Abstract
The shikimate pathway resulting in three aromatic amino acids is initiated in different organisms by two and three 3-deoxy-d-arabino-heptulosonate-7-phosphate synthases, respectively. Aro3p and Aro4p are the yeast enzymes feedback-inhibited by phenylalanine and tyrosine, respectively. A yeast strain deficient in the general control transcriptional regulatory system of amino acid biosynthesis is unable to live in the presence of high amounts of phenylalanine and tyrosine. Here, we show that this yeast strain can be rescued by the expression of aroH from Escherichia coli encoding the tryptophan-regulated AroH as third isoenzyme. Yeast carrying Ec AroH as the only enzyme for the initial step of the shikimate pathway can grow in the absence of tryptophan. Without aromatic amino acids, this yeast strain survives only when the yeast ARO3 promoter instead of the ARO4 promoter drives E. coli aroH. The detailed analysis of Aro3p and Aro4p revealed a triple feedback control by tyrosine/phenylalanine and tryptophan. Dissecting this control allowed engineering of Aro4p S195A as an enzyme, which is inhibited like AroH only by tryptophan. In addition, Aro4p variants were constructed that show an equally strong inhibition by tyrosine and tryptophan (Aro4p P165G Q302R) and in which the regulation by tyrosine and tryptophan was reversed (Aro4p P165G). Our data suggest that yeast possesses only two instead of three isogenes encoding 3-deoxy-d-arabino-heptulosonate-7-phosphate synthases because both isoenzymes can be fine tuned by tryptophan as additional effector and because transcriptional regulation by the general control system can be induced as backup when aromatic amino acids in the environment are imbalanced.
Keywords: general control, Aro4p, AroH
The shikimate pathway starts with the stereo specific condensation of erythrose-4-phosphate and phosphoenolpyruvate to 3-deoxy-d-arabino-heptulosonate-7-phosphate (DAHP) (1) and is catalyzed by the DAHP synthase (DAHPS, EC 4.1.2.15). DAHP is converted to the branch point compound chorismate (2–5), which is a precursor for the aromatic amino acids tyrosine (Tyr), phenylalanine (Phe), and tryptophan (Trp). The latter two amino acids are synthesized only by microorganisms and plants (6) and are assimilated by animals with the diet. Mammals convert Phe to Tyr and use aromatic amino acids as precursors for numerous important substances including neurotransmitters like serotonin (7). In Escherichia coli, the three isogenes aroF (8), aroG (9) and aroH (10) code for three DAHPS isoenzymes, each feedback-regulated by one of the aromatic amino acids Tyr (AroF), Phe (AroG), and Trp (AroH), respectively (11). Total cellular DAHPS activities are unequally distributed to the three isoenzymes. In E. coli, the Phe-inhibited AroG (79%) and the Tyr-inhibited AroF (20%) deliver nearly all of the DAHPS activity for the cell. The Trp-inhibited AroH contributes only 1% of the cellular enzyme activity (12). Inhibition of AroG (Pheinhibited) and AroF (Tyr-inhibited) is highly effective and abolishes >95% of their specific enzyme activity (13, 14), whereas the Trp inhibition of AroH is only ≈60%, thereby resulting in a 40% constitutive AroH activity (15).
Various fungi such as the yeast Saccharomyces cerevisiae and the filamentous fungi Aspergillus nidulans and Neurospora crassa express two differently regulated DAHPSs. The ARO3-encoded DAHPS is inhibited by Phe, whereas the ARO4 gene product is inhibited by Tyr (16–20). In A. nidulans, aroF and aroG code for two DAHPSs that are differentially regulated by Tyr (AROF) and Phe (AROG) (21). In addition to the feedback control of yeast Aro3p and Aro4p, the transcription of the corresponding genes is part of a complex regulatory network that couples transcriptional derepression of >500 genes to the availability of amino acids (22). This regulatory network is conserved between several fungi and is known as “cross-pathway control” in molds or as “general control” of amino acid biosynthesis in yeast (23–25). A key factor is the transcriptional activator Gcn4p of yeast and its interchangeable counterparts including, e.g., CPCA of A. nidulans (24). Gcn4p binds to the promoter region of target genes to a well characterized upstream activation site. These upstream sites are present in both ARO3 and ARO4 of yeast as well as in the corresponding genes of A. nidulans. During amino acid deprivation, ARO3 and ARO4 transcription is increased by a factor of two and four, respectively (17, 18).
Alignment of amino acid sequences of DAHPSs from E. coli and S. cerevisiae showed the sequence identity of 63% between Trp-regulated AroH and Phe-regulated Aro3p and of 64% between AroH and Tyr-regulated Aro4p. To date, the crystal structures of the aroG gene product (Phe-inhibited) from E. coli (26, 27) and the ARO4 gene product (Tyr-inhibited) from S. cerevisiae (28, 29) have been determined showing that the overall structures of bacterial and yeast DAHPSs are highly conserved.
