Abstract
The type III secretion system (T3SS) of Pseudomonas aeruginosa is an important virulence determinant. Transcription of the T3SS is highly regulated and intimately coupled to the activity of the type III secretion channel. The secretion channel is generally closed, and transcription is repressed. Inducing signals such as calcium depletion, however, open the secretion channel and derepress transcription of the T3SS. The coupling of transcription with secretion requires three previously identified cytoplasmic regulatory proteins. ExsA is a DNA-binding protein required for transcriptional activation of the entire T3SS. The second regulatory protein, ExsD, functions as anti-activator by directly binding to ExsA. Finally, ExsC functions as an anti-anti-activator by directly binding to and inhibiting ExsD. Although the regulatory roles of ExsC, ExsD, and ExsA were defined through these previous studies, the mechanism of coupling transcription to secretion was unclear. We now report the identification of ExsE as a secreted regulator of the T3SS and provide evidence that ExsE functions as a direct inhibitor of ExsC. When the secretion channel is closed, ExsE is complexed with ExsC in the cytoplasm, and transcription of the T3SS is repressed by sequestration of ExsA by ExsD. We propose that the secretion of ExsE provides an initiating signal that results in an equilibrium shift whereby ExsC becomes preferentially bound to ExsD, thus allowing liberated ExsA to activate transcription of the T3SS. The presence of ExsE homologs in the T3SSs of other bacterial species suggests that this mechanism of coupling transcription to secretion may be commonly used.
Keywords: anti-activator, anti-anti-activator, ExsE
Pseudomonas aeruginosa is a Gram-negative opportunistic pathogen and a leading cause of ventilator-associated pneumonia and urinary tract infections in the intensive care unit (1, 2). Individuals most susceptible to P. aeruginosa infection include those with immunodeficiency, cystic fibrosis, or severe burns (1). The type III secretion system (T3SS) of P. aeruginosa is a major virulence determinant and is used to translocate bacterial cytotoxins directly into the cytoplasm of eukaryotic host cells. The translocated cytotoxins play a vital role in pathogenesis by inhibiting phagocytosis and damaging host tissues (3, 4). Mutants lacking expression of the T3SS are significantly attenuated in animal models of P. aeruginosa pathogenesis, and expression of the T3SS in clinical isolates has been correlated to a poorer outcome in cases of human infection (5–7).
The P. aeruginosa T3SS consists of 38 coordinately regulated genes required for the biosynthesis, secretion, and translocation of the cytotoxins into host cells (8). Expression of the T3SS is highly regulated and induced by at least three environmental signals: (i) calcium chelation (hereafter referred to as low Ca2+), (ii) the presence of serum, and (iii) contact with eukaryotic host cells (8, 9). Induction by low Ca2+ has been studied most extensively and involves a mechanism of converting the type III secretion channel from a closed (inactive) state to an open (active) state (10). Transcription of the T3SS is directly coupled to the state of the secretion channel; transcription is repressed when the secretion channel is closed and derepressed when the secretion channel is open. Although the mechanism by which low Ca2+ opens the secretion channel is unknown, mutants lacking functional secretion machinery are repressed for transcription irrespective of Ca2+ levels, suggesting that the relationship between secretion and transcription is direct (10, 11).
The direct coupling of transcription to secretion is a feature common to many T3SSs and was first described for the flagellar assembly pathway (12). Flagellar biosynthesis is a complex and energetically consumptive process that requires upwards of 50 gene products. The genes required for flagellar assembly (including a T3SS) are expressed as an integrated hierarchy and cluster into one of three classes based on their timing of expression (13). Expression of the late genes (including the flagellin subunit) is positively controlled by the FliA σ factor and is negatively regulated by the FlgM anti-σ factor. FlgM sequesters FliA, repressing late gene expression before completion of the basal body. Upon completion of the basal body, FlgM is secreted via the flagellar T3SS enabling FliA-mediated transcription of the late genes. The coupling of transcription to secretion by FlgM provides a mechanism of limiting gene expression to the precise time in which the gene products are required in the assembly process, thus conserving energy.
Transcription of the P. aeruginosa T3SS is controlled by a regulatory cascade involving three cytoplasmic proteins (ExsC, ExsD, and ExsA). At the bottom of the cascade is ExsA, a positive transcriptional regulator required for expression of all T3SS genes (8, 14). ExsA is a member of the AraC family of transcriptional activators and binds to a defined consensus sequence in each of the T3SS promoters (15, 16). The second protein in the regulatory cascade, ExsD, functions just upstream of ExsA and acts as an anti-activator by directly binding to and inhibiting the activity of ExsA (10). Finally, ExsC is positioned just upstream of ExsD and functions as an anti-anti-activator by directly binding to and inhibiting ExsD (17).
