Abstract
Tumor necrosis factor (TNF) is a pro-inflammatory cytokine that plays an important role in a variety of infectious and autoimmune disorders. Its transcription is regulated in a stimulus- and cell-type-specific manner via the recruitment of distinct DNA/activator complexes forming secondary structures or enhanceosomes. NFATp, a member of the nuclear factor of activated T cells (NFAT) family of transcription factors, plays a critical role in TNF gene regulation under a variety of conditions. In this study, we show that NFAT5, the most recently described NFAT family member, binds to the TNF promoter in a manner distinct from other NFAT proteins and is a key mediator in the activation of TNF gene transcription during hypertonic stress alone.
INTRODUCTION
Regulation of the mammalian immune system involves complex mechanisms of transcriptional control. One model gene system in which cell-type- and inducer-specific transcriptional programs have been studied is the human tumor necrosis factor (TNF) gene. In vitro and in vivo studies have demonstrated that the mechanisms of transcriptional activation and enhanceosome formation involved in TNF gene expression are inducer- and cell-type-specific in T cells stimulated through the T-cell receptor (TCR) or infected with virus (1–4), in monocytic cells stimulated with lipopolysaccharide or Mycobacterium tuberculosis (5,6), and in fibroblasts stimulated with recombinant TNF (7). Members of the nuclear factor of activated T cells (NFAT), Sp and Ets families of transcription factors are recruited to promoter binding sites that are capable of binding multiple factors. The stimulus, cell type and the subsequent nuclear concentrations of transcriptional activators (1–4) induce a unique pattern of activator recruitment to the promoter.
TNF, a potent pro-inflammatory cytokine, plays a critical role in the immune response that leads to effective clearance of a variety of pathogens or to immunopathology and autoimmune disease (8). The transcription factor NFATp (also called NFAT1 or NFATc2) is a key factor involved in TNF gene regulation in T and B cells activated via their antigen receptors, calcium flux or virus (1,3,5,9,10). NFATp (11) is a member of a family of proteins that has five members including NFATc (also called NFAT2 or NFATc1) (12), NFAT3 (also called NFATc4), NFAT4 (also called NFATx or NFATc3) (13) and NFAT5 (14) [also called TonEBP (15), NFATL1 (16), OREBP (17) or NFATz (18)].
NFAT5 is distinct from the other four NFAT family members in that (i) it is not regulated by intracellular calcium levels, (ii) its activity cannot be blocked by calcineurin inhibitors, such as Cyclosporine A (CsA) and (iii) it has been implicated in the production of genes required for the cellular response to hypertonic stress (14–18). Furthermore, its crystal structure has revealed that, in contrast to NFATp, NFAT5 binds DNA as an obligate homodimer, similar to NF-κB (19).
Although there is considerable variation in the literature about the effects of hypertonic stress on cytokine production (20–22), hypertonic stress alone has been shown to activate cytokine production, including TNF in macrophage and B lymphocyte cell lines (23). In Jurkat T cells stimulated with phorbol ester (PMA) and hypertonic saline, TNF mRNA levels were increased. Using chromatin immunoprecipitation (ChIP) binding assays, NFAT5 was shown to be recruited to the TNF promoter, suggesting that NFAT5 plays a role in TNF gene regulation in PMA-activated T cells under hypertonic conditions (24). However, PMA alone activates TNF gene transcription in Jurkat cells (25). Thus, the role of NFAT5 in TNF induction by hypertonicity alone remains unclear. Here, we show that NFAT5 binds to the TNF promoter in a pattern distinct from that of other NFAT family members and is a key member of the enhancer complex that is recruited to the TNF promoter under hypertonic conditions.
MATERIALS AND METHODS
Plasmids
The −200 TNF-Luc reporter and isogenic TNF-Luc mutant constructs were made as described previously (3). The small hairpin (sh)RNA-expressing constructs were made by cloning the first 315 bp of the murine U6 promoter up to the base before the G marking the transcriptional start site. The promoter was amplified from mouse genomic DNA by PCR using the following primers (with restriction sites on the ends). Sense: –gcagaattcGATCCGACGCCGCCATCTCT–; antisense: –acaggtaccAAACAAGGCTTTTCTCCAAGGGATA–. The fragment was cloned between EcoRI and BamHI in pBluescript (Stratagene). This construct was used as a template for PCR of a U6-shRNA construct whose hairpin targeted exon 8 of the NFAT5 gene. A U6-shRNA construct with a scrambled hairpin was also designed. These PCRs used a common primer for the 5′ end. Sense: –gcagaattcGATCCGACGCCGCCATCTCT–. The antisense primers for the 3′ ends were as follows: gcagctagcCTCGAGAAAAAAGCAATGTCAGAGTAGAGCCCTACACAAAGGCTCTACTCT GACATTGCAAACAAGGCTTTTCTCCAAGGGATA– (shRNA targeting NFAT5 exon 8); gcagctagcCTCGAGAAAAAAGAACGTTCGATAATGGATCCTACACAAAGATCCATTATC GAACGTTCAAACAAGGCTTTTCTCCAAGGGATA– (scrambled non-silencing shRNA).
The NFAT5-dominant negative (N5-DN) expression plasmid was generated by cloning the NFAT5 DNA binding domain (DBD) into pcDNA3 vector (Invitrogen). The DBD fragment was amplified from the pEGFP-N1 vector (Clontech) containing full-length NFAT5 (24) by PCR using primers with restriction enzymes EcoRI and XbaI on the ends and ligated into pcDNA3 vector.
Cell culture and transfection
The murine fibroblast cell line L929 was grown at 37°C, 5% CO2, in DMEM supplemented with 10% fetal bovine serum (FBS) and 2 mM glutamine. The cells were lightly trypsinized with 0.05% trypsin in HBSS and EDTA and subcultured as necessary. The T-cell hybridoma 68–41 cell line was grown at 37°C, 5% CO2, in RPMI-1640 supplemented with 10% FBS and 2 mM glutamine. Transfections with the U6-shRNA constructs were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. All other transfections were performed using DEAE-dextran as previously described (5). Stimulation was performed with raffinose (Sigma) as described in the Results section. Small molecule inhibitor SB203580 was diluted in dimethyl sulfoxide. It was purchased from Calbiochem. Luciferase assays were carried out according to the manufacturer's instructions (Dual Luciferase Reporter Assay System, Promega) with a Dynex luminometer, using Renilla luciferase (pRL-TK) as a control for transfection efficiency.