Here, we started with the analysis of the influence of GCN4p and a heterologously expressed AroH, respectively, on yeast growing on an imbalanced amino acid diet. These experiments resulted in the discovery of an additional residual Trp regulation of Aro3p and Aro4p. Subsequently, the engineering of Aro4p variants with a distinct dual regulation by Tyr and/or Trp verified this subtle regulation of two isoenzymes by three effector amino acids.
Materials and Methods
Yeast Strains, Plasmids, Media, and Growth Conditions. Minimal vitamins (MV) and synthetic complete (SC) medium for the cultivation of yeast have been described (30). Transformation of S. cerevisiae was performed by the LiOAc method as described (31). For purification of Aro3p and Aro4p, for overexpression of yeast ARO4 alleles and as recipient for the E. coli aroH alleles, yeast strain RH2424 (MATa, can1-100, GAL+, aro3Δ::HIS3, aro4Δ::LEU2, ura3-1, his3–, leu2–) was used. Strain RH2424 (aro3Δ::HIS3, aro4Δ::LEU2) was transformed with a 3.1-kb BamHI/XhoI gcn4Δknockout cassette amplified from the gcn4Δ Euroscarf Strain ES249 (web.uni-frankfurt.de/fb15/mikro/euroscarf/). Transformants were selected on SC media supplemented with 150 μg/ml geneticin. Correct transformation was verified by Southern hybridization analysis and resulted in strain RH2804 (gcn4Δ::KanR, aro3Δ::HIS3, aro4Δ::LEU2). Strain RH2424 lacking any DAHPS activity was transformed with the aroH gene flanked by the promoter and terminator regions of ARO3 and ARO4, respectively, so that aroH was integrated and driven by the ARO3 or ARO4 promoter, respectively. The resulting strains RH2487 (MATa, can1-100, GAL+, aro3ΔaroH, aro4Δ::LEU2, ura3-1, his3–, leu2–) and RH2803 (MATa, can1-100, GAL+, aro3Δ::HIS3, aro4Δ aroH, ura3-1, his3–, leu2–) were verified by Southern hybridization. MV medium was supplemented with arginine, histidine, leucine, isoleucine, and valine to exclude other growth phenotypes produced by the knock-out cassettes used in these strains. For growth tests, strains RH1408 (MATa, ura3-52, gcn4-103) (32), RH730 (MATα WT) (33), RH1416 (MATa, ura3-52) (34), RH1313 (MATα, aro4-1) (18), RH1316 (MATa, aro3-2) (18) were used as controls.
Mutagenesis of ARO4. Site-directed mutations of ARO4 were introduced by using PCR with specific oligonucleotides carrying single nucleotide exchanges. Mutated alleles were cloned into p426MET25 (pRS426 carrying yeast MET25 promoter and CYC1 terminator, URA3, 2 μm, AmpR) and pME1513 (p426MET25 (URA3, 2 μm, AmpR) with altered MCS: SacIpMET25-XbaI SpeI BamHI SalI SfiI NotI XhoI-tCYC1-KpnI), respectively (35, 36). The DNA sequence of all vectors described in this work was determined by sequencing to rule out second-site mutations (37). After transformation of the allelic vectors into RH2424, transformants were selected on SC medium lacking aromatic amino acids to screen for functionally intact DAHPSs.
Enzyme Assays. The DAHPS activity was determined by the stop assay described by Takahashi and Chan (38) with the modifications of Schnappauf et al. (20). Measurements of enzyme activities were performed at 550 nm by using an Ultrospec 4000 spectrophotometer (Amersham Pharmacia Biotech). Enzyme assays with crude extracts were performed at least three times at 30°C for 1 min by using 0.5 mM of each substrate and effector. Assays were performed with pretested amounts of protein so that the absorbance of the product was proportional to the amount of enzyme added. Total protein was between 72 μg and 158 μg per reaction, yielding specific activities between 89 and 708 milliunits/mg. In effector titration assays, enzymatic activities were determined at least three times at 30°C after 1 min of reaction time with 150 μg of total protein. Substrate concentrations used were 0.4 mM erythrose-4-phosphate and 0.1 mM phosphoenolpyruvate. Specific activities measured without effector equaled between 53 and 464 milliunits/mg and were set 100% for each enzyme variant. In the enzyme assays with purified Aro3p or Aro4p, activities were measured with 160 ng and 250 ng of protein, respectively, 0.5 mM of each substrate, and the indicated amount of effector after a reaction time of 10 min. Protein contents of crude extracts were measured by using the Bradford assay (39).