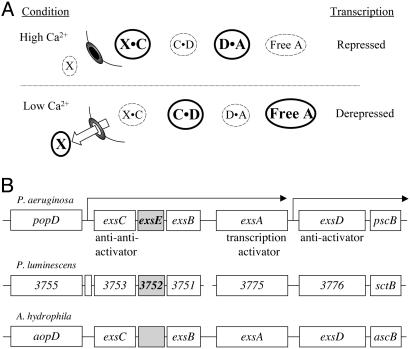
Lacking from the ExsC-D-A regulatory cascade, however, is a mechanism of coupling transcription to secretion. An important clue came from the observation that ExsC has biochemical properties characteristic of a type III secretion chaperone (17). Members of this chaperone family facilitate secretion of type III substrates and in some cases contribute to transcriptional regulation (18). Based upon the premise that ExsC was indeed a type III chaperone, we previously proposed that the regulatory activity of ExsC is controlled by its cognate secretion substrate (hypothetical protein X) (10, 17). The model predicted that the intracellular concentration of protein X would serve as a signal of whether the secretion channel was closed or open. When the secretion channel is closed, cytoplasmic protein X would sequester ExsC, and the binding of ExsD to ExsA would prevent transcription of the T3SS (Fig. 1A, high Ca2+). Conversely, when the secretion channel is open protein X is released from cells (Fig. 1 A, low Ca2+). The reduced intracellular pool of protein X increases the levels of ExsC available to bind ExsD and frees ExsA to activate transcription of the T3SS. In the current study, we report the identification of protein X as being ExsE and demonstrate that ExsE is a secreted regulator of the ExsC anti-anti-activator.
Fig. 1.
Proposed regulatory scheme for bacteria encoding ExsCDE homologs. (A) Model for regulation of the T3SS. Under high Ca2+ conditions, protein X (later identified as ExsE) remains cytoplasmic. This favors formation of the protein X·ExsC- and ExsD·ExsA-binding complexes (indicated by larger text and a larger dark circle) resulting in repression of the T3SS. Under low Ca2+ conditions, protein X is secreted. The decrease in intracellular protein X shifts the binding equilibrium in favor of forming the ExsC·ExsD complex and free ExsA. (B) exsE homologs are present in similar genetic contexts in other bacterial species. Comparison of the genetic organization of regions encoding putative homologs of the P. aeruginosa T3SS regulators ExsC, ExsA, and ExsD. The exsCEBA and the beginning of the exsD, pscB-L operons are indicated by arrows. Transcription of both of these operons is ExsA-dependent. The functions of ExsC, ExsA, and ExsD are indicated. ExsB is thought to be a chaperone involved in assembly of the PscC secretin and has no known role in transcriptional regulation. P.a., P. aeruginosa PA01; P.l., P. luminescens TT01; A.h., A. hydrophila. GenBank accession nos.: P.a., AE004091; P.l., BX571871; A.h., AY528667.
Materials and Methods
Strains, Growth Conditions, and Reagents. The ΔexsC and ΔexsD mutants, derivatives of P. aeruginosa strain PA103, were described previously (10, 17). For the T3SS expression studies, strains were grown at 30°C in trypticase soy broth supplemented with 1% glycerol, 100 mM monosodium glutamate and, where indicated, 2 mM EGTA. Antibiotics were used at the following concentrations: 300 μg/ml carbenicillin, 100 μg/ml tetracycline, 100 μg/ml gentamycin for P. aeruginosa; 100 μg/ml ampicillin, 10 μg/ml tetracycline, and 10 μg/ml gentamycin for Escherichia coli.
Mutant and Plasmid Construction. DNA fragments flanking the upstream (1,190 bp) and downstream (1,130 bp) regions of exsE were amplified by the PCR and sequentially ligated into pEX18Tc (19), resulting in an in-frame deletion of codons 4–78. The resulting plasmid (pEX18TcΔexsE) was mobilized to wild-type PA103, and mutants were isolated as described (10). The ΔexsCE double mutant was similarly constructed by using the previously described upstream flank for exsC (17) and the exsE downstream flank described above. Mutants were confirmed by PCR by using primers outside of the cloned regions.
pJexsE was constructed by PCR amplification of exsE from P. aeruginosa strain 388. The PCR product was cloned into the corresponding sites of pJN105 under the transcriptional control of an arabinose-inducible promoter (20). pJexsEHis-6 (encoding a C-terminal hexahistidine tag on ExsE) was similarly constructed by PCR. The pJexsD and pexsC expression plasmids were previously described (17).
Two-Hybrid Assay. The exsC and exsD two-hybrid expression vectors and controls for the two-hybrid assay have been described (10, 17, 21). The exsE two-hybrid expression plasmids were constructed by PCR amplification of exsE. The PCR product was cloned into the SacI and XbaI restriction sites of pSR658 and pSR659 (21). The E. coli SU202 reporter strain was transformed with the expression vectors as indicated and assayed for β-galactosidase activity as described (17).
ExsE and ExsC Purification and Ni-NTA Copurification Assays. For expression in E. coli and purification, exsE was PCR-amplified with primers incorporating NdeI and XhoI restriction sites at the 5′ and 3′ termini of the gene, respectively, and cloned into the corresponding sites of pET24a (Novagen). The resulting clone incorporates a hexahistidine epitope tag at the C terminus of ExsE (ExsEHis-6). ExsEHis-6 and ExsCHis-6 were purified by Ni2+-NTA affinity chromatography as described (17). For the copurification studies, extracts from E. coli BL21(DE3) overexpressing native ExsC or ExsD were prepared by French pressure cell disruption. Clarified extracts (200 mg) were incubated with 20 μg of purified ExsEHis-6 or ExsCHis-6 on ice and processed for Ni2+-NTA chromatography as described (17).
