI] [PubMed] [Google Scholar]
- 12.Lamartine, J., Seri, M., Cinti, R., Heitzmann, F., Creaven, M., Radomski, N., Jost, E., Lenoir, G. M., Romeo, G. & Sylla, B. S. (1997) Cytogenet. Cell Genet. 78, 31–35. [DOI] [PubMed] [Google Scholar]
- 13.Radomski, N. & Jost, E. (1995) Exp. Cell Res. 220, 434–445. [DOI] [PubMed] [Google Scholar]
- 14.Yoo, J. Y., Wang, X. W., Rishi, A. K., Lessor, T., Xia, X. M., Gustafson, T. A. & Hamburger, A. W. (2000) Br. J. Cancer 82, 683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, Y. X., Fondell, J. D., Wang, Q. B., Xia, X. M., Cheng, A. W., Lu, M. L. & Hamburger, A. W. (2002) Oncogene 21, 5609–5618. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, Y. & Hamburger, A. W. (2005) Br. J. Cancer 92, 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang, Y. X., Woodford, N., Xia, X. M. & Hamburger, A. W. (2003) Nucleic Acids Res. 31, 2168–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lang, R., Patel, D., Morris, J. J., Rutschman, R. L. & Murray, P. J. (2002) J. Immunol. 169, 2253–2263. [DOI] [PubMed] [Google Scholar]
- 19.Nakanishi, T., Karp, J. E., Tan, M., Doyle, L. A., Peters, T., Yang, W., Wei, D. & Ross, D. D. (2003) Clin. Cancer Res. 9, 3320–3328. [PubMed] [Google Scholar]
- 20.Nikitakis, N. G., Hamburger, A. W. & Sauk, J. J. (2002) Cancer Res. 62, 1004–1007. [PubMed] [Google Scholar]
- 21.Xia, X., Lessor, T. J., Zhang, Y., Woodford, N. & Hamburger, A. W. (2001) Biochem. Biophys. Res. Commun. 289, 240–244. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, Y. & Hamburger, A. W. (2004) J. Biol. Chem. 279, 26126–26133. [DOI] [PubMed] [Google Scholar]
- 23.Lessor, T. J., Yoo, J. Y., Xia, X., Woodford, N. & Hamburger, A. W. (2000) J. Cell. Physiol. 183, 321–329. [DOI] [PubMed] [Google Scholar]
- 24.Long, B. J., Grigoryev, D. N., Nnane, I. P., Liu, Y., Ling, Y. Z. & Brodie, A. M. (2000) Cancer Res. 60, 6630–6640. [PubMed] [Google Scholar]
- 25.Partin, A. W., Catalona, W. J., Finlay, J. A., Darte, C., Tindall, D. J., Young, C. Y., Klee, G. G., Chan, D. W., Rittenhouse, H. G., Wolfert, R. L. et al. (1999) Urology 54, 839–845. [DOI] [PubMed] [Google Scholar]
- 26.Babu, E., Kanai, Y., Chairoungdua, A., Kim, d. K., Iribe, Y., Tangtrongsup, S., Jutabha, P., Li, Y., Ahmed, N., Sakamoto, S., et al. (2003) J. Biol. Chem. 278, 43838–43845. [DOI] [PubMed] [Google Scholar]
- 27.Chuaqui, R. F., Englert, C. R., Strup, S. E., Vocke, C. D., Zhuang, Z., Duray, P. H., Bostwick, D. G., Linehan, W. M., Liotta, L. A. & Emmert-Buck, M. R. (1997) Urology 50, 302–307. [DOI] [PubMed] [Google Scholar]
- 28.Afar, D. E., Vivanco, I., Hubert, R. S., Kuo, J., Chen, E., Saffran, D. C., Raitano, A. B. & Jakobovits, A. (2001) Cancer Res. 61, 1686–1692. [PubMed] [Google Scholar]