RNase protection assay
L929 cells were left unstimulated or stimulated with 200 mM raffinose for 18 h. Total RNA was purified using TRIzol Reagent (GibcoBRL). RNA was hybridized overnight to [α-32P]UTP-labeled antisense RNA probes for TNF and L-32, which had been synthesized in vitro from separate templates according to standard techniques (9,26,27). After hybridization, the samples were digested with RNase A to remove single-stranded ends of hybridized RNA. Proteinase K was added to stop the digestion reaction, and double-stranded RNA was purified by phenol/chloroform extraction followed by ethanol precipitation. Protected RNA pieces were resolved on an 8% denaturing polyacrylamide gel and exposed overnight on film.
Western blot analysis
Whole cell extracts were collected with lysis buffer containing 150 mM NaCl, 50 mM Tris–HCl, pH 7.5, 1% Triton, 10% glycerol and 1 tablet of Complete EDTA-free Protease Inhibitor Cocktail (Roche) per 25 ml of buffer. Extracts were stored at −70°C until ready for use. Extracts were boiled for 5 min in 1× Laemmli sample buffer with 5% v/v 2-mercaptoethanol. Extracts were run at 100 V on 5% Tris–HCl polyacrylamide 10-well pre-cast gels with a 4% stacking gel (BioRad) with a Kaleidoscope molecular weight marker. The gel was transferred to nitrocellulose Trans-Blot Transfer Membrane (BioRad) at 40 V for 1.5 h in a tank transfer apparatus. The blot was then blocked for 1 h at 37°C in a solution of 4% BSA (Sigma) and 0.1% Tween-20 (BioRad) in Tris-buffered saline with 50 mM Tris and 150 mM NaCl at pH 7.6 (BSA/TBST). Primary incubation was carried out with 1:200 rabbit-anti-NFAT5 antibody (Santa Cruz, sc-13035) and 1:500 goat-anti-Lamin B1 antibody (Santa Cruz, sc-6217) in BSA/TBST for 2 h at room temperature. The blot was washed 3 × 5 min in TBST and incubated in 1:6000 donkey-anti-goat-HRP (Santa Cruz) or goat-anti-rabbit-HRP (BioRad) as appropriate for 1 h. The blot was again washed 3 × 5 min in TBST and developed with SuperSignal West Pico Chemiluminescent Reagent (Pierce). Anti-NFATp antibody was generated in the laboratory of Anjana Rao as described previously (11).
DNase I footprinting and electrophoretic mobility shift assay (EMSA)
DNase I footprinting of the human TNF promoter was carried out with recombinant DBD of NFAT5 (24), as described previously (3). Briefly, the −200 to +87 fragment of the wild-type TNF promoter was end-labeled with [γ-32P]ATP on one end using T4 polynucleotide kinase and incubated with increasing amounts of recombinant proteins as labeled in Figure 2. After 30 min of incubation, the samples were digested with 0.3 U of DNase I for exactly 1 min, resolved on an 8% denaturing polyacrylamide gel and exposed overnight on film. EMSA was performed, as described previously (28), using oligonucleotides end-labeled with [γ-32P]ATP and DBD5 and DBD5-GST recombinant proteins or nuclear extracts from L929 cells left untreated or treated with 200 mM raffinose for 18 h. In supershift assay, the appropriate antibodies (Santa Cruz) were pre-incubated with nuclear extracts for 30 min before adding the labeled probe. The sequence of the oligos used is shown in Figure 3. Recombinant proteins were a kind gift from Tim Hoey, Dimitris Thanos and Anjana Rao. Escherichia coli expression vectors for each protein were constructed and proteins were expressed using T7 polymerase expression system in the strain BL21 (Invitrogen) and purified by DNA affinity chromatography as described in details (13). The sizes of the different proteins are as follows: NFATp, 353 amino acids [the residues homologous to 185–537 according to (11)]; NFATc, 309 amino acids [residues 408–716 according to (12)]; NFAT3, 345 amino acids (residues 400–744) and NFAT4, 316 amino acids (residues 393–708) according to (13).
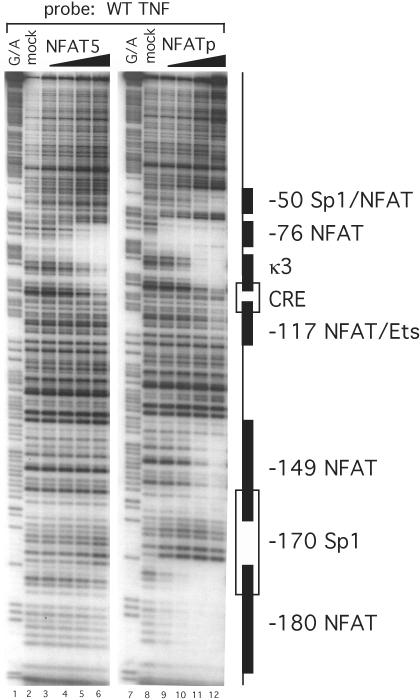
Figure 2.
NFAT5 binds to the proximal TNF promoter in a pattern distinct from NFATp. DNase I footprinting assay of the TNF promoter with NFAT5. The DBA of recombinant human NFAT5 (DBD5) binds to two sites on the proximal TNF promoter between −120 and +87 nt relative to the transcription start site. Of the two sites, the −76 NFAT binding site binds at lower protein concentrations than the κ3 site. Increasing concentrations of DBD5 (25 ng, 125 ng, 500 ng and 2.5 μg) are represented by the wedge over lanes 3–6. Lighter bands indicate regions of reduced cleavage by DNase I due to protein binding. Several known transcription factor binding sites and their locations are indicated to the right of the gel. Recombinant NFATp binds six sites in the proximal promoter region (see lanes 9–12).
