Abstract
Background
Emerging perspectives on tumor metabolism reveal its heterogeneity, a characteristic yet to be fully explored in gliomas. To advance therapies targeting metabolic processes, it is crucial to uncover metabolic differences and identify distinct metabolic subtypes. Therefore, we aimed to develop a classification system for gliomas based on the enrichment levels of four key metabolic pathways: glutaminolysis, glycolysis, the pentose phosphate pathway, and fatty acid oxidation.
Methods
Energy-related features of glioma were characterized through integrative analyses of multiple datasets, including bulk, single-cell, and spatial transcriptome profiling. The glioma energy metabolic subtypes were constructed using the R package ConsensusClusterPlus. Kaplan–Meier analysis was conducted to compare clinical outcomes between different metabolic groups. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were employed to elucidate the biological functions of genes of interest. Cell-cell communication analysis was performed at single-cell resolution using the R package CellChat and at spatial resolution using the standard stLearn pipeline.
Results
Glioma samples were stratified into two prognostic subtypes. Group 1, enriched in the glutaminolysis pathway, had better clinical outcomes. In contrast, Group 2 exhibited high activities in glycolysis, the pentose phosphate pathway, and fatty acid oxidation, correlating with decreased survival time. Group 1 samples were predominantly located in the peripheral region and had a high composition of neuron cells. Group 2, however, had increased infiltration of tumor-promoting immune cells, such as M2 macrophages, and was characterized by traits of invasion, hypoxia, and immunity. Lastly, cell-cell communications were compared across different tumor regions, and the CX3CL1/CX3CR1 ligand-receptor pair was validated using spatial transcriptomic data.
Conclusions
Our work revealed the metabolic heterogeneity in glioma by developing a new classification system with significant prognostic and therapeutic value. Single-cell transcriptional profiles offer novel insights into tumor metabolic reprogramming, which could enhance therapies tailored to cell- or patient-specific metabolic patterns.
Keywords: Glioma, Metabolic heterogeneity, Subtype, Single cell transcriptomic, Spatial transcriptomic
Highlights
-
•
Glioma samples were stratified into two prognostic subtypes based on their metabolic profiles.
-
•
Group 1 glioma, enrichment in the glutaminolysis pathway, exhibited better clinical outcomes.
-
•
Group 1 gliomas were mainly located in peripheral region and had a high proportion of neuron cells.
-
•
Group 2 gliomas were characterized with invasion, hypoxic condition, and heightened immunity.
Abbreviations:
| CGGA | Chinese Glioma Genome Atlas |
| CI | Confidence interval |
| CL | Classical |
| DEG | Differentially expressed gene |
| EMT | Epithelial-mesenchymal transition |
| FAO | Fatty acid oxidation |
| GBM | Glioblastoma |
| GEO | Gene Expression Omnibus |
| GO | Gene Ontology |
| HR | Hazard ratio |
| IGAP | Ivy Glioblastoma Atlas Project |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| L-R | Ligand-receptor |
| ME | Mesenchymal |
| MSigDB | Molecular Signatures Database |
| NE | Neural |
| O2 | Oxygen |
| OPC | Oligodendrocyte precursor cell |
| PN | Proneural |
| PPP | Pentose phosphate pathway |
| scRNA-seq | Single-Cell RNA sequencing |
| ssGSEA | Single sample gene set enrichment analysis |
| ST | Spatial transcriptomic |
| TAM | Tumor-Associated macrophage |
| TCGA | The Cancer Genome Atlas |
| TME | Tumor microenvironment |
| tSNE | T-Distributed stochastic neighbor embedding |
1. Introduction
Glioma is the most common primary brain tumor in adults. It arises from glial cells, specifically astrocytes, and is characterized by rapid, infiltrative growth that makes complete surgical removal challenging [1]. The standard treatment is maximal surgical resection, followed by radiation, with concurrent adjuvant chemotherapy. Glioblastoma (GBM, WHO 4) is the most aggressive type of glioma, and prognosis remains poor despite aggressive treatment, with a median survival of 12–15 months [2]. Recent high-throughput genomics, transcriptomics, proteomics and epigenetic data have offered a comprehensive view of the molecular mechanisms underlying glioma oncogenesis and progression, and have been used to classify tumors so that targeted therapy could be developed for personal subtype [3]. Many attempts have tried to characterize glioma phenotypes through somatic mutation pattern [4], RNA expression profile [5] and protein level [6], to define predictive or prognostic subgroups which could serve as pharmacological targets. Fan and others constructed a classifier system based on metabolic gene expression profiles, which highlighted metabolic heterogeneity in low grade glioma [7]. Glioma shows high heterogeneity, including metabolism, and there is rare research exploring glioma metabolic heterogeneity at pathway level [7].
Metabolic reprogramming is a hallmark of cancer, which plays a vital role in tumor malignancy progression. Metabolic changes have been noticed several years before clinical presentation in glioma, suggesting that metabolic reprogramming can be the early event in gliomagenesis [8]. Exploration of metabolic alterations among different grades of glioma may indicate potential opportunities for tumor prevention, diagnosis and therapy [9]. Some key metabolites, such as glutamate metabolism, choline metabolism and cysteine metabolism, are related to glioma grading [10]. While there are increasing researches studying metabolites associated with glioma progression, the results for metabolic changes are still equivocal. One possible explanation for this is that methodologies and protocols employed in different studies are different [11]. And different grades of glioma tissues are not easily available resulting in a small cohort which may also contribute to this [11]. Furthermore, most studies usually concentrate on a particular metabolic perturbation and explore it in isolation [12]. It is urgent to disclose the metabolic pattern and recognize metabolic subtypes in glioma, which will promote the development of therapy targeting metabolism.