Results
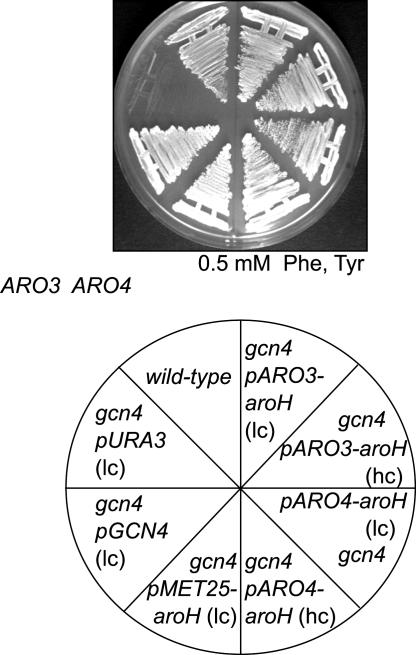
A Trp-Inhibited DAHP Synthase Regenerates Growth of a gcn4 Mutant Yeast Strain in the Presence of Phe and Tyr. We wanted to know how a gcn4 S. cerevisiae mutant strain carrying the WT ARO3 and ARO4 genes responds to imbalanced amino acid supply. The mutant strain RH1408 (gcn4-104) is unable to activate the general control of amino acid biosynthesis due to the lack of the transcriptional activator Gcn4p. As we observed, this yeast is unable to grow on MV medium that is supplemented with 5 mM Phe and 5 mM Tyr to inhibit both DAHPSs (Fig. 1). Addition of Trp regenerates growth of strain RH1408, suggesting that, under these conditions, this strain is starving for Trp. In the presence of only Phe and Tyr, strain RH1408 can be rescued by the expression of a plasmid carrying a copy of the GCN4 gene. In response to Trp starvation, Gcn4p increases the expression of ARO3 and ARO4, as well as that of the other genes of the general control network, and therefore regenerates growth (Fig. 1).
Fig. 1.
Suppression of the growth defect of a gcn4-103 mutation of S. cerevisiae on Phe- and Tyr-supplemented medium. Strain RH1408 (gcn4-103) was transformed with the low-copy GCN4-carrying vector (pME1083), with the low-copy URA3-carrying control vector (pRS416), with high-copy (hc) and low-copy (lc) number vectors carrying E. coli aroH fused to the ARO3 promoter [pARO3-aroH (pME1874; pME1873)], by vectors carrying E. coli aroH fused to the ARO4 promoter [pARO4-aroH (pME2391; pME2390)], and streaked out on MV medium. RH1408 was also transformed with aroH regulated by the MET25 promoter (pME1877). The WT strain RH730 was used as positive control.
Similarly, the E. coli aroH gene encoding the Trp-regulated DAHPS was heterologously expressed in yeast to prevent Trp starvation. The aroH-encoding sequence was driven either by the ARO3, by the ARO4, or by the MET25 promoter and expressed on low-copy or high-copy number plasmids in yeast strain RH1408. Expression of all aroH constructs rescued growth of strain RH1408 on media supplemented with 5 mM Phe and 5 mM Tyr. Therefore, Ec AroH suppresses this specific yeast gcn4 phenotype by regenerating the metabolic flux into the shikimate pathway (Fig. 1). Thus, in the presence of a specific unbalanced amino acid diet and in the absence of a functional general control system as backup, the lack of a third DAHPS enzyme is of disadvantage for yeast.
Heterologously Expressed DAHP SynthaseTrp Is Sufficient for Aromatic Amino Acid Biosynthesis in Yeast. The result that a Trp-inhibited third DAHPS isoenzyme improves the physiological potential of yeast gcn4Δ mutant strain in the presence of an unbalanced aromatic amino acid diet prompted us to construct a yeast strain with a Trp-regulated enzyme as the only enzyme for the initial step of the shikimate biosynthesis. Strain RH2804 (gcn4Δ::KanR, aro3Δ::HIS3, aro4Δ::LEU2) has no DAHPS activity and no possibility to adapt to imbalances in the amino acid nutrition. Strains RH2804 and as control RH2424 (aro3Δ::HIS3, aro4Δ::LEU2) were transformed with high-copy and low-copy number vectors, respectively, expressing the E. coli aroH gene driven by the ARO3 promoter (pARO3) and the ARO4 promoter (pARO4), respectively. As control, the E. coli aroH gene was fused to the MET25 promoter (pMET25) that is constitutively expressed on media lacking methionine. A growth test on supplemented MV media demonstrated that a yeast strain expressing high amounts of the Trp-inhibited DAHPS was able to survive on media without aromatic amino acids (Fig. 2A). Even without the transcriptional regulator Gcn4p, these artificial yeasts channeled enough flux into the shikimate pathway and were able to grow (Fig. 2 A).
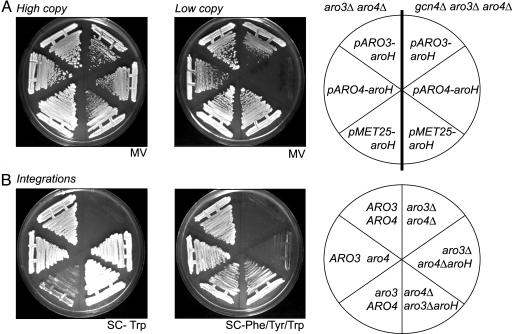
Fig. 2.