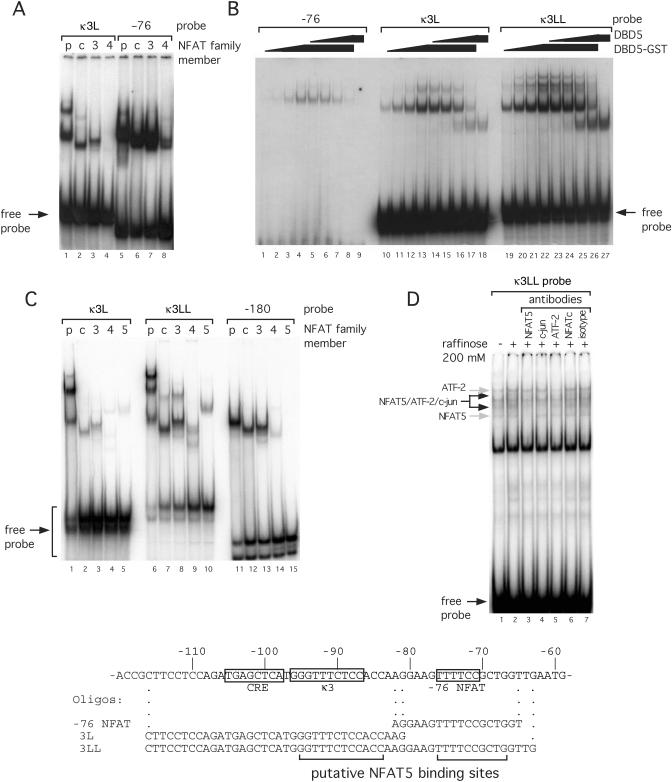
Figure 3.
NFAT family members bind portions of the TNF promoter with different affinities. (A) EMSA using recombinant NFATp, NFATc, NFAT3 and NFAT4 with portions of the TNF promoter. Three probes (−76, κ3L and κ3LL) were end-labeled with 32P and incubated with recombinant protein, as indicated. Free probe marked at the bottom of the gel indicates equal loading. (B) EMSA using recombinant DBD5 or DBD5-GST and probes with portions of the TNF promoter. A diagram of the proximal human TNF promoter with probes for EMSA is just below. Known binding elements are boxed. Putative NFAT5 binding sites, based on the known consensus sequence, are bracketed. (C) EMSA using recombinant NFATp, NFATc, NFAT3, NFAT4 and NFAT5 with portions of the TNF promoter. Three probes (κ3L, κ3LL and −180) were end-labeled with 32P and incubated with recombinant protein, as indicated. Free probe marked at the bottom of the gel indicates equal loading. (D) EMSA using raffinose-stimulated L929 nuclear extracts and the κ3LL probe. Nuclear extracts were prepared from L929 cells unstimulated (−) or stimulated (+) with raffinose (200 mM) for 18 h. An oligonucleotide probe containing the −76 NFAT, κ3-NFAT and the CRE binding sites (κ3LL) was used and the binding assay was performed in the presence or absence of antibodies to NFAT5, NFATc, ATF-2, c-jun or isotype control (normal rabbit serum) as indicated in the figure. The complexes containing NFAT5 or ATF-2 alone are indicated by the gray arrows, and the composite NFAT5/ATF-2/c-jun complexes are indicated by the black arrows in the figure.
RESULTS
Induction of TNF mRNA and NFAT5 protein in L929 cells under hypertonic stress
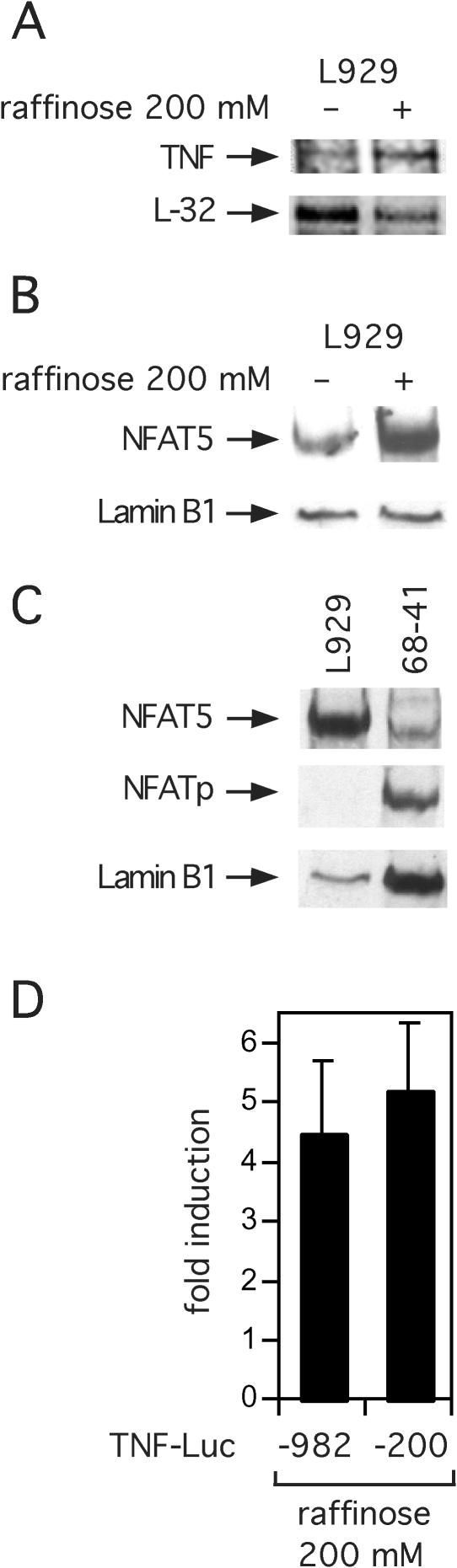
We sought first to establish that hypertonic stress alone is sufficient to activate TNF gene transcription and to validate L929 murine fibroblast cells as a model system to probe the role of NFAT5 in TNF transcription. We exposed L929 cells to 200 mM raffinose, a large membrane-impermeable sugar causing intracellular hypertonic stress (29) for 18 h, harvested RNA and performed an RNase protection assay. As shown in Figure 1A, endogenous TNF mRNA levels as measured by RNase protection assay were significantly increased by raffinose treatment of L929 cells at 18 h, and we detected this increase at 3 h post stimulation (data not shown). We also examined NFAT5 protein levels in stimulated cells by western blot analysis. Hypertonic stress induced a several-fold increase in NFAT5 protein levels compared with untreated cells (Figure 1B). Lamin B1, a nuclear structural protein, was used as a loading control. As shown in Figure 1C, L929 cells do not express NFATp, whereas the T cell line 68–41 that we used as a positive control does express both NFATp and NFAT5 proteins constitutively. The absence of NFATp thus simplifies the analysis of the role of NFAT5 in hypertonic induction of TNF gene expression since high levels of NFATp could potentially interfere with NFAT5 binding to shared sites within the promoter.
Figure 1.