Cell Fractionation, SDS/PAGE, and Immunoblots. Supernatant fractions were concentrated by precipitation with trichloroacetic acid as described (10). Cell-associated fractions were prepared by pelleting 1.25 ml of cell culture (A540 = 1.0), suspending in 300 μl of SDS/PAGE sample buffer, and sonicating for 10 s. Supernatant and cell-associated samples were heated for 5 min at 95°C and electrophoresed on Hi-Tris (to visualize ExsC and ExsE) or 15% SDS–polyacrylamide gels. Immunoblots were developed by using an anti-His-horseradish peroxidase (HRP) conjugate (Qiagen, Valencia, CA) or by using anti-ExoU and anti-ExsC with detection by secondary anti-IgG-HRP conjugate and enhanced chemiluminescence (Amersham Pharmacia Biosciences).
For the fractionation studies, cells were sedimented by centrifugation (4 min, 12,500 × g) and suspended in ice-cold 300-μl PBS containing 10 μg/ml leupeptin, 5 mM 1,10-phenanthroline, 10 μg/ml pepstatin A, and 1 mM phenylmethyl sulfonyl fluoride. Cells were lysed by sonication for 20 s and centrifuged (12,500 × g) for 4 min at 4°C to remove unbroken cells. The cell extract was transferred to a fresh tube and centrifuged at 100,000 × g for 20 min at 4°C to separate the soluble fraction from the insoluble membrane fraction (pellet).
β-Galactosidase Assays. The PexsD-lacZ transcriptional reporter was reported previously (10). Strains carrying the reporter were grown under the indicated conditions until the absorbance (A600) reached 1.0, and β-galactosidase activity was then measured by using the substrate p-nitrophenyl β-d-galactopyranoside, as described (17). The reported values (Miller units) represent the average of at least three independent experiments.
Results
Identification of ExsE. Homologs of ExsC and ExsD appear in the recently released genome sequences of Aeromonas hydrophila and Photorhabdus luminescens (22–24). In each of these organisms, the ExsC and ExsD homologs are encoded near one another on the chromosome and are associated with a cluster of genes encoding a T3SS (Fig. 1B). The ExsC homologs possess 49% amino acid identity (80% similarity), whereas the ExsD homologs are less conserved at 27% identity (59% similarity). Examination of the genomic sequences in the vicinity of exsC revealed the presence of a small conserved ORF located immediately downstream of exsC in each of the organisms (Fig. 1B). This small ORF is annotated in P. aeruginosa (PA1711) and P. luminescens (3752) but not in A. hydrophila (23, 25). Although an amino acid alignment of the PA1711 homologs is not striking (21% identity, 43% similarity), the overall lengths of the predicted proteins (ranging from 77 to 81 amino acids) are similar. PA1711 encodes an 8.7-kDa basic protein and will hereafter be referred to as exsE.
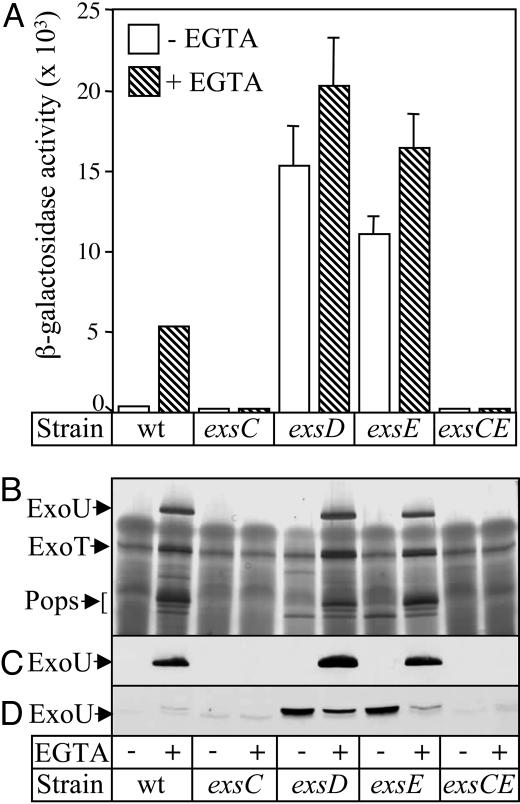
ExsE Is a Regulator of the T3SS. Based on homology searches, the presence of exsC, exsD, and exsE homologs within any given bacterial species appear to be 100% concordant, i.e., organisms either have all three or none of the homologs. Because of this relationship, we hypothesized that ExsE may be involved in the ExsC/ExsD regulatory cascade. To test this idea, an unmarked in-frame deletion (codons 4–78) of exsE was constructed and returned to the chromosome of P. aeruginosa strain PA103 by allelic exchange. We also integrated a transcriptional reporter (PexsD-lacZ) into the CTX phage attachment site of the ΔexsE mutant. This reporter consists of the promoter for exsD fused to lacZ and has been shown to be ExsA-dependent and inducible by low Ca2+ growth conditions in wild-type PA103 and serves as a reliable surrogate for monitoring transcription of the T3SS (Fig. 2A) (10, 17).
Fig. 2.