The importance of glucose, amino acids, and fatty acids in tumor progression has been investigated for a long time. High aerobic glycolysis (Warburg effect) and a high rate of glutamine consumption for energetic and anaplerotic purposes (glutaminolysis) are essential features of cancer metabolic reprogramming [13,14]. Pentose phosphate pathway (PPP) is known to be upregulated in cancers and plays a critical role in cancer cell division by supplying cells with not only ribose-5-phosphate but also NADPH for ribose biogenesis, reductive biosynthesis, and detoxification of intracellular reactive oxygen species [15,16]. Fatty acid oxidation (FAO) is the mitochondrial aerobic process of breaking down fatty acids into acetyl-CoA units and increasing evidence reveal the role of lipid metabolism in tumorigenesis and tumor progression [17]. Because none of the above-mentioned substrates fulfills all the needs of tumor growth, it may be vital for tumor cells to coordinate metabolic pathways that process different ingredients to achieve the largest output from the same raw nutrients. Furthermore, metabolism reprogramming is considered one of the hallmarks of cancer as it is different from normal cells and mainly consists of glycolysis, glutaminolysis, PPP and FAO [18]. Herein, we defined a new energy metabolic classifier based on unsupervised clustering of these 4 key metabolism pathways, namely glutaminolysis, glycolysis, PPP and FAO, enrichment level. The stability and reproducibility of our subtype system were estimated in three independent glioma cohorts. Each of the two metabolic classifiers was associated with distinct clinical prognosis, immune infiltration, cell type composition and spatial distribution. In addition, we compared cell-cell interactions between different tumor regions using a glioma single-cell RNA sequencing (scRNA-seq) dataset. And one ligand-receptor (L-R) pair was further verified by glioma spatial transcriptomics (ST) data. These findings unraveled metabolic heterogeneity in glioma at bulk, single-cell and spatial levels, which may drive the development of metabolism-targeted treatments.
2. Materials and methods
2.1. Bulk RNA sequencing datasets
Transcriptional profiles with corresponding clinical information for human gliomas were obtained from the Chinese Glioma Genome Atlas (CGGA) [19], The Cancer Genome Atlas (TCGA) [20] and the Rembrandt brain cancer dataset [21] respectively. A total of 693 glioma samples from the CGGA, 702 glioma samples from TCGA and 475 glioma samples from the Rembrandt were used in our study. RNA expression data of 122 glioblastoma (GBM) samples with anatomic features from the Ivy Glioblastoma Atlas Project (IGAP) were also utilized in this study [22]. Because these datasets are from different commercial platforms (Affymetrix, Agilent, Illumina), we adopt a strategy to integrate all glioma samples as follows to remove batch effect: the expression matrices were firstly scaled; then the batch effect of the scaled matrices was corrected by the function ComBat in the package sva (3.46.0); finally, the PCA analysis result confirmed the batch effect was removed well. The pathological classification on glioma is the fourth WHO's classification of tumors of the central nervous system in this research [23]. Galina Zakharova and others have reclassified TCGA diffuse glioma according to the 2021 WHO Central Nervous System Tumor Classification [24], and we utilized their reclassification results to compare the difference of glioma metabolism subtype between the 2021 and 2016 WHO classification of tumors of the central nervous system.
2.2. Construction of the energy metabolic subtypes
To delineate glioma metabolism, we collected and tailored the metabolic gene sets into 4 core pathways: glutaminolysis, glycolysis, PPP and FAO, by browsing the Molecular Signatures Database (MSigDB) [25] and searching for papers [[26], [27], [28], [29], [30]]. Then, we performed single sample gene set enrichment analysis (ssGSEA) by using the R package GSVA (v.1.44) to evaluate the enrichment level of each metabolic pathway in each glioma sample through transcriptomics [31]. The former metabolic enrichment levels were selected for consensus clustering by using the R package ConsensusClusterPlus (v.1.60) to reveal robust subtypes. The optimal K = 2 was accessed through the consensus heatmap and cumulative distribution function [32]. A consensus classifier was performed using 1000 iterations and a resampling of 80 %. To better identify the variations between metabolic subtype 1 and 2, a heatmap was also applied.
2.3. Survival analysis
Kaplan–Meier analysis was done to compare the clinical outcome between different metabolic subtypes, which were tested for significance using the Mantel-Cox log-rank test. Hazard ratio (HR) and confidence interval (CI) were also computed to verify the survival difference in glioma metabolic subtypes. A p-value <0.05 was recognized as statistically significant [33].
2.4. Screening for differentially expressed genes (DEGs)
Gene differential analysis was performed between metabolic subtype 1 and 2 using R package limma (v.3.52) [34]. DEGs were considered to be statistically significant when |logFC| > 1 and p.adj <0.05 [35]. We kept a list of intersected DEGs among the CGGA, TCGA and the Rembrandt glioma samples for subsequent enrichment analysis.
2.5. Enrichment analysis
Gene Ontology (GO) [36] and Kyoto Encyclopedia of Genes and Genomes (KEGG) [37] analyses were applied to discover the biological function of the former DEGs. Furthermore, biological pathway differences between metabolic subtype 1 and 2 were carried out by R package GSVA, and method = ‘ssgsea’ was set.