Hypertonicity induces TNF transcription and NFAT5 expression. (A) Induction of TNF mRNA by hypertonic stress. L929 cells were cultured in medium supplemented with 200 mM raffinose for 18 h. RNA was harvested and submitted to RNase protection assay. (B) Hypertonic stress up-regulates NFAT5 expression. L929 cells were cultured to ∼95% confluence in a 6-well plate and stimulated with complete medium or medium supplemented with 200 mM raffinose for 18 h, as indicated. Whole cell extracts were collected and submitted to western blot analysis. Results are representative of four independent experiments. (C) L929 cells do not express NFATp. Whole cell extract (30 μg) from 68 to 41 cells were submitted to western blot analysis as a positive control for NFATp expression. L929 cells were stimulated with hypertonic stress as above. Results are representative of two independent experiments. Lamin B1 was used as a loading control. (D) Induction of TNF luciferase activity by hypertonic stress in L929 fibroblasts. A total of 2 × 106 L929 cells were transfected with 1 μg −200 TNF-Luc or −982 TNF-Luc and 0.4 μg pTK-RL control vectors by DEAE-dextran. Cells were stimulated with complete medium supplemented with 200 mM raffinose 24 h after transfection. Cells were harvested 18 h after stimulation and luciferase activities were assayed. Results are from four independent experiments. Error bars are the standard deviation.
To begin to determine the TNF promoter sequences that were required for hypertonic induction of TNF, we next transfected two human TNF promoter reporter genes with either −982 or −200 nt relative to the transcription start site linked to luciferase reporter genes. Transfection of these constructs into L929 cells that were then treated with raffinose demonstrated that −200 nt was sufficient for maximal induction of TNF by hypertonic stress (Figure 1D). These results are consistent with these sequences being sufficient for TNF induction using a variety of stimuli and cell types (1,3,5–7,9,30). We note that endogenous levels of TNF mRNA have correlated closely with the activity of the TNF luciferase reporter in all cell lines tested to date (Goldfeld lab, unpublished data) (Figure 1).
NFAT5 and NFATp bind the TNF promoter distinctly
The NFAT5 consensus sequence is 5′-TGGAAANYNY-3′ (14,19). Visual inspection of the TNF promoter reveals putative NFAT5 consensus binding sequences in both the −76 and κ3-NFAT binding regions, which are two of the six TNF promoter sites previously shown to bind NFATp. To determine if and where NFAT5 binds to TNF promoter sequences, we performed DNase I in vitro footprinting. A radiolabeled fragment of the human TNF promoter spanning the region from −200 to +87 nt was incubated with increasing amounts of recombinant human NFAT5 DBD5 protein and treated with DNase I. This analysis demonstrated that NFAT5 bound to the κ3 site and to the abutting −76 NFAT site, but did not bind to the four other TNF NFAT binding sites (−55, −117, −149 and −180), motifs that bind to NFATp, (2,3,31) (Figure 2, lanes 1–6). The pattern of binding of NFAT5 is distinct from that of NFATp at all concentrations of both factors. Using increasing amounts of recombinant human DBD5 protein fused to GST (DBD5-GST) in footprinting analysis gave us identical results with DBD5 protein alone, suggesting that the presence of GST did not affect DBD5 binding to the TNF promoter (Figure 2) (data not shown).
Recombinant NFAT5 binds distinctly to TNF NFAT sites as compared with NFATp, c, 3 and 4 in vitro
Consistent with the footprinting results presented above using recombinant NFATp and other footprinting studies using recombinant NFATc, NFAT3 and NFAT4 (data not shown), equal amounts of recombinant protein in every case resulted in stronger binding of these four proteins to the −76 NFAT site as compared with the κ3 site in an EMSA (Figure 3A). To further compare the binding of NFAT5 with other NFAT family members, we performed EMSAs with fragments of the human TNF promoter containing either the −76 NFAT site, the κ3 site (κ3L) or both the −76 NFAT site and the κ3 site (κ3LL) with NFAT5 (DBD5 or/and a DBD5-GST fusion protein). Since the sizes of these proteins can be differentiated on the gel, we were able to demonstrate that both the DBD5 and DBD5-GST bound the probe containing both the −76 NFAT site and κ3-NFAT sites alone. Moreover, the proteins bound to the probe containing both sites more strongly than to either site alone (Figure 3B), consistent with the DNase I footprinting results demonstrating the occupation of both sites at the same concentration of NFAT5 (Figure 2). We note that higher molecular weight complexes are visible when DBD5-GST is used in EMSA (Figure 3B, lanes 13–16 and lanes 20–26), consistent with the ability of GST to oligomerize.
To further investigate the specificity of NFAT5 binding to the κ3 and −76 NFAT sites but not to other sites in the TNF promoter, we performed an EMSA with all five recombinant NFAT proteins and a probe spanning the −180 NFAT sequence in addition to the κ3L and κ3LL probes. As shown in Figure 3C, NFATp, c, 3 and 4 all bind similarly to all three probes. In contrast, NFAT5 does not bind to the −180 NFAT site. These data further support our hypothesis that NFAT5 has a unique pattern of binding to the TNF promoter that is distinct from the other NFAT family members.
NFAT5 is recruited to the κ3-NFAT binding site in osmotically stressed L929 cells
We next performed an EMSA using the strongest NFAT5 binding probe (κ3LL) and nuclear extracts from osmotically stressed L929 cells to demonstrate that NFAT5 indeed can specifically bind this site using physiologically stimulated L929 cells. As shown in Figure 3D, raffinose stimulation of L929 cells results in the binding of two additional complexes that form on κ3LL probe as compared with what we observed using nuclear extracts from unstimulated L929 cells (compare lanes 1 and 2). Furthermore, anti-NFAT5 antibodies inhibit the formation of the two inducible complexes and a constitutive complex (Figure 3D, lane 3) whereas antibodies to NFATc and isotype control do not (lanes 6 and 7), demonstrating that these three complexes contain NFAT5. Moreover, the addition of antibodies against ATF-2 and c-jun also inhibits the formation of the same two inducible complexes that contain NFAT5 in raffinose-stimulated L292 nuclear extracts consistent with the two complexes being formed by NFAT5 together with ATF-2/c-jun dimer bound to the CRE sequence included in the κ3LL probe (Figure 3D, compare lanes 3–5).