Characterization of type III secretion in the exsE deletion mutant. The indicated strains carrying the PexsD-lacZ reporter were grown under noninducing (–EGTA) or inducing (+EGTA) conditions. (A) β-Galactosidase activity (Miller units) from the PexsD-lacZ reporter. The reported values represent the average from three independent experiments. The standard deviation is indicated by the error bars. (B) Silver-stained gel of concentrated culture supernatants. The secreted effectors ExoU and ExoT are indicated, whereas the Pops represent the closely migrating proteins PopD, PopN, and PcrV. (C and D) Anti-ExoU immunoblot of concentrated culture supernatants (C) and whole-cell lysates (D).
Transcription of the PexsD-lacZ reporter was assessed by growing strains under noninducing (high Ca2+, –EGTA) and inducing (low Ca2+, +EGTA) conditions. Samples were harvested for β-galactosidase assay, and supernatant and cell-associated protein fractions were analyzed by SDS/PAGE and immunoblot. As shown previously, regulation of the PexsD-lacZ reporter is altered in mutants lacking either the ExsD anti-activator (resulting in derepression) or the ExsC anti-anti-activator (resulting in constitutive repression) (Fig. 2 A) (10, 17). The phenotype of the ΔexsE mutant resembles that of an ΔexsD mutant, i.e., transcription of the PexsD-lacZ reporter is derepressed irrespective of growth conditions, although a residual (<2-fold) response to low Ca2+ is still seen in both mutants (Fig. 2 A). As reported, secretion of ExoU, ExoT, and other type III-related exoproducts from wild-type PA103 and the ΔexsD mutant depends upon low Ca2+ growth conditions (10). Similar results are seen with the ΔexsE mutant, indicating that ExsE is not required for secretion (Fig. 2 B and C). We also performed anti-ExoU immunoblots of the cell lysate fractions. In both the ΔexsE and ΔexsD mutants, ExoU was detected in the lysate fraction when cells were grown under high Ca2+ conditions (Fig. 2D). As reported for the ΔexsD mutant (10), the cytoplasmic accumulation of ExoU in the ΔexsE mutant likely results from the combined effect of derepressed expression of the T3SS and the inhibition of secretion by high Ca2+.
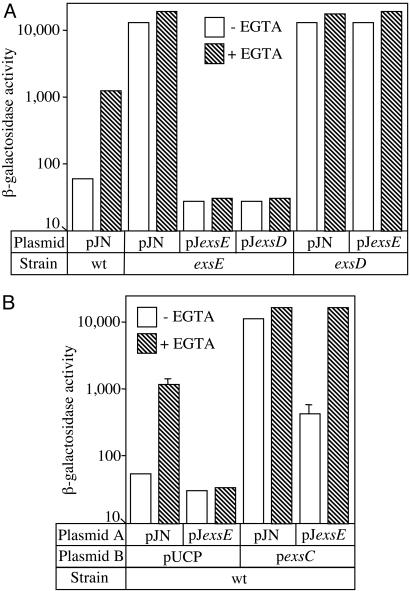
To confirm that the derepressed phenotype of the ΔexsE mutant results from loss of ExsE expression, the mutant was transformed with an arabinose-inducible ExsE expression vector (pJexsE) or a vector control (pJN). Whereas transcription of the PexsD-lacZ reporter remained derepressed with the vector control, pJexsE strongly repressed expression of the lacZ reporter under both high and low Ca2+ culture conditions (Fig. 3A). Similar results were seen in wild-type PA103 whereby pJexsE also repressed expression of the PexsD-lacZ reporter (Fig. 3B). A previous report found that ExsD overexpression also results in strong repression of the T3SS irrespective of Ca2+ levels (10). Based on the observations that transcription of the PexsD-lacZ reporter and ExoU expression are derepressed in the ΔexsE mutant and repressed by plasmid-encoded ExsE, we conclude that ExsE functions as a negative regulator of the entire T3SS.
Fig. 3.
The negative regulatory activity of ExsE depends on a functional ExsC/ExsD regulatory system. Strains carrying the PexsD-lacZ reporter were transformed with either a vector control (pJN) or ExsE (pJexsE), ExsD (pJexsD), or ExsC (pexsC) expression vectors. The transformants were grown in the presence of the appropriate antibiotics under either noninducing (–EGTA) or inducing (+EGTA) conditions in the presence of 0.5% arabinose, and β-galactosidase activity (Miller units) was determined. The reported values represent the average from three independent experiments. When plotted on a log scale, some of the errors bars are not visible.
The Negative Regulatory Activity of ExsE Is ExsD-Dependent and Suppressed by ExsC. To characterize the mechanism of negative regulation by ExsE, a complementation approach was taken. Because ExsE and ExsD are both negative regulators of the T3SS, we asked whether their activities depend upon one another. The ΔexsE and ΔexsD mutants were transformed with either pJexsE or an ExsD expression vector (pJexsD) and assayed for expression of the PexsD-lacZ reporter. Plasmid expressed ExsD strongly repressed transcription of the PexsD-lacZ reporter in an ΔexsE mutant. In the converse experiment, however, plasmid expressed ExsE was unable to repress transcription in an ΔexsD mutant (Fig. 3A). From these data, we conclude that the regulatory activity of ExsE requires ExsD, whereas the activity of ExsD functions independently of ExsE.