- 29.Paralkar, V. M., Vail, A. L., Grasser, W. A., Brown, T. A., Xu, H., Vukicevic, S., Ke, H. Z., Qi, H., Owen, T. A. & Thompson, D. D. (1998) J. Biol. Chem. 273, 13760–13767. [DOI] [PubMed] [Google Scholar]
- 30.Shang, Y., Myers, M. & Brown, M. (2002) Mol. Cell 9, 601–610. [DOI] [PubMed] [Google Scholar]
- 31.Igawa, T., Lin, F. F., Lee, M. S., Karan, D., Batra, S. K. & Lin, M. F. (2002) Prostate 50, 222–235. [DOI] [PubMed] [Google Scholar]
- 32.Klein, K. A., Reiter, R. E., Redula, J., Moradi, H., Zhu, X. L., Brothman, A. R., Lamb, D. J., Marcelli, M., Belldegrun, A., Witte, O. N., et al. (1997) Nat. Med. 3, 402–408. [DOI] [PubMed] [Google Scholar]
- 33.Dotzlaw, H., Moehren, U., Mink, S., Cato, A. C., Iniguez Lluhi, J. A. & Baniahmad, A. (2002) Mol. Endocrinol. 16, 661–673. [DOI] [PubMed] [Google Scholar]
- 34.Squatrito, M., Mancino, M., Donzelli, M., Areces, L. B. & Draetta, G. F. (2004) Oncogene 23, 4454–4465. [DOI] [PubMed] [Google Scholar]
- 35.He, B., Kemppainen, J. A., Voegel, J. J., Gronemeyer, H. & Wilson, E. M. (1999) J. Biol. Chem. 274, 37219–37225. [DOI] [PubMed] [Google Scholar]
- 36.Aarnisalo, P., Santti, H., Poukka, H., Palvimo, J. J. & Janne, O. A. (1999) Endocrinology 140, 3097–3105. [DOI] [PubMed] [Google Scholar]
- 37.Alen, P., Claessens, F., Verhoeven, G., Rombauts, W. & Peeters, B. (1999) Mol. Cell. Biol. 19, 6085–6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hodgson, M. C., Astapova, I., Cheng, S., Lee, L. J., Verhoeven, M. C., Choi, E., Balk, S. P. & Hollenberg, A. N. (2005) J. Biol. Chem. 280, 6511–6519. [DOI] [PubMed] [Google Scholar]
- 39.Lyne, J. C., Melhem, M. F., Finley, G. G., Wen, D., Liu, N., Deng, D. H. & Salup, R. (1997) Cancer J. Sci. Am. 3, 21–30. [PubMed] [Google Scholar]
- 40.Qiu, Y., Ravi, L. & Kung, H. J. (1998) Nature 393, 83–85. [DOI] [PubMed] [Google Scholar]
- 41.Pompeia, C., Hodge, D. R., Plass, C., Wu, Y. Z., Marquez, V. E., Kelley, J. A. & Farrar, W. L. (2004) Cancer Res. 64, 3465–3473. [DOI] [PubMed] [Google Scholar]
- 42.Veldscholte, J., Ris-Stalpers, C., Kuiper, G. G., Jenster, G., Berrevoets, C., Claassen, E., van Rooij, H. C., Trapman, J., Brinkmann, A. O. & Mulder, E. (1990) Biochem. Biophys. Res. Commun. 173, 534–540. [DOI] [PubMed] [Google Scholar]
- 43.Feldman, B. J. & Feldman, D. (2001) Nat. Rev. Cancer 1, 34–45. [DOI] [PubMed] [Google Scholar]
- 44.Xia, X., Cheng, A., Lessor, T., Zhang, Y. & Hamburger, A. W. (2001) J. Cell. Physiol. 187, 209–217. [DOI] [PubMed] [Google Scholar]
- 45.Nesslinger, N. J., Shi, X. B. & DeVere White, R. W. (2003) Cancer Res. 63, 2228–2233. [PubMed] [Google Scholar]
- 46.Isaacs, J. T. & Isaacs, W. B. (2004) Nat. Med. 10, 26–27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.