Mutations in the −76 or κ3-NFAT sites abrogate binding of recombinant NFAT5 and TNF transcriptional induction by hypertonic stress
To establish that NFAT5 can bind to each site individually, we next performed a quantitative DNase I footprinting analysis with increasing amounts of DBD5-GST. As shown in Figure 4, mutation of the κ3-NFAT site (3′M or 5′M) abrogates NFAT5 binding to the κ3 site but does not abolish binding to the −76 site (Figure 4A, compare lanes 4–6 with 10–12 and 16–18). Conversely, mutation of the −76 site does not abolish binding of NFAT5 to the κ3-NFAT site, but abrogates binding of NFAT5 to the −76 site. We note that the κ3-NFAT site appears darker at the highest levels of DBD5-GST than the corresponding bands in the WT control only because the −76 mutant gel has a darker exposure overall (Figure 4A, compare lanes 4–6 with 22–24). In contrast, using a template with a mutation in the −84-Ets site that is located between the κ3 and −76 NFAT sites, binding of NFAT5 is unaffected (Figure 4A, lanes 27–30).
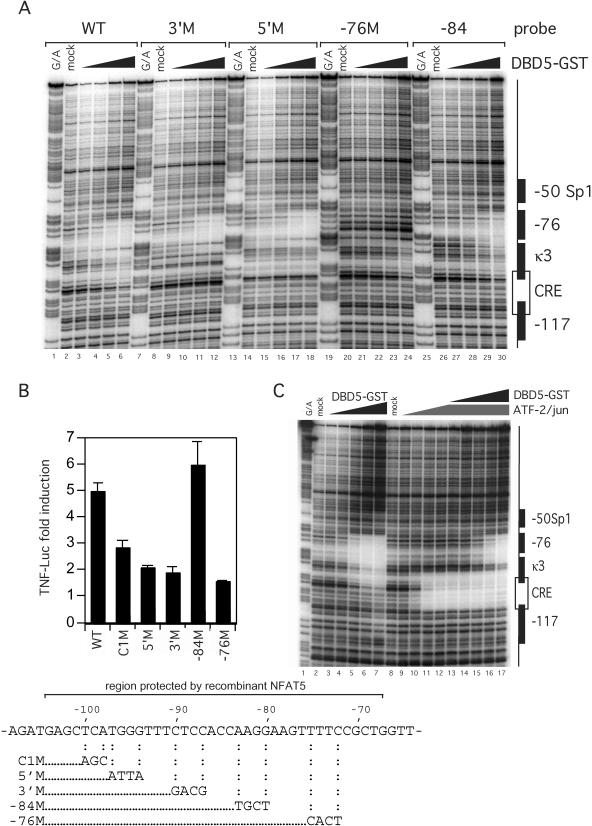
Figure 4.
Mutations in the −76 NFAT and κ3 site, but not in the −84-Ets site, decrease binding of recombinant NFAT5 and hypertonic induction of TNF. (A) DNase I footprinting assay of mutated TNF promoter with NFAT5 (DBD5-GST). The 3′M and 5′M sites are in the κ3 element and the −76 mutation is in the −76 NFAT site. The −84 mutation is between these two sites. For a diagram, see the bottom of the figure. (B) Relative activity of TNF luciferase constructs containing mutations in known binding sites of transcriptional activators. A total of 5 × 106 L929 cells were transfected with 1 μg −200 TNF-Luc and 0.2 μg pTK-RL control vectors by DEAE-dextran. Cells were stimulated with complete medium supplemented with 200 mM raffinose 24 h after transfection. Cells were harvested 18 h after stimulation and Photinus luciferase and Renilla activities were assayed. (C) DNase I footprinting assay of competitive binding of NFAT5 and ATF-2/c-jun. The binding of ANF-2/c-jun to the CRE element was not blocked by increasing amounts of DBD5-GST (see lanes 13–17).
To correlate the above binding studies with promoter elements required for the induction of the TNF gene by hypertonic stress, we next transfected L929 cells with the wild-type (WT) −200 human TNF-Luc reporter gene or isogenic constructs bearing mutations in the κ3-NFAT (5′M and 3′M), −76 NFAT (−76M), CRE (C1M) and −84-Ets (−84M) sites (as shown at the bottom of Figure 4) and stimulated them with raffinose (Figure 4B). Mutation of the κ3 or −76 NFAT sites significantly inhibited induction of the reporter genes by hypertonic stress, whereas the −84 mutation had no effect (Figure 4B). We also note that hypertonic induction of TNF in L929 cells is not sensitive to CsA (data not shown). Thus, the inhibition of raffinose-stimulated TNF gene expression by the −76 and κ3 site mutations is consistent with their interference with NFAT5 binding to the −76 and κ3 sites and demonstrates the importance of both of these sites in the regulation of TNF by hypertonic stress.
The fact that the CRE mutant, which disrupts binding of ATF-2/c-jun (28), also had a significant impact upon raffinose induction of TNF indicates that ATF-2/c-jun is also involved in the enhancer complex together with NFAT5. To formally prove that both ATF-2/c-jun and NFAT5 proteins could simultaneously bind to the TNF promoter, given the proximity of the sites, we next performed a quantitative DNase I footprinting analysis with ATF-2/c-jun and increasing amounts of NFAT5. As shown in Figure 4C, ATF-2/c-jun binding to the CRE is not blocked by NFAT5 at increasing concentrations. Thus, the CRE/κ3/−76 sites form a composite binding element for ATF-2/c-jun/NFAT5 required for the regulation of TNF by hypertonic stress.
Blocking NFAT5 with a construct expressing an shRNA or a dominant negative NFAT5 protein inhibits TNF reporter activity in response to hypertonic stress
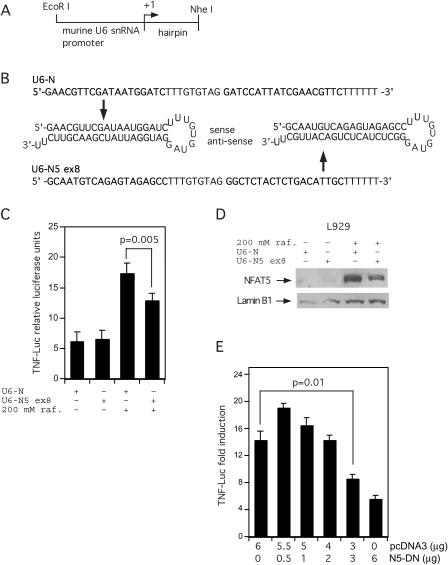
To demonstrate that NFAT5 is a critical activator in hypertonic induction of the TNF gene, we next employed two approaches: knockdown of NFAT5 mRNA and co-transfection of a dominant negative form of NFAT5 (24). Although NFAT5 has been successfully knocked down using synthetic small interfering (si)RNA oligonucleotides (32), this approach was not successful with NFAT5 in L929 cells (data not shown). We made shRNA-expressing construct targeting exon 8 of the murine NFAT5 gene (U6-N5 ex8) under the control of the murine U6 small nuclear (sn)RNA promoter (Figure 5A). Furthermore, a construct expressing a scrambled shRNA was also made to serve as a negative control (U6-N). See Figure 5A and B for diagrams of the transcribed DNA region and shRNAs from these two constructs. The constructs were co-transfected into L929 cells with the −200 TNF-Luc vector and the cells were stimulated with 200 mM raffinose 36 h after transfection and harvested 18 h after stimulation. As shown in Figure 5C, there was a statistically significant inhibition of TNF reporter activity in response to hypertonicity in the cells transfected with U6-N5 ex8 vector as compared with the scrambled control vector, U6-N. As a control, we performed a western blot analysis of extracts from cells transfected with the U6-N5 ex8 or U6-N vector and stimulated with hypertonic stress. We observed a ∼50% knockdown of NFAT5 protein levels in cells transfected with U6-N5 ex8 compared with NFAT5 in cells transfected with the control U6-N vector (Figure 5D).