ExsC positively regulates transcription of the T3SS by binding to and inhibiting ExsD activity (17). To determine whether ExsC regulates ExsE activity, an ΔexsCE double mutant was constructed. Although transcription of the PexsD-lacZ reporter was derepressed in the ΔexsE mutant, transcription was repressed in the ΔexsCE double mutant, indicating that the repressed phenotype of the ΔexsC mutant is dominant to that of the ΔexsE mutant (Fig. 2 A). Importantly, the repressed phenotype of the ΔexsCE double mutant is reversed by plasmid-encoded ExsC indicating that the repressed phenotype results from the lack of ExsC (data not shown). The dominance of the ΔexsC mutant phenotype is consistent with a model in which ExsE may function as a regulator of ExsC. This interpretation is further supported by the finding that pJexsE-mediated repression of the PexsD-lacZ reporter is suppressed by coexpression of plasmid-encoded ExsC (Fig. 3B).
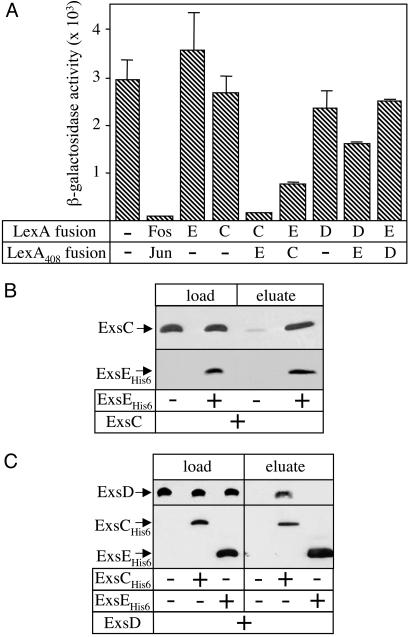
ExsE Exerts Its Regulatory Activity by Directly Binding ExsC. The data presented above indicate that ExsE-mediated repression depends on a functional ExsC-ExsD regulatory system. We hypothesized that the negative regulatory activity of ExsE might result from the disruption of the ExsC–ExsD complex through a binding interaction with either ExsC or ExsD. To test for these potential interactions, we used the LexA bacterial two-hybrid system (21). This system makes use of the dimeric LexA transcriptional repressor, a LexA mutant (LexA408) having altered DNA binding specificity, and an E. coli lacZ reporter strain. Transcription of the LexA-repressible lacZ reporter is regulated by an operator sequence with half-sites specific for LexA and LexA408, and therefore only LexA-LexA408 heterodimers are able to bind the operator and repress lacZ transcription. Upon coexpression of LexA and LexA408 lacking dimerization domains, there is no repression of lacZ expression, whereas coexpression of the control fusion proteins LexA-Fos and LexA408-Jun results in strong repression of lacZ expression (Fos and Jun are eukaryotic transcription factors known to dimerize) (Fig. 4A).
Fig. 4.
ExsE interacts with ExsC. (A) Two-hybrid interaction assay. The E. coli reporter strain SU202 carries plasmids expressing only the LexA or LexA408 DNA-binding domain (–) or fusions of the LexA or LexA408 DNA-binding domains fused to ExsE (E), ExsC (C), ExsD (D), or the control proteins Fos and Jun. Cells were grown in LB medium and assayed for β-galactosidase activity (Miller units). (B) ExsC copurification assay. A cell extract of E. coli overexpressing ExsC was mixed with either purified ExsEHis-6 (+) or with buffer (–); the mixtures were then subjected to Ni-NTA affinity chromatography. Aliquots of the mixture (load) and the column eluate were subjected to anti-ExsC and anti-His-6 immunoblot analyses. (C) ExsD copurification assay. A cell extract of E. coli overproducing ExsD was mixed with either purified ExsCHis-6 or purified ExsEHis-6 as indicated (+) or mixed with buffer (–). The mixtures were then subjected to Ni-NTA affinity chromatography and subjected to anti-ExsD and anti-His-6 immunoblot analyses.
To test for specific interactions with ExsE, the DNA-binding domains of LexA and LexA408 were genetically fused to ExsE, ExsC, or ExsD. Plasmids encoding the chimeric LexA fusions were introduced into the E. coli lacZ reporter strain and assayed for β-galactosidase activity. Expression of the individual LexA/LexA408 fusions to ExsE, ExsC, or ExsD did not significantly repress expression of the lacZ reporter when compared with the negative control (Fig. 4A). The combination of LexA-ExsC with LexA408-ExsE resulted in strong repression of lacZ expression, whereas the opposite combination (LexA-ExsE and LexA408-ExsC) resulted in more modest repression. The direct interaction of ExsE with ExsC is consistent with a model in which ExsE regulates the activity of ExsC. Conversely, neither of the LexA-ExsD fusion combinations showed significant repression, suggesting that ExsE and ExsD do not interact.
To corroborate the two-hybrid data, we performed copurification studies using purified hexahistidine-tagged ExsE (ExsEHis-6). An extract was prepared from E. coli engineered to overexpress native ExsC and was incubated in the absence or presence of purified ExsEHis-6. The mixtures were applied to a Ni-NTA affinity column to capture ExsEHis-6, the columns were washed of unbound material, and bound material was then eluted with imidazole. Analysis of the bound material by anti-His and anti-ExsC immunoblot analyses showed that native ExsC bound to the column only in the presence of ExsEHis-6, confirming that ExsE interacts with ExsC (Fig. 4B).