Growth of different S. cerevisiae strains carrying various plasmids expressing the E. coli aroH gene or carrying E. coli aroH integrated into the genome. (A) High-copy and low-copy: Strains RH2424 (aro3Δ, aro4Δ), and RH2804 (gcn4Δ, aro3Δ, aro4Δ) lacking any DAHPS activity were transformed with high-copy and low-copy number vectors carrying E. coli aroH fused to the ARO3 promoter [pARO3-aroH (pME1874, pME1873)], ARO4 promoter [pARO4-aroH (pME2391, pME2390)], and MET25 promoter [pMET25-aroH (pME1878, pME1877)], respectively. Vector-carrying yeast strains were streaked out on supplemented MV media lacking all three aromatic amino acids. (B) Integrations: aroH was integrated at the ARO3 locus in strain RH2487 (aro3ΔaroH), or at the ARO4 locus in strain RH2803 (aro4ΔaroH). As controls, a yeast WT strain (RH1416), an ARO3, aro4-1 mutant strain (RH1313), an aro3-2, ARO4 mutant strain (RH1316), and an aro3Δ, aro4Δ mutant strain (RH2424) were plated out. All strains were plated on SC medium lacking Trp or on SC medium lacking Phe, Tyr, and Trp, respectively.
When expressed from pARO3 and pARO4 on low-copy vectors, E. coli AroH was able to maintain the flux into the shikimate pathway, when the yeast still possessed the master regulator of amino acid biosynthesis, Gcn4p. In a yeast gcn4Δ mutant strain, however, only the pARO3-aroH fusion but not the pARO4-aroH fusion resulted in normal growth (Fig. 2 A). These data suggested that the basal promoter of ARO3 was stronger than the basal promoter of ARO4 and is counteracted by the fact that Aro4 can be induced 2-fold more by the general control than ARO3 (17, 18).
We constructed yeasts where single copies of pARO3-aroH or pARO4-aroH fusions were integrated into the yeast genome to verify these results. The E. coli aroH gene integrated at the ARO4 locus and driven by the ARO4 promoter (RH2803) resulted in a specific DAHPS activity that was hardly sufficient to rescue the aro3Δ/aro4Δ double deletion on SC media lacking all three aromatic amino acids (Fig. 2B). This yeast could grow only if Trp was the only amino acid missing. Integration of aroH at the ARO3 locus (RH2487), however, resulted in normal growth under all conditions (Fig. 2B). Similarly, a yeast strain with intact ARO4 and deficient in ARO3 (RH1316, aro3-2) was retarded in growth on SC media lacking Trp because the Tyr-inhibited Aro4p did not produce enough DAHP for sufficient Trp, although growth was like the WT control on SC medium without aromatic amino acids when Aro4p was not inhibited. A strain carrying an intact ARO3, but a defective ARO4 (RH1313, aro4-1) showed normal growth under all conditions because there were obviously more Aro3p enzymes in the cells to channel sufficient substrate into the shikimate pathway (Fig. 2B). Thus, in the presence or absence of Gcn4p, a DAHPS-encoding gene was more efficiently transcribed when expressed from the yeast ARO3 promoter than from the ARO4 promoter. The yeast cell thus possesses more Aro3p than Aro4p enzymes as it was described for the DAHPSPhe and DAHPSTyr in E. coli (12). All these data suggest that DAHPSTrp was sufficient for production of aromatic amino acids in yeast, if the enzyme amounts were at least as high as the endogenous basal DAHPSPhe pool or could be further supported by the general control.
The Yeast DAHPSs Are Regulated by Trp. The high degree of sequence identity and the structural similarities between Aro3p and Aro4p and between the yeast and the E. coli enzymes prompted us to ask whether the yeast Phe- and Tyr-regulated DAHPSs might be regulated to a less extent by the other aromatic amino acids. In enzyme activity assays performed with purified DAHPS, the Phe-inhibited DAHPS (Aro3p) exhibited a specific activity of 44 milliunits/mg in the presence of 0.1 mM Trp (97% of unregulated activity) but with 10.5 milliunits/mg only 22% of specific activity at 1.0 mM Trp (Fig. 3). The Trp inhibition was even more obvious for the Tyr-inhibited DAHPS (Aro4p). In the presence of 0.1 mM Trp, the DAPHTyr synthase showed a specific activity of 5.5 milliunits/mg (46%), whereas it displayed a specific activity of 2 milliunits/mg (18%) at 1.0 mM Trp (Fig. 3). At 0.1 mM of Trp the Aro4p enzyme was reduced to half activity, whereas the isoenzyme Aro3p displayed nearly full activity. Similarly, Aro4p was inhibited by the third aromatic amino acid (Phe) more strongly than Aro3p by Tyr at the lowest concentration tested. These results indicate that, in addition to their known major control by Phe (Aro3p) and Tyr (Aro4p), respectively, both enzymes can be down-regulated when high amounts of Trp or Tyr/Phe are present in the diet. Aro4p is more sensible and responds to the effectors in lower concentrations than Aro3p. Thus, the weak triple feedback-regulation of the two yeast isoenzymes might counteract the lack of the third enzyme as long as the Gcn4p-dependent general control is functional as a further backup system.
Fig. 3.
Inhibition of Aro3p and Aro4p by aromatic amino acids. The specific Phe-inhibited (Aro3p) and Tyr-inhibited (Aro4p) DAHPS activities of purified enzymes were measured without effector (w/o aa), and with the indicated amount of Phe, Tyr and Trp, respectively.