2.6. Estimating immune infiltration
The infiltration level of stromal cells and immune cells in glioma samples was estimated by the method ESTIMATE [38]. CIBERSORTx portal using a set of 22 human immune cell reference profiles was adopted to infer immune cell compositions within individual glioma samples [39]. The relative composition ratio of immune cells was compared between different subtypes, checked by t-test, and was used for heatmap. We also explored the IGAP dataset by CIBERSORTx, and its result was further clustered by the function vegdist in the R package vegan (v.2.6.8) with default parameters. The tumor microenvironment (TME) plays a vital role in cancer response to therapy and clinical prognosis. Thus, we further explored the glioma TME characteristics by comparing the enrichment level of 28 gene sets involved in 12 cancer features, which are detailed in the work by Bagaev and others [40].
2.7. Single cell RNA sequencing (scRNA-seq) data processing
The human GBM scRNA-seq data were obtained from the Gene Expression Omnibus (GEO) under accession number GSE84465 [41]. The expression matrices were analyzed by R package Seurat (v.4.3.0) [42], and cell type annotated by Darmanis and others were also adopted in this work [41]. We removed the cells from downstream analysis where >10 % transcripts aligned to the mitochondrial genome, >50 % mapped to ribosomal gene, or which had either <400 or >10,000 RNA counts. In addition, the gene was filtered out when it expressed in <3 cells among the 4 GBM patients. The remained 23,230 genes in 3589 cells passed quality control and were used in downstream analysis. The enrichment level of a specific gene set at the single cell level was evaluated by utilizing the function AddModuleScore in Seurat.
Glioma metabolic activity was also evaluated at single-cell resolution. Firstly, the enrichment level of the former 4 metabolic pathways was done to explore the metabolic heterogeneity among different cell types. Subsequently, we took a computational pipeline as previously described [43] to characterize the glioma metabolic landscape. As malignant cells are heterogeneous, we compared the top 10 pathways contributing to the metabolic heterogeneities among malignant cells from different glioma samples. Metabolic pathways with the highest contribution to the metabolic heterogeneities among glioma stromal cells were also explored.
Cell-cell communication analysis of tumor core and periphery was performed by R package CellChat (v.1.6.1) by integrating scRNA-seq data with CellChatDB, an L-R interaction database [44]. We first focused on the overall signaling changes on cell population levels and then drilled down to the dysregulated signaling pathways and L-R pairs. The output intercellular communication networks for each L-R pair, where the calculated communication probability was assigned as edge weight to quantify L-R interaction strength. Intercellular communication at the signaling pathway level was evaluated by summarizing the communication probabilities of all related L-R pairs.
2.8. Spatial transcriptomics (ST) data analysis
The human GBM ST data (UKF_243, UKF_266 and UKF_313) were downloaded from https://doi.org/10.5061/dryad.h70rxwdmj [45]. As a quality control step, the Python package SCANPY (v.1.9.1) was utilized [46]. Spots with fewer than 100 UMIs, fewer than 50 genes or more than 50 % mitochondrial reads were filtered out. Then, we mapped the fine-annotated GBM cell types from the former scRNA-seq data in UKF_243, UKF_266 ST and UKF_313 slides through the standard Cell2location analysis pipeline [47]. The cell abundance output matrices were used in further analysis, and we chose the top value as the final annotated cell type. To decipher metabolic differences among spots, the R package SPATA2 (v.0.1.0) was utilized to evaluate the enrichment level of the former 4 core metabolic pathways. Finally, cell-cell interaction at spot resolution was accomplished by using the standard stLearn pipeline [48]. This analysis calls significant spots of L-R interactions from a database of candidate L-R (connectomeDB2020). The L-R of interest expressing/co-expressing simultaneously with the dominant spot cell type was visualized. The inner spot was colored by the dominant spot cell type, and the outer spot showed the expression of ligand, receptor and co-expression.