The copurification experiment was repeated by using an E. coli extract overexpressing native ExsD. The extract was mixed with ExsEHis-6 or ExsCHis-6 and processed as described above. As reported (17), ExsD copurifies with ExsCHis-6 in this assay, indicating that the ExsD protein in the extract is in a native conformation (Fig. 4C). Nevertheless, ExsD did not copurify with ExsEHis-6. These data are consistent with the two-hybrid data and indicate that ExsE does not interact with ExsD.
ExsE Requires ExsC for Stability and Secretion. ExsC is a member of the CesT-like family of chaperones and has biochemical characteristics of a type III chaperone including a C-terminal amphipathic α-helix, low molecular weight, and acidic pI (17, 18). CesT-like chaperones are found in most bacterial species possessing a T3SS and share a common role in facilitating substrate secretion via the T3SS (18). Based on the conserved role of these chaperones in secretion and on the observed interaction of ExsE with ExsC, we hypothesized that ExsE is secreted by the T3SS in an ExsC-dependent manner.
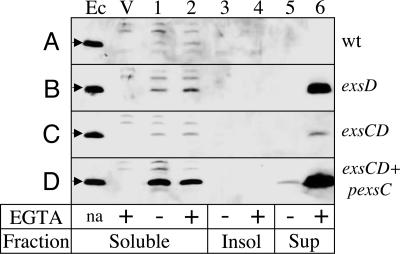
To test this hypothesis, a C-terminal hexahistidine-tagged derivative (ExsEHis-6) was expressed from an arabinose-inducible expression plasmid. When expressed in wild-type P. aeruginosa, ExsEHis-6 inhibits expression of the T3SS to a level similar to that seen with untagged ExsE (data not shown). Under these conditions, ExsEHis-6 was not detected in the culture supernatants (Fig. 5A, lanes 5 and 6), but a small amount could be seen in soluble cellular fractions (lanes 1 and 2). This result would be expected if the T3SS is required for ExsE secretion, because expression of the T3SS in this transformant is severely repressed (Fig. 3B). ExsEHis-6 was then expressed in an exsD mutant where expression of the T3SS is constitutive. In this background, a large amount of ExsEHis-6 was secreted into the culture supernatant when cells were grown in the presence but not in the absence of EGTA (Fig. 5B, lanes 5 and 6). In addition, there was a slight increase in the amount of ExsEHis-6 found in the soluble cell fractions of the exsD mutant when compared with the wild-type strain (lanes 1 and 2). The combined findings that secretion of ExsEHis-6 requires expression of the T3SS and is EGTA-dependent indicate that ExsE is indeed secreted by the T3SS.
Fig. 5.
ExsE is secreted in an ExsC-dependent response to low Ca2+. Wild-type PA103 and the indicated mutants carrying the pexsEHis-6 expression plasmid were grown under either noninducing (–EGTA) or inducing (+EGTA) conditions for type III secretion in the presence of 0.5% arabinose. Culture supernatants (sup) (derived from 4.5 × 108 cells), and cell-associated soluble and insoluble (insol) fractions (derived from 1.15 × 108 cells) were immunoblotted with anti-His-6. Lanes A1–6, PA103; lanes B1–6, ΔexsD; lanes C1–6, ΔexsCD; and lanes D1–6, ΔexsCD additionally transformed with an ExsC expression plasmid (pexsC). Control lanes: Ec, soluble cell-associated fraction from E. coli overexpressing ExsEHis-6 (arrows); V, soluble cell-associated fraction from an ΔexsD mutant carrying the plasmid control pJN105.
We next examined whether the putative chaperone activity of ExsC is required for ExsE secretion. For these studies, ExsEHis-6 was expressed in an exsC, exsD double mutant where expression of the T3SS is constitutive but ExsC is lacking. In the absence of ExsC, the amount of ExsEHis-6 in the concentrated culture supernatant was reduced 16-fold when compared with the exsD single mutant (Fig. 5 B vs. C, lane 6). The amount of ExsEHis-6 in the soluble cell fraction was also slightly diminished in the absence of ExsC (Fig. 5 B vs. C, lanes 1 and 2). These data demonstrate that ExsC is required for efficient secretion of ExsE and suggest that ExsC may increase ExsEHis-6 stability before secretion. This idea is supported by the finding that ExsC overexpressed from a plasmid results in a substantial increase in the amount of ExsEHis-6 in both the supernatant and soluble cell fractions (Fig. 5D, lanes 1, 2, and 6).
Discussion
The identification of ExsE as a secreted regulator of the P. aeruginosa T3SS provides a vital link toward understanding the mechanism of coupling the activity of the type III secretory channel with transcription of the T3SS. Rather than exerting its negative regulatory activity directly at the level of transcription, ExsE functions indirectly through a signaling cascade that ultimately controls the ExsA transcriptional activator. We show here that ExsE is positioned at the top of this signaling cascade and functions as an inhibitor of the ExsC anti-anti-activator through a direct protein–protein interaction. ExsE is also secreted by the T3SS, and it is this feature that provides the mechanism of coupling transcription with secretion. The experimental findings for ExsE are in complete agreement with the predicted role for protein X in our model of regulation (Fig. 1 A). A second prediction from our model is that ExsC functions as type III secretion chaperone for ExsE (protein X). The previously reported chaperone-like properties of ExsC (17), combined with the experimental findings that ExsC interacts with ExsE, is required for efficient secretion of ExsE, and increases the stability of cytoplasmic ExsE, confirm that ExsC does indeed fit the classical definition of a type III secretion chaperone (26). It remains possible, however, that other mechanisms (perhaps regulatory in nature) influence the stability of ExsE.