A Single S195A Substitution Results in an Aro4p Yeast DAHPS Inhibited by Trp. We aimed to dissect the enzyme regulation to verify the triple control of the two yeast DAHPSs. Recent experiments demonstrated that the yeast Tyr-inhibited DAHPS could be turned into a Phe-regulated enzyme by a single amino acid substitution and vice versa (29). Therefore, we performed in vitro evolution of Aro4p to change regulation of the Tyr-inhibited DAHPS toward a stronger inhibition by Trp. We applied an amino acid sequence alignment of E. coli AroH (Trp-inhibited) and yeast Aro4p (Tyr-inhibited), a comparison of the 3-dimensional structures of E. coli AroG (26, 40) and Aro4p (28, 29), as well as results from genetic screens (29). By site-directed mutagenesis, the codons of glycine 193, serine 195, proline 165, alanine 234, and glutamine 302 in the yeast ARO4-encoded DAHPTyr synthase, respectively, were exchanged, and the resulting enzyme variants were examined regarding Tyr and Trp regulation. These residues are mainly located at the putative effector-binding cavity of Aro4p (Fig. 5, which is published as supporting information on the PNAS web site) (29). The structure of this cavity is very similar to the structure published for the Phe-inhibited aroG from E. coli. The hydrophobic and hydrophilic residues G193 and S195, respectively, are located in the connector region between the half-sites of the α/β barrel and were substituted by a polar lysine (GGT→AAG) and a hydrophobic alanine (TCT→GCT) residue, respectively. Proline 165 in helix α3 is located in the bottom of the cavity and was substituted by a glycine (CCT→GGT), which is present in the Trp-regulated AroH at this position. The hydrophobic residue alanine 234 in the internal β strand β6b, which forms the side of the cavity, was exchanged against a polar threonine residue (GCT→ACT). In addition, glutamine 302 was substituted by arginine (CAA→CGA). This amino acid residue is located in helix α7, which is remote from the putative effector binding cavity.
Specific enzyme activities from crude extract measurements with effector concentrations of 0.5 mM revealed that the exchange of an A234T did not result in significant alteration of Aro4p regulation. The enzyme was still completely inhibited by Tyr, and activity was reduced to half by Trp (data not shown). Therefore, this amino acid residue seemed not to be involved in the regulation of the yeast DAHPTyr. Exchange of G193K in Aro4p abolished most of DAHPS activity and thus obviously disturbed the overall structure of Aro4p (data not shown).
The exchange of P165G in the yeast Tyr-inhibited DAHPS resulted in an enzyme for which regulation was shifted more toward Trp inhibition compared with Aro4p wild type, but which maintained a small degree of Tyr inhibition (Fig. 4A). In fact, the regulation pattern by Tyr and Trp was reversed in this Aro4p variant. An additional substitution of Q302R in the P165G variant, however, reversed the effect on Tyr regulation, resulting in an enzyme variant that was still Tyr inhibited and equally strongly inhibited by Trp (Fig. 4A). Therefore, this enzyme constituted a hybrid of a DAHPSTyr and DAHPSTrp. In contrast, the exchange of S195A in the yeast Tyr-inhibited DAHPS resulted in an enzyme that no longer was inhibited by its main effector Tyr, but was inhibited by Trp as single effector (Fig. 4A). By removing the polar hydroxyl group of serine at the side of the cavity, the yeast DAHPSTyr became DAHPSTrp although there is also a serine at the respective position in the Trp-regulated AroH. Thus, a single amino acid substitution in the putative effector binding site is sufficient to turn Aro4p into a Trp-inhibited DAHPS.
Fig. 4.
Inhibition of WT and mutant Aro4p and AroH activity by Tyr and Trp. DAHPS-encoding genes were expressed from the MET25 promoter on high-copy vectors in strain RH2424 (aro3Δ, aro4Δ). The vectors carried the WT ARO4 gene (pME2385), the alleles encoding Aro4p P165G (pME2395), Aro4p P165G Q302R (pME2878), Aro4p S195A (pME2396), and the WT aroH gene (pME1878). (A) Enzyme assays were performed with 0.5 mM each substrate and effector. (B and C) Tyr and Trp titration reactions were carried out by using 0.4 mM erythrose-4-phosphate, 0.1 mM phosphoenolpyruvate, and the indicated effector concentrations.
Effector titration of crude extracts of RH2424 expressing the different Aro4p alleles was performed to measure the sensitivity to Tyr and Trp more precisely. Wild-type Aro4p was inhibited to a maximum of below 9% of original activity above 100 μM Tyr (Fig. 4B). A sharp decrease of activity was observed at low effector concentration, so that 50% inhibition was achieved at 10 μM Tyr. The Aro4p P165G and Aro4p P165G/Q203R enzymes were inhibited to a similar maximum of 15% and 7% activity, respectively, but showed only partial inhibition at lower concentrations. At the physiological concentration of 0.5 mM (41), Tyr caused only partial inhibition of the P165G DAHPS (33%) compared with nearly complete inhibition of the Aro4p wild type and P165G/Q302R variant. In comparison, WT AroH and the AroH-like Aro4p S195A variant were not inhibited by Tyr, but were slightly activated instead.