3. Results
3.1. Glioma presents two metabolic statuses according to energy metabolic classifier
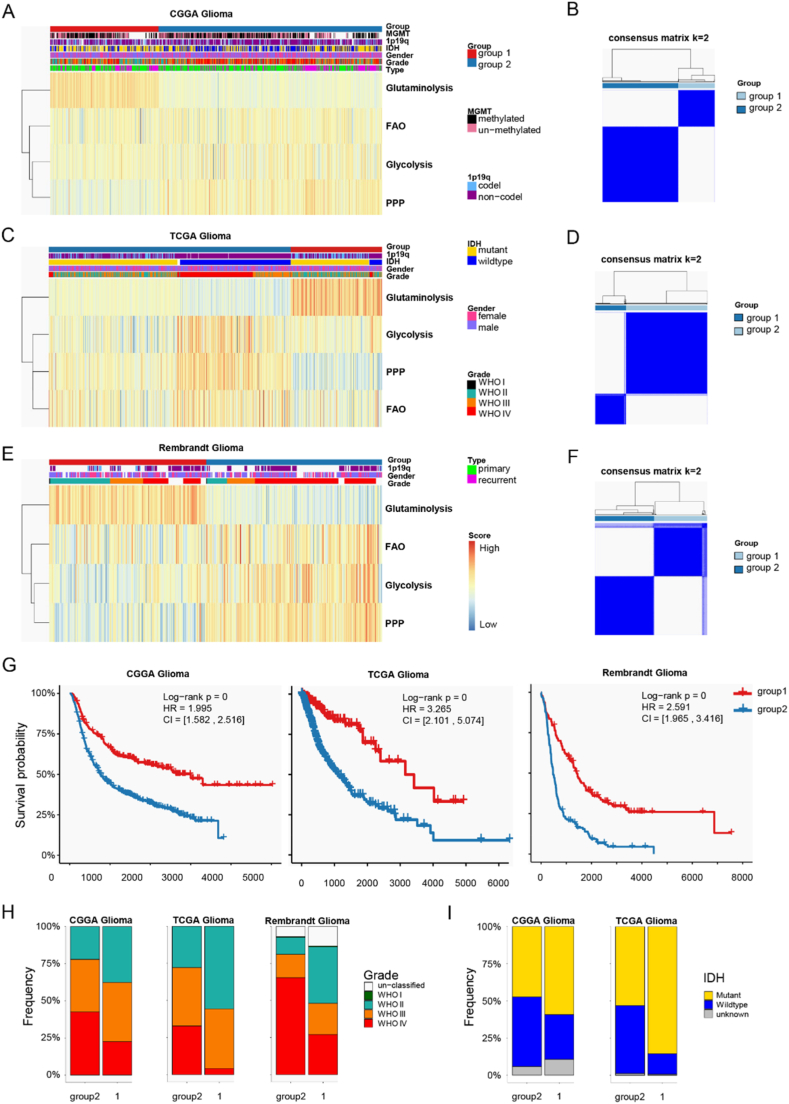
To explore metabolic differences in glioma, we focused on four central metabolic pathways: glutaminolysis, glycolysis, the pentose phosphate pathway (PPP), and fatty acid oxidation (FAO). We evaluated the ssGSEA scores to assess the activities of these pathways in each glioma sample. Glioma samples from the CGGA database were classified into two distinct metabolic phenotypes (Fig. 1A), which was further supported by optimal and robust clustering (Fig. 1B). This classification identified two glioma energy metabolic patterns: (1) glutaminolysis-dependent and (2) enriched in glycolysis, PPP, and FAO (Fig. 1A). Additionally, analysis of the TCGA and Rembrandt glioma cohorts externally validated the reproducibility and robustness of our metabolic classification (Fig. 1C–F). Survival analysis indicated that glioma samples in group 2 were associated with poorer clinical outcomes, while those in group 1 had better prognoses. The univariate Cox proportional hazards model confirmed that group 2 predicted unfavorable prognosis in glioma patients (hazard ratios (HR) 1.995 in the CGGA, 3.265 in TCGA, and 2.591 in the Rembrandt glioma database) (Fig. 1G).
Fig. 1.
Identification of distinct metabolic subtypes in glioma. A k-means clustering of glioma metabolism relying on 4 metabolic pathways evaluated by a ssGSEA method of the CGGA cohort. B Heatmap showing consensus clustering with the robust classification (k = 2) in the CGGA cohort. And the similar results were discovered in TCGA cohort (C, D) and the Rembrandt glioma cohort (E, F). G Kaplan-Meier curves of prognosis between metabolic subtypes in the CGGA, TCGA and the Rembrandt glioma cohorts. Log-rank test p values and HRs were shown. H Overlay of the energy metabolic classifier with glioma pathological grades in three independent cohorts. I Overlay of the energy metabolic classifier with IDH mutation status in the CGGA and TCGA cohort.
PPP, pentose phosphate pathway; FAO, fatty acid oxidation.
Previous studies have explored biologically discrete subsets associated with progression and prognosis [20,49]. To determine whether expression profiles based on metabolic classification could underlie differences between previously established phenotypes, we evaluated these glioma molecular subtypes for each glioma sample. Firstly, we compared the pathological grade distribution between group 1 and 2. It disclosed that group 2 contained a higher frequency of WHO IV tumors, whereas group 1 predominantly included WHO II samples (Fig. 1H). This was consistent with former survival analysis results as WHO IV glioma was one of the most malignant brain tumors. In addition, group 2 had more Classical (CL), Mesenchymal (ME) and Proneural (PN) subtypes, while group 1 mainly involved Neural (NE) subtype. As the expression pattern of glioma NE phenotype was recognized as the most similar to normal brain samples than CL, ME and PN subtype [20], group 1 was more similar to normal brain tissues than group 2. Then, we uncovered the difference between the metabolic classifiers and pan-glioma transcriptome subtypes established by Ceccarelli and others [49]. Group 2 had LGr1, LGr3 and LGr4 subtypes, but LGr2 was unique to group 1. As LGr2 glioma samples showed longer survival time [50], it matched that glioma metabolic phenotype 1 had better clinical outcome. IDH mutations occur most frequently in low grade glioma, and the reliance on glutaminolysis can be altered by IDH mutation status in glioma, since IDH1/2 mutated enzymes use α-ketoglutarate as a substrate to produce the oncometabolite 2-hydroxyglutarate, therefore a higher commitment of glutamine-derived glutamate is needed to produce α-ketoglutarate [51]. Thus, glutaminolysis is activated in gliomas with IDH mutations. And our results also revealed that group 1 with enhanced glutaminolysis level had more IDH mutations than group 2 (Fig. 1I). These results confirmed our metabolic classification of glioma was distinct from former established phenotypes and it had prognostic significance.