ExsE is encoded by the exsCEBA operon and is under the transcriptional control of ExsA, whereas ExsD is expressed from a separate ExsA-dependent transcript (Fig. 1B). A previous study found that the steady-state levels of ExsC, ExsA, and ExsD increase proportionately under low Ca2+ growth conditions when compared with high Ca2+ conditions (17). This finding suggested that a change in the relative ratio of ExsC-ExsA-ExsD was an unlikely explanation for transcriptional induction by low Ca2+. We expect the steady-state levels of ExsE to follow this same pattern; however, confirmation will require an anti-ExsE antibody. We propose that the normal equilibrium between the ExsE·ExsC, ExsC·ExsD, and ExsD·ExsA complexes and unbound ExsA favors the ExsE·ExsC and ExsD·ExsA bound states, resulting in repression of transcription (Fig. 1 A, high Ca2+). Assuming the relative ratio of ExsE to ExsC-ExsA-ExsD remains constant under both high and low Ca2+ conditions, the simple act of secreting ExsE under low Ca2+ conditions would shift the binding equilibria of the cascade in favor of unbound ExsA, resulting in transcriptional activation (Fig. 1B, low Ca2+). The identification of ExsE homologs suggests that a similar means of regulation may also be involved in the regulation of other T3SSs.
Compared with the flagellar system, where secretion of an anti-σ factor couples late gene transcription to secretion, the mechanism described herein appears more complex. An obvious question is: why use the more complicated ExsE-ExsC system when secretion of ExsD would appear to serve the same purpose? One possibility is that the increased number of regulatory proteins represented by the ExsECDA pathway expands the number of potential targets for regulation by additional signaling networks. If this is true, then additional regulatory pathways influencing expression of the T3SS may be discovered that function at the level of the ExsECDA signaling cascade. A second possibility is that the T3SS is likely expressed at a low constitutive level that strikes a balance between the requirements to sense and rapidly respond to inducing stimuli and to conserve energy. The ExsE-ExsC regulatory system may provide more flexible control over the coupling of transcription to secretion and allow for both rapid induction and repression as dictated by environmental conditions.
Although our studies have focused on the coupling of transcription to secretion in response to low Ca2+, we speculate that attachment of P. aeruginosa to host cells may trigger translocation of ExsE into the host cytoplasm. Whether by translocation into a host cell or secretion into the medium, reduced intracellular concentrations of ExsE would lead to derepression of the T3SS. Precedence for such a model comes from studies of the Yersinia pseudotuberculosis and Yersinia enterocolitca T3SSs. Similar to the P. aeruginosa T3SS, transcription of the Yersinia sp. T3SSs is induced by low Ca2+ growth conditions or after contact with host cells and is intimately linked to secretion (27). Despite these similarities, the mechanism of coupling transcription to secretion is not conserved between these two organisms. Y. pseudotuberculosis and Y. enterocilitca lack homologs of ExsC, ExsD, and ExsE and instead use a secreted negative regulator LcrQ (also called YscM) of the T3SS. LcrQ is secreted under low Ca2+ conditions or injected into host cells after contact, thus relieving the block on transcription through an unknown mechanism (28, 29). Whether ExsE is translocated into host cells and, if so, exerts cytotoxic activity toward the host remain to be determined.
Acknowledgments
We thank Mark Pallen for discussions concerning the identity of protein X and Thomas Bair for efforts to identify protein X through a bioinformatics approach. Support for these studies was provided by the Howard Hughes Medical Institute Biomedical Research Support Faculty Start-up Program, the University of Iowa W. M. Keck Microbial Communities and Cell Signaling Program, and the National Institutes of Health (RO1-AI055042).
Author contributions: M.L.U. and T.L.Y. designed research; M.L.U., G.L.L., and T.L.Y. performed research; M.L.U., G.L.L., and T.L.Y. analyzed data; and M.L.U. and T.L.Y. wrote the paper.
Abbreviation: T3SS, type III secretion system.
Note Added in Proof. After submission of this work, Rietsch et al. (30) published a manuscript that also reports the identification of ExsE as a secreted regulator of the T3SS in P. aeruginosa strain PAK.