When Trp was used as an effector, maximum inhibition was nearly the same for the AroH wild type, the Aro4p wild type, and the AroH-like S195A variant (21–32% activity), whereas inhibition was nearly complete for Aro4p P165G/Q302R and P165G above 500 μM Trp (Fig. 4C). At the physiological Trp concentration in yeast cells of ≈20 μM (41), AroH was slightly activated, whereas Aro4p and Aro4p S195A were not affected. For none of the enzymes tested, a rapid and complete inhibition like the Tyr inhibition of Aro4p was observed with Trp, but all enzymes showed a moderate decrease in activity, with half-maximum activity between 120 and 270 μM Trp. These experiments strongly suggest that Aro4p exhibits a rudimental Trp inhibition that strongly resembles the AroH WT inhibition and can be amplified by specific amino acid substitutions that, however, in part have to be paid for by reduced Tyr inhibition.
Discussion
Evolution of DAHPS enzymes in prokaryotes has been described in detail (42–47). The evolution of prokaryotic DAHPSs presumably started at the level of an allosterically insensitive DAHPS0 and a tyr-sensitive DAHPSTyr, and evolutionary processes changed the DAHPS0 enzyme toward a Trp-inhibited DAHPSTrp enzyme. In secondary evolutionary processes, some prokaryotes presumably have lost the DAHPSTyr, carrying only one, highly efficient DAHPSTrp enzyme. Among those microbial representatives with two enzymes, a wide variability of additional chorismate sensitivity of DAHPSTrp was found (42, 45). In the course of evolution, a loss of the chorismate sensitivity of the DAHPSTrp seemed to occur at the same time when microorganisms acquired a third Phe-regulated enzyme activity (DAHPSPhe), presumably by a gene duplication event of a DAHPSTrp-encoding gene (47, 48). Only those prokaryotes carried three DAHPSs, which had a DAHPSTrp enzyme that was inhibited only by Trp but not by chorismate, whereas the allosterical chorismate-dependent regulations of DAHPSPhe or DAHPSTyr were variable (45).
It is unknown whether S. cerevisiae has lost a third DAHPSTrp activity or never acquired one, but the known yeast genome definitely contains only two DAHPS loci (49). We have recently shown that a single amino acid substitution (S219G) was sufficient to switch the ARO3-encoded DAHPSPhe into an Aro4p-like DAHPSTyr, and vice versa the exchange of G226S of the ARO4-encoded DAHPSTyr resulted in an Aro3p-like DAHPSPhe (29). We now extended this study and showed here that the single amino acid substitution S195A turns Aro4p into a DAHPSTrp enzyme very similar to the homologous AroH from E. coli. The most important residues controlling binding of Tyr or Trp proved to be those at the bottom and the side of the putative effector binding site. Obviously, the polar S195 is important for contact to Tyr, whereas a more hydrophobic environment or a sterical adjustment due to the loss of proline in helix α3 favors Trp binding. The fact that E. coli AroH also possessed a serine at the corresponding position 179 and that substitutions to amino acid residues not present at respective positions in the bacterial enzyme also produced Trp inhibition showed that Trp inhibition in Aro4p S195A was achieved by a different intramolecular signal transduction pathway between allosteric and catalytic site, which depends on further differences in the amino acid sequence. These results suggest that evolution of yeast DAHPS might be different from that described in prokaryotes.
Because we found weak Trp regulation for both fungal isoenzymes, fungal evolution might have started with a Trp-inhibited DAHPS precursor enzyme. Gene duplication and the transcriptional regulation of the general control network of amino acid biosynthesis finally resulted in two Gcn4p-regulated genes encoding for highly effective and closely related DAHPS activities, with additional regulation by Tyr and Phe, DAHPSTyr (Aro4p) and DAHPSPhe (Aro3p), respectively. Tryptophan regulation in yeast was shifted downstream in the pathway to the first branch point because chorismate mutase evolved as an important key regulator that was activated by Trp and inhibited by Tyr (50). At this strongly regulated branch point, anthranilate synthase encoded by TRP2 and TRP3, which is transcriptionally regulated by the general control (41), competes with chorismate mutase for chorismate. The anthranilate synthase complex has a lower Km for chorismate than chorismate mutase. When TRP2 and TRP3 are transcriptionally induced, the gene products direct the metabolic flux toward Trp production, which ultimately results in their feedback-inhibition by Trp (41, 51). At intracellular Trp levels of 20 μM, at which anthranilate synthase is not yet strongly inhibited by Trp (Ki 56 μM), this amino acid already activates chorismate mutase (Ka, 1.5 μM) leading to increased Phe and Tyr synthesis. When Tyr and Phe are being synthesized, the DAHPSs at the beginning of the pathway (Ki, 10 μM and 1 μM) as well as chorismate mutase (Ki, 50 μM) are inhibited, making an additional Trp control dispensable (19, 20, 41, 52).