3.2. Metabolic classifier-specific biological functions of glioma
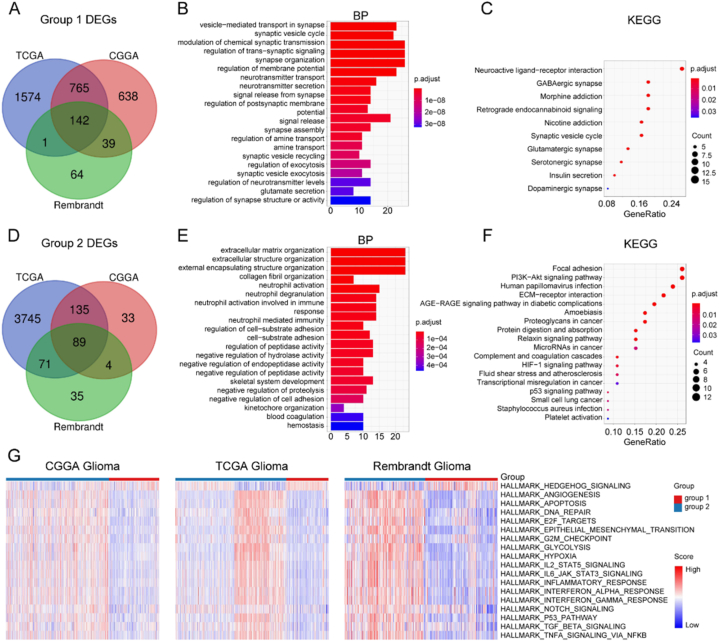
Then we finished DEG and enrichment analysis to further understand biological functions of different glioma metabolic phenotypes. The intersected DEGs among TCGA, the CGGA and the Rembrandt glioma cohort were 142 in metabolic group 1 and 89 in group2 (Fig. 2A–D, and detailed in, which were used in subsequent enrichment analysis. We found group 1 was related to synapse function, such as glutamate secretion, glutamatergic synapse et alia (Fig. 2B and C). This was identical to that metabolic group 1 was dependent on glutaminolysis (Fig. 1A–C, E). However, group 2 was associated with migration, immunity and hypoxia, such as extracellular matrix organization, extracellular structure organization, neutrophil activation involved in immune response, neutrophil-mediated immunity, HIF−1 signaling pathway et alia (Fig. 2E and F). Furthermore, the enrichment level of HALLMARK gene sets was compared between metabolic classifiers. While group 1 was enriched in the Hedgehog signaling pathway, group 2 was characterized by angiogenesis, apoptosis, proliferation, epithelial-mesenchymal transition (EMT), hypoxia, glycolysis and immunity (Fig. 2G).
Fig. 2.
Distinct functional features of the metabolic classifier. A Venn diagram showing 142 shared DEGs of metabolic subtype 1 among three independent cohorts. GO (B) and KEGG (C) analysis results of metabolic subtype 1 DEGs. D Venn diagram showing 89 shared DEGs of metabolic subtype 2 among three independent cohorts. GO (E) and KEGG (F) analysis results of metabolic subtype 2 DEGs. G Heatmap depicting normalized enrichment scores of HALLMARK pathways between metabolic subtypes.
DEG, differentially expressed gene; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
3.3. Metabolic classifier defines specific immune characteristics of glioma
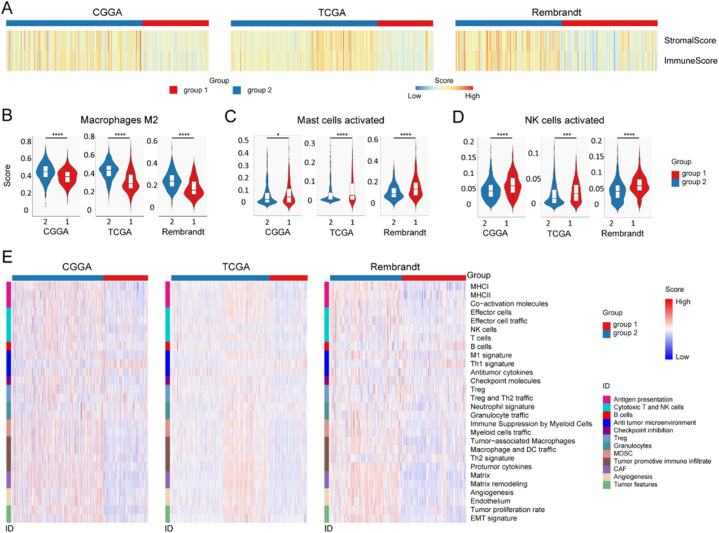
Because TME is a crucial mediator of cancer progression and therapeutic response, and metabolic classifier 2 was related to immunity, we explored the infiltrated immune cells and enrichment level of 29 TME signatures in glioma. The results indicated that metabolic group 1 had lower stromal and immune scores, but group 2 showed higher scores (Fig. 3A). In addition, the standard CIBERSORTx pipeline was utilized to further evaluate the relationship between glioma metabolism classifier and infiltration of 22 immune cells in the glioma microenvironment. Among the CGGA, TCGA and the Rembrandt glioma cohort, group 1 showed more activated mast cells and activated NK cells infiltration, but group 2 was featured with more infiltrating M2 type tumor-associated macrophages (TAMs) (Fig. 2B ∼ D). Because M2 TAM elicits a pro-tumorigenic property [33], glioma metabolic group 2 was related to tumor immunosuppression. Then, the TME features of our glioma metabolic classifier were studied. Group 1 demonstrated anti-tumor microenvironment, such as high enrichment level of Th1 signature (Fig. 3E). Conversely, group 2 was featured with immune suppression and tumor promotive immune infiltrate, for example myeloid cells traffic, TAM, macrophage and DC traffic, Th2 signature, and protumor cytokines (Fig. 3E). Furthermore, group 2 was also enriched in EMT, tumor proliferation rate, angiogenesis and matrix remodeling (Fig. 3E), which can promote tumor progression [52]. These results indicated group 2 had an immunosuppressive microenvironment.