Figure 5.
A vector expressing a shRNA targeting NFAT5 and a dominant negative NFAT5 down-regulate hypertonic induction of TNF luciferase activity. (A) Diagram of the shRNA-expressing construct. The murine U6 small nuclear RNA (snRNA) promoter was cloned into pBluescript followed by a sequence coding for the shRNA targeting murine NFAT5. See Materials and Methods for details. (B) Diagram of the shRNAs expressed from the scrambled sequence (U6-N) and the sequence targeting exon 8 of murine NFAT5 (U6-N5 ex8). (C and D) A vector expressing an shRNA against NFAT5 depresses hypertonic induction of TNF-Luc activity in L929 cells and lowers NFAT5 levels. A total of 3 × 105 cells were plated in 6-wells plates (∼95% confluent) and were transfected the next day with 0.7 μg TNF-Luc, 0.3 μg of TK-RL and 1 μg of either the U6-N or the U6-N5 ex8 vector. The cells were stimulated as indicated 36 h later with complete medium or medium supplemented with 200 mM raffinose for 18 h. Cells were harvested and assayed as above. Error bars are the SD. The P-value was calculated with an unpaired two-tailed t-test. Results are from four independent experiments. (D) Nuclear extracts from mock or raffinose treated L929 cells transfected with the U6 constructs were also analyzed by western blot analysis with an antibody to NFAT5, which demonstrates specific knockdown of NFAT5 protein levels by the U6-N5 ex8 vector as shown. (E) A vector expressing a dominant negative NFAT5 decreases hypertonic induction of TNF-Luc activity. L929 cells were co-transfected by DEAE-dextran with TNF-Luc, pTK-RL, and different amounts of either pcDNA3, N5-DN or both vectors as indicated. Cells were stimulated and extracts were assayed as above. The P-value was calculated with an unpaired two-tailed t-test. Results are from four independent experiments.
To further establish a role for NFAT5 in TNF transcription during hyptertonic stress, increasing amounts of a dominant negative version of NFAT5 (NFAT5-DN) were transiently transfected together with the TNF-Luc reporter gene into L929 cells before hypertonic stimulation. NFAT5-DN suppressed TNF production during hypertonic stress in a dose-dependent manner (Figure 5E). When the highest concentration of NFAT5-DN was transfected (6 μg) into L929 cells, TNF-Luc activity was reduced by ∼75% as compared with co-transfection of the same amount of the pcDNA3 vector alone. Even at half this concentration of NFAT5-DN (3 μg) the repression of TNF compared with the pcDNA3 control (6 μg) was highly significant (P = 0.01) (Figure 5E). Taken together with the NFAT5 knockdown experiments, these studies demonstrate that NFAT5 is a critical activator in TNF induction by hypertonic stress.
DISCUSSION
TNF gene transcription is controlled by enhanceosome formation in a stimulus- and cell-type-specific manner (3–6). Previous work has shown that NFAT family members play a critical role in TNF gene expression in T cells stimulated by TCR ligands, ionophore, virus and superantigen (2,3). This work broadens the role that NFAT family members play in TNF transcription by showing that NFAT5 activates TNF transcription in response to hypertonic stress alone and that NFAT5 binds to the TNF promoter in a pattern distinct from other NFAT family members. Thus, a single family of transcription factors achieves specificity in TNF gene activation via the diversity of binding patterns of its members in response to various activation stimuli.
NFAT5 is the only member of the NFAT family to bind the −76 NFAT and the κ3 site, but not to other NFAT sites in the TNF promoter. We note that the −76 site contains a better match for the core of the consensus sequence (GGAAA) (see Figure 3) than does the κ3. However, the affinity of NFAT5 for a site may be strengthened when the second half of the site is similar to the core consensus sequence (19). This observation may explain as to why NFAT5 binds to the κ3-NFAT site despite the lack of a perfect core consensus sequence. Nevertheless, the precise reasons, why NFAT5 has high affinity for −76 and κ3 but not to other NFAT sites in the proximal promoter, remain to be elucidated.
ATF-2 is a known downstream target of p38 MAP kinase, which is activated in mammalian cells in response to hypertonic stress (33). We note that treatment with SB203580, a specific inhibitor of p38 MAP kinase, reduces the hypertonic induction of TNF-Luc activity in L929 cells (data not shown). Inhibition of p38 has also been shown to block NFAT5 activity (34). ATF-2/c-jun/NFATp binds to the CRE/κ3 composite site to form the core of the enhanceosome in T cells activated by TCR ligands or viruses. Our data here suggest that during hypertonic stress, a new composite element comprising two NFAT5 dimers binding to −76 and κ3-NFAT and ATF-2/c-jun at the CRE is formed. These data are consistent with the recruitment of a distinct enhanceosome to the TNF promoter under hypertonic conditions. TNF was first described as a target of NFAT5 on the basis of a ChIP assay that showed that NFAT5 is recruited to the TNF promoter in T cells after stimulation with hypertonic stress and PMA (24). Our study gives further evidence that hypertonic stress alone, in the absence of NFATp (since L929 cells do not contain NFATp [Figure 1C and see (1)], activates TNF transcription via NFAT5.