In E. coli, AroH as third DAHPS together with the residual activity of the other DAHPS, ensures a small but steady flux into the shikimate pathway because it has 40% constitutive activity. Here, we demonstrated that the expression of a Trp-inhibited DAHPS from E. coli had the same effect on Trp production as the general control exerted by GCN4p. Thus, the general control evolved for this pathway in yeast as protection from shutting down the shikimate pathway and preventing Trp starvation through transcriptional GCN4p dependent up-regulation of transcription of ARO3/ARO4 and TRP genes (41) when Aro3p and Aro4p are inhibited. A single Trp-inhibited DAHPS in sufficient amounts or even less DAHPSTrp combined with a functional general control can save yeast from amino acid starvation by unbalanced diets. Thus, the end product Trp can be used as the only feedback regulator of this biosynthetic pathway in an artificial yeast with a DAHPSTrp as first enzyme. The observed mixed inhibition by Tyr and Trp most pronounced in Aro4p reflects a complex evolution and allows yeast cells a subtle fine tuning of the branched pathway even under conditions of very unbalanced amino acid diet, including high Trp concentrations.
Supplementary Material
Acknowledgments
We thank Gabriele Heinrich for excellent technical assistance and Daniel Schulz for his help during the initial phase of this work. We are grateful to Ron Bauerle (Department of Biology, University of Virginia, Charlottesville) for providing the plasmid pCHA3 and to Andrea Pfeil (Molekulare Mikrobiologie und Genetik, Institut für Mikrobiologie und Genetik) for providing purified Aro3p enzyme. We thank Hans-Ulrich Mösch and Sven Krappmann for helpful discussions and critical reading of this manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG), the Volkswagen-Stiftung, the Fonds der Chemischen Industrie, and the DFG Research Center for Molecular Physiology of the Brain (CMPB). W.N.L. acknowledges support by National Institutes of Health Grant GM 06920.
Author contributions: K.H., A.S., W.N.L., and G.H.B. designed research; and K.H. and A.S. performed research.
Abbreviations: DAHP, 3-deoxy-d-arabino-heptulosonate-7-phosphate; DAHPS, DAHP thase; SC, synthetic complete; MV, minimal vitamins.
References
- 1.Herrmann, K. M. & Poling, M. D. (1975) J. Biol. Chem. 250, 6817–6821. [PubMed] [Google Scholar]
- 2.Herrmann, K. M. (1995) Plant Cell 7, 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knaggs, A. R. (1999) Nat. Prod. Rep. 16, 525–560. [DOI] [PubMed] [Google Scholar]
- 4.Knaggs, A. R. (2000) Nat. Prod. Rep. 17, 269–292. [DOI] [PubMed] [Google Scholar]
- 5.Knaggs, A. R. (2001) Nat. Prod. Rep. 18, 334–355. [DOI] [PubMed] [Google Scholar]
- 6.Haslam, E. (1974) The Shikimate Pathway (Butterworth, London).
- 7.Fernstrom, J. D. (1981) Annu. Rev. Med. 32, 413–425. [DOI] [PubMed] [Google Scholar]
- 8.Shultz, J., Hermodson, M. A., Garner, C. C. & Herrmann, K. M. (1984) J. Biol. Chem. 259, 9655–9661. [PubMed] [Google Scholar]
- 9.Davies, W. D. & Davidson, B. E. (1982) Nucleic Acids Res. 10, 4045–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zurawski, G., Gunsalus, R. P., Brown, K. D. & Yanofsky, C. (1981) J. Mol. Biol. 145, 47–73. [DOI] [PubMed] [Google Scholar]
- 11.Byng, G. S., Johnson, J. L., Whitaker, R. J., Gherna, R. L. & Jensen, R. A. (1983) J. Mol. Evol. 19, 272–282. [DOI] [PubMed] [Google Scholar]
- 12.Tribe, D. E., Camakaris, H. & Pittard, J. (1976) J. Bacteriol. 127, 1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoner, R. & Herrmann, K. M. (1976) J. Biol. Chem. 251, 5440–5447. [PubMed] [Google Scholar]
- 14.McCandliss, R. J., Poling, M. D. & Herrmann, K. M. (1978) J. Biol. Chem. 253, 4259–4265. [PubMed] [Google Scholar]
- 15.Pittard, J., Camakaris, J. & Wallace, B. J. (1969) J. Bacteriol. 97, 1242–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teshiba, S., Furter, R., Niederberger, P., Braus, G., Paravicini, G. & Hütter, R. (1986) Mol. Gen. Genet. 205, 353–357. [DOI] [PubMed] [Google Scholar]
- 17.Künzler, M., Paravicini, G., Egli, C. M., Irniger, S. & Braus, G. H. (1992) Gene 113, 67–74. [DOI] [PubMed] [Google Scholar]
- 18.Paravicini, G., Mösch, H. U., Schmidheini, T. & Braus, G. (1989) Mol. Cell. Biol. 9, 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paravicini, G., Schmidheini, T. & Braus, G. (1989) Eur. J. Biochem. 186, 361–366. [DOI] [PubMed] [Google Scholar]