References
- 1.Richards, M. J., Edwards, J. R., Culver, D. H. & Gaynes, R. P. (1999) Crit. Care Med. 27, 887–892. [DOI] [PubMed] [Google Scholar]
- 2.Torres, A., Aznar, R., Gatell, J. M., Jimenez, P., Gonzalez, J., Ferrer, A., Celis, R. & Rodriguez-Roisin, R. (1990) Am. Rev. Respir. Dis. 142, 523–528. [DOI] [PubMed] [Google Scholar]
- 3.Barbieri, J. T. & Sun, J. (2004) Rev Physiol. Biochem. Pharmacol. 152, 79–92. [DOI] [PubMed] [Google Scholar]
- 4.Finck-Barbancon, V., Goranson, J., Zhu, L., Sawa, T., Wiener-Kronish, J. P., Fleiszig, S. M. J., Wu, C., Mende-Mueller, L. & Frank, D. W. (1997) Mol. Microbiol. 25, 547–557. [DOI] [PubMed] [Google Scholar]
- 5.Holder, I. A., Neely, A. N. & Frank, D. W. (2001) Burns 27, 129–130. [DOI] [PubMed] [Google Scholar]
- 6.Roy-Burman, A., Savel, R. H., Racine, S., Swanson, B. L., Revadigar, N. S., Fujimoto, J., Sawa, T., Frank, D. W. & Wiener-Kronish, J. P. (2001) J. Infect. Dis. 183, 1767–1774. [DOI] [PubMed] [Google Scholar]
- 7.Sawa, T., Yahr, T. L., Ohara, M., Kurahashi, K., Gropper, M. A., Wiener-Kronish, J. P. & Frank, D. W. (1999) Nat. Med. 5, 392–398. [DOI] [PubMed] [Google Scholar]
- 8.Frank, D. W. (1997) Mol. Microbiol. 26, 621–629. [DOI] [PubMed] [Google Scholar]
- 9.Vallis, A. J., Yahr, T. L., Barbieri, J. T. & Frank, D. W. (1999) Infect. Immun. 67, 914–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCaw, M. L., Lykken, G. L., Singh, P. K. & Yahr, T. L. (2002) Mol. Microbiol. 46, 1123–1133. [DOI] [PubMed] [Google Scholar]
- 11.Yahr, T. L., Goranson, J. & Frank, D. W. (1996) Mol. Microbiol. 22, 991–1003. [DOI] [PubMed] [Google Scholar]
- 12.Miller, V. L. (2002) Curr. Opin. Microbiol. 5, 211–215. [DOI] [PubMed] [Google Scholar]
- 13.Aldridge, P. & Hughes, K. T. (2002) Curr. Opin. Microbiol. 5, 160–165. [DOI] [PubMed] [Google Scholar]
- 14.Yahr, T. L. & Frank, D. W. (1994) J. Bacteriol. 176, 3832–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hovey, A. K. & Frank, D. W. (1995) J. Bacteriol. 177, 4427–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yahr, T. L., Hovey, A. K., Kulich, S. M. & Frank, D. W. (1995) J. Bacteriol. 177, 1169–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dasgupta, N., Lykken, G. L., Wolfgang, M. C. & Yahr, T. L. (2004) Mol. Microbiol. 53, 297–308. [DOI] [PubMed] [Google Scholar]
- 18.Pallen, M. J., Beatson, S. A., Bailey, C. M. (2005) FEMS Microbiol. Rev. 29, 201–229. [DOI] [PubMed] [Google Scholar]
- 19.Hoang, T. T., Karkhoff-Schweizer, R. R., Kutchma, A. J. & Schweizer, H. P. (1998) Gene 212, 77–86. [DOI] [PubMed] [Google Scholar]
- 20.Newman, J. R. & Fuqua, C. (1999) Gene 227, 197–203. [DOI] [PubMed] [Google Scholar]
- 21.Daines, D. A. & Silver, R. P. (2000) Methods Enzymol. 182, 5267–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vilches, S., Urgell, C., Merino, S., Chacon, M. R., Soler L., Castro-Escarpulli, G., Figueras, M. J. & Tomas, J. M. (2004) Appl. Environ. Microbiol. 70, 6914–6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duchaud, E., Rusniok, C., Frangeul, L., Buchrieser, C., Givaudan, A., Taourit, S., Bocs, S., Boursaux-Eude, C., Chandler, M. & Charles, J. F., et al. (2003) Nat. Biotechnol. 11, 1307–1313. [DOI] [PubMed] [Google Scholar]
- 24.Waterfield, N. R., Daborn, P. J. & ffrench-Constant, R. H. (2002) Trends Microbiol. 10, 541–545. [DOI] [PubMed] [Google Scholar]
- 25.Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., Brinkman, F. S., Hufnagle, W. O., Kowalik, D. J., Lagrou, M., et al. (2000) Nature 406, 959–964. [DOI] [PubMed] [Google Scholar]
- 26.Feldman, M. F. & Cornelis, G. R. (2003) FEMS Microbiol. Lett. 219, 151–158. [DOI] [PubMed] [Google Scholar]
- 27.Francis, M. S., Wolf-Watz, H. & Forsberg, A. (2002) Curr. Opin. Microbiol. 5, 166–172. [DOI] [PubMed] [Google Scholar]
- 28.Cambronne, E. D., Cheng, L. W. & Schneewind, O. (2000) Mol. Microbiol. 37, 263–273. [DOI] [PubMed] [Google Scholar]
- 29.Pettersson, J., Nordfelth, R., Dubinina, E., Bergman, T., Gustafsson, M., Magnusson, K. E. & Wolf-Watz, H. (1996) Science 273, 1231–1233. [DOI] [PubMed] [Google Scholar]
- 30.Rietsch, A., Vallet-Gely, I., Dove, S. L. & Mekalanos, J. J. (2005) Proc. Natl. Acad. Sci. USA 102, 8006–8011. [DOI] [PMC free article] [PubMed] [Google Scholar]