Notably, NFAT5 has been shown to be involved in T lymphocyte development and survival (16). Murine lymphoid tissues are hypertonic, and heterozygous NFAT5 knockout mice show defective proliferation of lymphocytes under hypertonic conditions (35). These results indicate that NFAT5 could play a role in the regulation of TNF during lymphocyte proliferation, although to date there is no direct evidence that NFAT5 regulates TNF transcription in T lymphocytes under isotonic conditions. For example, TNF production was not affected in N5-DN transgenic primary Con A blasts activated with PMA and ionomycin (36). This result is consistent with the critical and non-redundant role of NFATp in immediate early TNF production in T cells (2). However, given that rapidly dividing cells become hypotonic relative to their extracellular environment due to rapid volume increase (37), it is intriguing to speculate that NFAT5 could co-regulate both cellular tonicity and TNF production at later time points while T cells are undergoing rapid clonal expansion during the immune response.
Acknowledgments
The authors thank Tim Hoey for the generous gift of recombinant NFATp, NFATc, NFAT3 and NFAT4 proteins and Dimitris Thanos for his generous gift of recombinant ATF-2/c-jun and NFATp and the CBR Institute of Biomedical Research for Institutional support. This work was supported by grants from the NIH to A.E.G. (GM056492) and to A.R. (CA42471), from the Harvard College Research Program and the Board of Tutors in Biochemical Sciences (Harvard University) to J.H.E, from the Arthritis National Research Foundation to A.V.T., and from the Cancer Research Institute and the Leukemia & Lymphoma Society Career Development Program to C.L.-R. Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Falvo J.V., Uglialoro A.M., Brinkman B.M., Merika M., Parekh B.S., Tsai E.Y., King H.C., Morielli A.D., Peralta E.G., Maniatis T., et al. Stimulus-specific assembly of enhancer complexes on the tumor necrosis factor alpha gene promoter. Mol. Cell. Biol. 2000;20:2239–2247. doi: 10.1128/mcb.20.6.2239-2247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsytsykova A.V., Goldfeld A.E. Nuclear factor of activated T cells transcription factor NFATp controls superantigen-induced lethal shock. J. Exp. Med. 2000;192:581–586. doi: 10.1084/jem.192.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsytsykova A.V., Goldfeld A.E. Inducer-specific enhanceosome formation controls tumor necrosis factor alpha gene expression in T lymphocytes. Mol. Cell. Biol. 2002;22:2620–2531. doi: 10.1128/MCB.22.8.2620-2631.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falvo J.V., Brinkman B.M., Tsytsykova A.V., Tsai E.Y., Yao T.P., Kung A.L., Goldfeld A.E. A stimulus-specific role for CREB-binding protein (CBP) in T cell receptor-activated tumor necrosis factor alpha gene expression. Proc. Natl Acad. Sci. USA. 2000;97:3925–3929. doi: 10.1073/pnas.97.8.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai E.Y., Falvo J.V., Tsytsykova A.V., Barczak A.K., Reimold A.M., Glimcher L.H., Fenton M.J., Gordon D.C., Dunn I.F., Goldfeld A.E. A lipopolysaccharide-specific enhancer complex involving Ets, Elk-1, Sp1, and CREB binding protein and p300 is recruited to the tumor necrosis factor alpha promoter in vivo. Mol. Cell. Biol. 2000;20:6084–6094. doi: 10.1128/mcb.20.16.6084-6094.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barthel R., Tsytsykova A.V., Barczak A.K., Tsai E.Y., Dascher C.C., Brenner M.B., Goldfeld A.E. Regulation of tumor necrosis factor alpha gene expression by mycobacteria involves the assembly of a unique enhanceosome dependent on the coactivator proteins CBP/p300. Mol. Cell. Biol. 2003;23:526–533. doi: 10.1128/MCB.23.2.526-533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinkman B.M., Telliez J.B., Schievella A.R., Lin L.L., Goldfeld A.E. Engagement of tumor necrosis factor (TNF) receptor 1 leads to ATF-2- and p38 mitogen-activated protein kinase-dependent TNF-alpha gene expression. J. Biol. Chem. 1999;274:30882–30886. doi: 10.1074/jbc.274.43.30882. [DOI] [PubMed] [Google Scholar]
- 8.Aggarwal B.B. Signalling pathways of the TNF superfamily: a double-edged sword. Nature Rev. Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 9.Goldfeld A.E., McCaffrey P.G., Strominger J.L., Rao A. Identification of a novel cyclosporin-sensitive element in the human tumor necrosis factor alpha gene promoter. J. Exp. Med. 1993;178:1365–1379. doi: 10.1084/jem.178.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCaffrey P.G., Goldfeld A.E., Rao A. The role of NFATp in cyclosporin A-sensitive tumor necrosis factor-alpha gene transcription. J. Biol. Chem. 1994;269:30445–30450. [PubMed] [Google Scholar]
- 11.McCaffrey P.G., Luo C., Kerppola T.K., Jain J., Badalian T.M., Ho A.M., Burgeon E., Lane W.S., Lambert J.N., Curran T., et al. Isolation of the cyclosporin-sensitive T cell transcription factor NFATp. Science. 1993;262:750–754. doi: 10.1126/science.8235597. [DOI] [PubMed] [Google Scholar]
- 12.Northrop J.P., Ho S.N., Chen L., Thomas D.J., Timmerman L.A., Nolan G.P., Admon A., Crabtree G.R. NF-AT components define a family of transcription factors targeted in T-cell activation. Nature. 1994;369:497–502. doi: 10.1038/369497a0. [DOI] [PubMed] [Google Scholar]
- 13.Hoey T., Sun Y.L., Williamson K., Xu X. Isolation of two new members of the NF-AT gene family and functional characterization of the NF-AT proteins. Immunity. 1995;2:461–472. doi: 10.1016/1074-7613(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Rodriguez C., Aramburu J., Rakeman A.S., Rao A. NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc. Natl Acad. Sci. USA. 1999;96:7214–7219. doi: 10.1073/pnas.96.13.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyakawa H., Woo S.K., Chen C.P., Dahl S.C., Handler J.S., Kwon H.M. Cis- and trans-acting factors regulating transcription of the BGT1 gene in response to hypertonicity. Am. J. Physiol. 1998;274:F753–F761. doi: 10.1152/ajprenal.1998.274.4.F753. [DOI] [PubMed] [Google Scholar]