- 20.Schnappauf, G., Hartmann, M., Künzler, M. & Braus, G. H. (1998) Arch. Microbiol. 169, 517–524. [DOI] [PubMed] [Google Scholar]
- 21.Hartmann, M., Heinrich, G. & Braus, G. H. (2001) Arch. Microbiol. 175, 112–121. [DOI] [PubMed] [Google Scholar]
- 22.Natarajan, K., Meyer, M. R., Jackson, B. M., Slade, D., Roberts, C., Hinnebusch, A. G. & Marton, M. J. (2001) Mol. Cell. Biol. 21, 4347–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinnebusch, A. G. (1997) J. Biol. Chem. 272, 21661–21664. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann, B., Valerius, O., Andermann, M. & Braus, G. H. (2001) Mol. Biol. Cell 12, 2846–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinnebusch, A. G. & Natarajan, K. (2002) Eukaryot. Cell 1, 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shumilin, I. A., Kretsinger, R. H. & Bauerle, R. H. (1999) Structure Fold Des. 7, 865–875. [DOI] [PubMed] [Google Scholar]
- 27.Shumilin, I. A., Kretsinger, R. H. & Bauerle, R. (1996) Proteins 24, 404–406. [DOI] [PubMed] [Google Scholar]
- 28.Schneider, T. R., Hartmann, M. & Braus, G. H. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 1586–1588. [DOI] [PubMed] [Google Scholar]
- 29.Hartmann, M., Schneider, T. R., Pfeil, A., Heinrich, G., Lipscomb, W. N. & Braus, G. H. (2003) Proc. Natl. Acad. Sci. USA 100, 862–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miozzari, G., Niederberger, P. & Hütter, R. (1978) J. Bacteriol. 134, 48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito, H., Fukuda, Y., Murata, K. & Kimura, A. (1983) J. Bacteriol. 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hinnebusch, A. G. (1985) Mol. Cell. Biol. 5, 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohlhaw, G. B., Hsu, Y. P., Lemmon, R. D. & Petes, T. D. (1980) J. Bacteriol. 144, 852–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berben, G., Legrain, M. & Hilger, F. (1988) Gene 66, 307–312. [DOI] [PubMed] [Google Scholar]
- 35.Mumberg, D., Müller, R. & Funk, M. (1994) Nucleic Acids Res. 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krappmann, S., Helmstaedt, K., Gerstberger, T., Eckert, S., Hoffmann, B., Hoppert, M., Schnappauf, G. & Braus, G. H. (1999) J. Biol. Chem. 274, 22275–22282. [DOI] [PubMed] [Google Scholar]
- 37.Rosenblum, B. B., Lee, L. G., Spurgeon, S. L., Khan, S. H., Menchen, S. M., Heiner, C. R. & Chen, S. M. (1997) Nucleic Acids Res. 25, 4500–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi, M. & Chan, W. W. (1971) Can. J. Biochem. 49, 1015–1025. [DOI] [PubMed] [Google Scholar]
- 39.Bradford, M. M. (1976) Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 40.Shumilin, I. A., Zhao, C., Bauerle, R. & Kretsinger, R. H. (2002) J. Mol. Biol. 320, 1147–1156. [DOI] [PubMed] [Google Scholar]
- 41.Braus, G. H. (1991) Microbiol. Rev 55, 349–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmad, S., Rightmire, B. & Jensen, R. A. (1986) J. Bacteriol. 165, 146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahmad, S., Johnson, J. L. & Jensen, R. A. (1987) J. Mol. Evol. 25, 159–167. [DOI] [PubMed] [Google Scholar]
- 44.Ahmad, S. & Jensen, R. A. (1988) Mol. Biol. Evol. 5, 282–297. [DOI] [PubMed] [Google Scholar]
- 45.Ahmad, S., Weisburg, W. G. & Jensen, R. A. (1990) J. Bacteriol. 172, 1051–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jensen, R. A. (1985) Mol. Biol. Evol. 2, 92–108. [DOI] [PubMed] [Google Scholar]
- 47.Jensen, R. A. & Ahmad, S. (1988) Microbiol. Sci. 5, 316–319. [PubMed] [Google Scholar]
- 48.Jensen, R. A. & Byng, G. S. (1981) Isozymes Curr. Top. Biol. Med. Res. 5, 143–174. [PubMed] [Google Scholar]
- 49.Mewes, H. W., Albermann, K., Bähr, M., Frishman, D., Gleissner, A., Hani, J., Heumann, K., Kleine, K., Maierl, A., Oliver, S. G., et al. (1997) Nature 387, 7–65. [DOI] [PubMed] [Google Scholar]
- 50.Helmstaedt, K., Krappmann, S. & Braus, G. H. (2001) Microbiol. Mol. Biol. Rev. 65, 404–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krappmann, S., Lipscomb, W. N. & Braus, G. H. (2000) Proc. Natl. Acad. Sci. USA 97, 13585–13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidheini, T., Mösch, H. U., Evans, J. N. & Braus, G. (1990) Biochemistry 29, 3660–3668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.