Fig. 3.
Distinct immune features of the metabolic classifier. A Heatmap depicting normalized stromal and immune scores in three independent cohorts. B Infiltration of M2 macrophages was compared between metabolic subtypes. C Infiltration of activated mast cells was compared between metabolic subtypes. D Infiltration of activated NK cells was compared between metabolic subtypes. E Heatmap depicting normalized scores of 28 gene sets describing 12 cancer features.
∗, p < 0.05; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001; NK cells, natural killer cells; MHC, major histocompatibility complex; Th1, type 1 T helper; Treg, regulatory T cell; Th2, type 2 T helper; DC, dendritic cell; EMT, epithelial–mesenchymal transition.
3.4. Metabolic features of malignant and stromal cells in glioma microenvironment
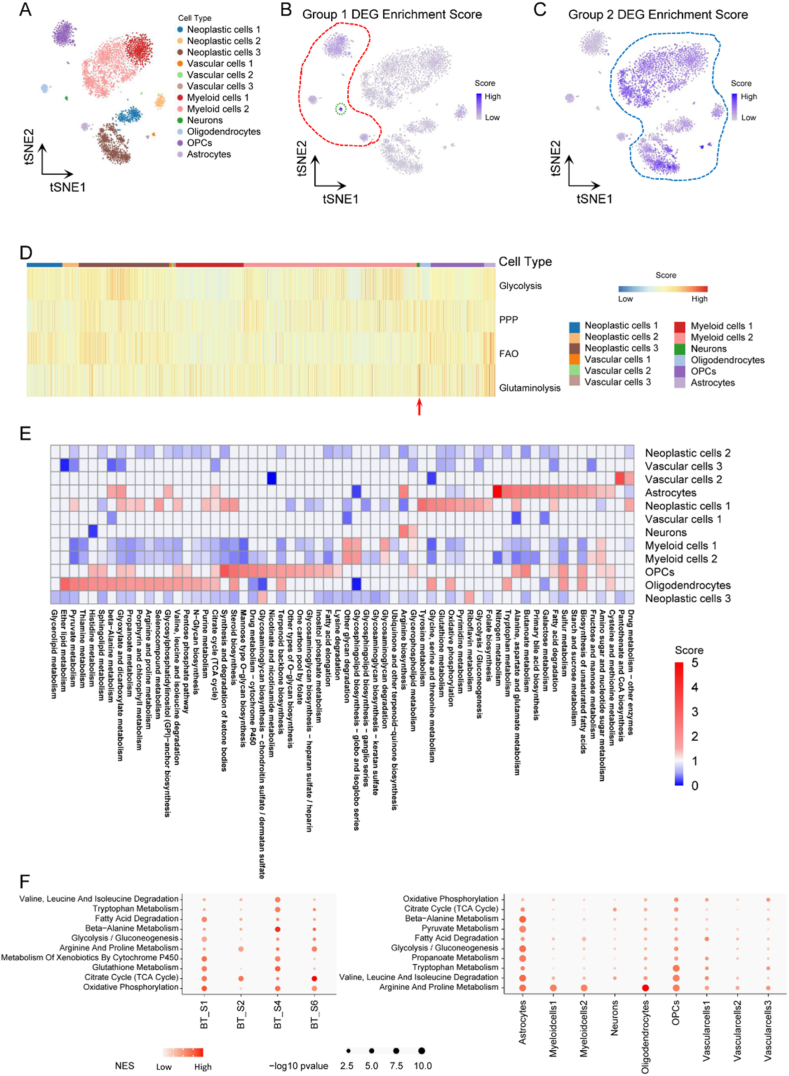
We then searched for whether the metabolic difference between the former two classifiers could be distinguished at single-cell resolution, which assisted in understanding the landscape of metabolic heterogeneity and TME. The GSE84465 database included tumor core and periphery region samples collected from 4 GBM patients. Twelve cell types were annotated and shown in tSNE (Fig. 4A). Moreover, metabolic classifier scores of each cell were estimated. It uncovered neurons, oligodendrocyte precursor cells (OPCs), astrocytes and oligodendrocytes had relatively higher metabolic group 1 DEG scores, among which neurons presented the highest scores (Fig. 4B). Interestingly, these four cell types were all the indigenously non-malignant brain cells, and it was identical to our former finding that metabolic classifier 1 was associated with synapse function (Fig. 2B and C). On the contrary, glioma neoplastic cells, tumor-associated vascular cells and tumor-associated myeloid cells appeared relatively higher metabolic group 2 DEG scores (Fig. 4C), which coincided that metabolic group 2 was related to migration, immunity, hypoxia and angiogenesis (Fig. 2E and F). Then, we estimated enrichment scores of the 4 key metabolic pathways in different cell types. The result confirmed that neurons relied on glutaminolysis (Fig. 4D).
Fig. 4.
Glioma metabolic heterogeneity at single-cell resolution. A tSNE of glioma cells from GEO: GSE84465. B tSNE of metabolic group 1 DEG enrichment score in glioma cells. C tSNE of metabolic group 2 DEG enrichment score in glioma cells. D Heatmap depicting normalized enrichment scores of 4 key metabolic pathways among glioma cells. E Metabolic pathway activities in cell types in the glioma dataset. F Metabolic pathways with highest contribution to the heterogeneity among malignant cells from different glioma samples in the glioma dataset (left column). Metabolic pathways with highest contribution to the heterogeneity among different cell types of non-malignant cells from the glioma dataset (right column).
tSNE, t-distributed stochastic neighbor embedding; OPC, oligodendrocyte precursor cell; DEG, differentially expressed gene; PPP, pentose phosphate pathway; FAO, fatty acid oxidation.