- 16.Trama J., Lu Q., Hawley R.G., Ho S.N. The NFAT-related protein NFATL1 (TonEBP/NFAT5) is induced upon T cell activation in a calcineurin-dependent manner. J. Immunol. 2000;165:4884–4894. doi: 10.4049/jimmunol.165.9.4884. [DOI] [PubMed] [Google Scholar]
- 17.Ko B.C., Turck C.W., Lee K.W., Yang Y., Chung S.S. Purification, identification, and characterization of an osmotic response element binding protein. Biochem. Biophys. Res. Commun. 2000;270:52–61. doi: 10.1006/bbrc.2000.2376. [DOI] [PubMed] [Google Scholar]
- 18.Pan S., Tsuruta R., Masuda E.S., Imamura R., Bazan F., Arai K., Arai N., Miyatake S. NFATz: a novel rel similarity domain containing protein. Biochem. Biophys. Res. Commun. 2000;272:765–776. doi: 10.1006/bbrc.2000.2831. [DOI] [PubMed] [Google Scholar]
- 19.Stroud J.C., Lopez-Rodriguez C., Rao A., Chen L. Structure of a TonEBP–DNA complex reveals DNA encircled by a transcription factor. Nature Struct. Biol. 2002;9:90–94. doi: 10.1038/nsb749. [DOI] [PubMed] [Google Scholar]
- 20.Shapiro L., Dinarello C.A. Hyperosmotic stress as a stimulant for proinflammatory cytokine production. Exp. Cell Res. 1997;231:354–362. doi: 10.1006/excr.1997.3476. [DOI] [PubMed] [Google Scholar]
- 21.Cuschieri J., Gourlay D., Garcia I., Jelacic S., Maier R.V. Hypertonic preconditioning inhibits macrophage responsiveness to endotoxin. J. Immunol. 2002;168:1389–1396. doi: 10.4049/jimmunol.168.3.1389. [DOI] [PubMed] [Google Scholar]
- 22.Junger W.G., Liu F.C., Loomis W.H., Hoyt D.B. Hypertonic saline enhances cellular immune function. Circ. Shock. 1994;42:190–196. [PubMed] [Google Scholar]
- 23.Lang K.S., Fillon S., Schneider D., Rammensee H.G., Lang F. Stimulation of TNF alpha expression by hyperosmotic stress. Pflugers Arch. 2002;443:798–803. doi: 10.1007/s00424-001-0768-7. [DOI] [PubMed] [Google Scholar]
- 24.Lopez-Rodriguez C., Aramburu J., Jin L., Rakeman A.S., Michino M., Rao A. Bridging the NFAT and NF-kappaB families: NFAT5 dimerization regulates cytokine gene transcription in response to osmotic stress. Immunity. 2001;15:47–58. doi: 10.1016/s1074-7613(01)00165-0. [DOI] [PubMed] [Google Scholar]
- 25.Goldfeld A.E., Strominger J.L., Doyle C. Human tumor necrosis factor alpha gene regulation in phorbol ester stimulated T and B cell lines. J. Exp. Med. 1991;174:73–81. doi: 10.1084/jem.174.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Y.J., Dissen G.A., Rage F., Ojeda S.R. RNase protection assay. Methods. 1996;10:273–278. doi: 10.1006/meth.1996.0102. [DOI] [PubMed] [Google Scholar]
- 27.Stalder A.K., Campbell I.L. Simultaneous analysis of multiple cytokine receptor mRNAs by RNase protection assay in LPS-induced endotoxemia. Lymphokine. Cytokine Res. 1994;13:107–112. [PubMed] [Google Scholar]
- 28.Tsai E.Y., Jain J., Pesavento P.A., Rao A., Goldfeld A.E. Tumor necrosis factor alpha gene regulation in activated T cells involves ATF-2/Jun and NFATp. Mol. Cell. Biol. 1996;16:459–467. doi: 10.1128/mcb.16.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaneko H., Yamada H., Mizuno S., Udagawa T., Kazumi Y., Sekikawa K., Sugawara I. Role of tumor necrosis factor-alpha in Mycobacterium-induced granuloma formation in tumor necrosis factor-alpha-deficient mice. Lab. Invest. 1999;79:379–386. [PubMed] [Google Scholar]
- 30.Goldfeld A.E., Doyle C., Maniatis T. Human tumor necrosis factor alpha gene regulation by virus and lipopolysaccharide. Proc. Natl Acad. Sci. USA. 1990;87:9769–9773. doi: 10.1073/pnas.87.24.9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai E.Y., Yie J., Thanos D., Goldfeld A.E. Cell-type-specific regulation of the human tumor necrosis factor alpha gene in B cells and T cells by NFATp and ATF-2/JUN. Mol. Cell. Biol. 1996b;16:5232–5244. doi: 10.1128/mcb.16.10.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Na K.Y., Woo S.K., Lee S.D., Kwon H.M. Silencing of TonEBP/NFAT5 transcriptional activator by RNA interference. J. Am. Soc. Nephrol. 2003;14:283–288. doi: 10.1097/01.asn.0000045050.19544.b2. [DOI] [PubMed] [Google Scholar]
- 33.Cowan K.J., Storey K.B. Mitogen-activated protein kinases: new signaling pathways functioning in cellular responses to environmental stress. J. Exp. Biol. 2003;206:1107–1115. doi: 10.1242/jeb.00220. [DOI] [PubMed] [Google Scholar]
- 34.Ko B.C., Lam A.K., Kapus A., Fan L., Chung S.K., Chung S.S. Fyn and p38 signaling are both required for maximal hypertonic activation of the osmotic response element-binding protein/tonicity-responsive enhancer-binding protein (OREBP/TonEBP) J. Biol. Chem. 2002;277:46085–46092. doi: 10.1074/jbc.M208138200. [DOI] [PubMed] [Google Scholar]
- 35.Go W.Y., Liu X., Roti M.A., Liu F., Ho S.N. NFAT5/TonEBP mutant mice define osmotic stress as a critical feature of the lymphoid microenvironment. Proc. Natl Acad. Sci. USA. 2004;101:10673–10678. doi: 10.1073/pnas.0403139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trama J., Go W.Y., Ho S.N. The osmoprotective function of the NFAT5 transcription factor in T cell development and activation. J. Immunol. 2002;169:5477–5488. doi: 10.4049/jimmunol.169.10.5477. [DOI] [PubMed] [Google Scholar]
- 37.Ho S.N. The role of NFAT5/TonEBP in establishing an optimal intracellular environment. Arch. Biochem. Biophys. 2003;413:151–157. doi: 10.1016/s0003-9861(03)00130-9. [DOI] [PubMed] [Google Scholar]