Furthermore, glioma metabolic landscape was depicted by taking a standard pipeline (Fig. 4E). It revealed that neurons relied on Glycerophospholipid metabolism and Arginine biosynthesis pathways; vascular cells 2 showed higher enrichment level at Drug metabolism−other enzymes and Pantothenate and CoA biosynthesis pathways; astrocytes relied on Biosynthesis of unsaturated fatty acids, Fatty acid degradation, Alanine, aspartate and glutamate metabolism, Nitrogen metabolism, Arginine biosynthesis, Citrate cycle (TCA cycle) et alia; OPCs were enriched in Biosynthesis of unsaturated fatty acids, Fatty acid degradation, Alanine, aspartate and glutamate metabolism, Fatty acid elongation, One carbon pool by folate, Citrate cycle (TCA cycle), Pentose phosphate pathway et alia; oligodendrocytes relied on Biosynthesis of unsaturated fatty acids, Oxidative phosphorylation, Citrate cycle (TCA cycle), Pentose phosphate pathway, Pyruvate metabolism and Ether lipid metabolism pathways et al. ; neoplastic cells 1 was enriched in Fatty acid degradation, Alanine, aspartate and glutamate metabolism, Glycolysis/Gluconeogenesis, Pyrimidine metabolism, Oxidative phosphorylation, Glutathione metabolism, Citrate cycle (TCA cycle), Glyoxylate and dicarboxylate metabolism and Pyruvate metabolism et alia; neoplastic cells 3 relied on Riboflavin metabolism and Glycosaminoglycan biosynthesis−chondroitin sulfate/dermatan sulfate pathways; Myeloid cells 1 was similar to Myeloid cells 2, and both of them showed high enrichment scores at Amino sugar and nucleotide sugar metabolism, Glycosaminoglycan degradation, Glycosphingolipid biosynthesis−globo and isoglobo series and Other glycan degradation pathways. However, neoplastic cells 2, vascular cells 1 and vascular cells 3 had lower metabolism levels than other cell types. Then, metabolic pathways enriched in genes with the highest contribution to the metabolic heterogeneities among malignant cells from different samples and among different cell types from all samples in glioma were also shown (Fig. 4F).
3.5. Glioma spatial heterogeneity at metabolic level
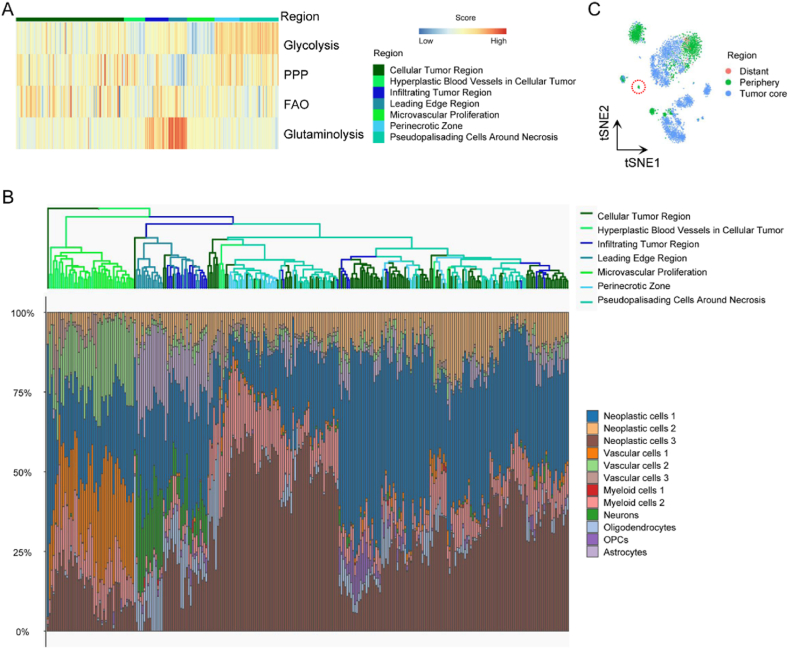
Spatial heterogeneity of glioma is the barrier for clinical treatment, and multiple TCGA transcriptional subtypes can coexist in close tumor regions [53]. Thus, we explored spatially metabolic heterogeneity in glioma. The key metabolic pathway enrichment levels were compared in different GBM tumor regions, and samples were clustered according to their cell composition. Necrotic foci with surrounded "pseudopalisading" cells is unique to malignant gliomas, and studies have demonstrated that pseudopalisades are severely hypoxic, overexpress HIF1, and secrete proangiogenic factors such as VEGF and IL8 [54]. Mechanisms underlying necrosis and hypoxia remain obscure, but necrosis is often accompanied by hypoxia [55]. While cancer glycolysis is reported as producing energy in an oxygen-independent manner, Robin and others reported that hypoxia increase the activities of all glycolytic enzymes and enhances glycolysis [56]. And our result demonstrated that the peri-necrotic zone and pseudo-palisading cells around necrosis had high enrichment levels in glycolysis (Fig. 5A). However, infiltrating tumor region and leading edge regions relied on the glutaminolysis pathway, and the leading edge region showed the highest glutaminolysis pathway enrichment score than other tumor regions (Fig. 5A). The glioma consisted of neoplastic cell, vascular cell, myeloid cell, neuron, oligodendrocyte, OPC and astrocyte (Fig. 4A), we further assessed cell type composition of GBM samples from the IGAP cohort. It uncovered an infiltrating tumor region and the leading edge region had more neurons, and the leading edge region showed the highest composition ratio of neurons (Fig. 5B). These revealed that neurons relied on glutaminolysis. In addition, cell composition in different glioma regions based on scRNA-seq data revealed that neuron was unique to periphery region (Fig. 5C). To sum up, neurons distributed in the glioma periphery region and were dependent on glutaminolysis, which also explained why glioma samples with high glutaminolysis pathway enrichment level exhibited better clinical outcome (Fig. 1G).
Fig. 5.
Distinct region feature of the metabolic classifier. A Heatmap depicting normalized enrichment scores of 4 key metabolic pathways among glioma samples with anatomic information. B Clustering of glioma samples from the IGAP dataset based on cell type compositions annotated by CIBERSORTx. C tSNE of glioma cells with anatomic information from GEO: GSE84465.
PPP, pentose phosphate pathway; FAO, fatty acid oxidation; OPC, oligodendrocyte precursor cell.
3.6. Altered signaling interaction between tumor core and periphery region in glioma
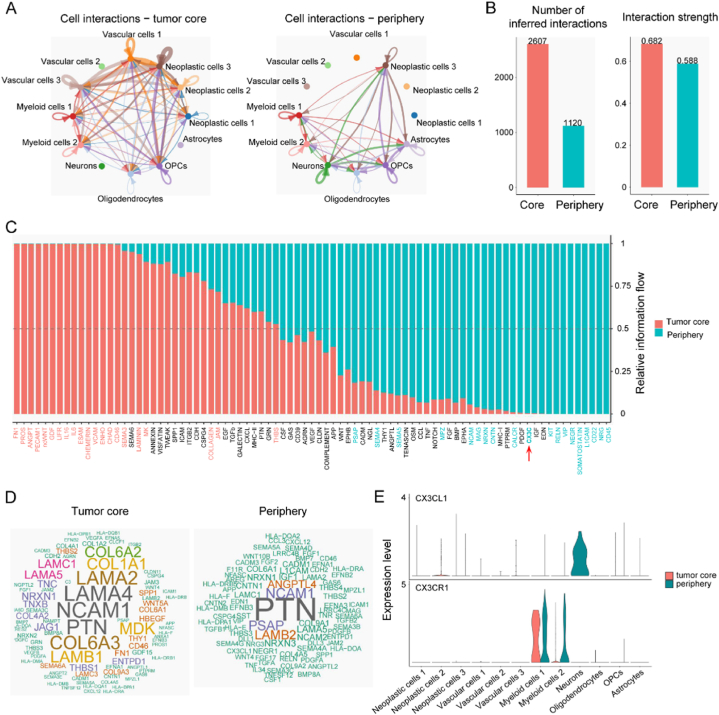
The former results revealed spatial metabolic heterogeneity in glioma which affected patient prognosis (Fig. 1, Fig. 5). We subsequently researched cell-cell interaction between tumor core and periphery region to study mechanisms underlying glioma prognosis differences affected by spatially metabolic heterogeneity. Interactions between cell types in the glioma core and periphery region were portrayed (Fig. 6A). The tumor core had more possible cell-cell interactions and stronger interaction strength than the periphery region (2607 vs. 1120, 0.682 vs. 0.588) (Fig. 6B). All communication probabilities in a given inferred network were summarized to calculate differences of overall interaction flows, and to rank significant signaling pathways. These colored red and green cell-cell interaction pathways were more enriched in tumor core and periphery region, respectively (Fig. 6C). Certain pathways, such as FN1, ANGPT, PECAM1, ncWNT, GDF, LIFR, IL16, IL6, ESAM, CHEMERIN, VCAM, SEMA3, LAMININ, COLLAGEN, JAM and THBS, prominently increased their information flow in tumor core region and promoted tumor progression leading to bleak prognosis. However, specific pathways, such as CD45, SOMATOSTATIN, VIP, RELN, CX3C, CALCR, NRXN, NCAM and SEMA5, predominantly functioned in the tumor periphery region and inhibited tumor prognosis, which resulted in better clinical outcomes. The CX3C pathway negatively controls glioma progression, for example CX3CR1/CX3CL1 axis inhibits cell invasion, and loss of CX3CR1 promotes gliomagenesis [57,58]. After obtaining an understanding of the general flow governing cell-cell interactions in the glioma core and periphery region, we studied molecular signaling mechanisms in detail to identify candidate pathways potentially underlying the altered cellular compositions and communications in different glioma regions. While LAMA4, LAMA2, COL6A3, COL1A1, LAMB1 and MDK were significantly up-regulated in the glioma core region, PSAP, ANGPTL4 and LAMB2 were increased in the glioma periphery region (Fig. 6D).
Fig. 6.
Difference of cell-cell communication between tumor core and periphery region. A Possible interactions among glioma cell types in tumor core (left column) and periphery region (right column). B Total number (left column) and strength (right column) of possible cell-cell interactions. C Significant signaling pathways were ranked based on their differences of overall information flow, calculated by summarizing all interaction probabilities. These colored red and green were more enriched in the tumor core and periphery region, respectively. D Word cloud showing upregulated signaling pathways in tumor core (left column) and periphery region (right column). Word size indicated the extent of enrichment. E Comparison of expression levels of CX3CL1 and CX3CR1 in glioma cells in tumor core and periphery region.
OPC, oligodendrocyte precursor cell.
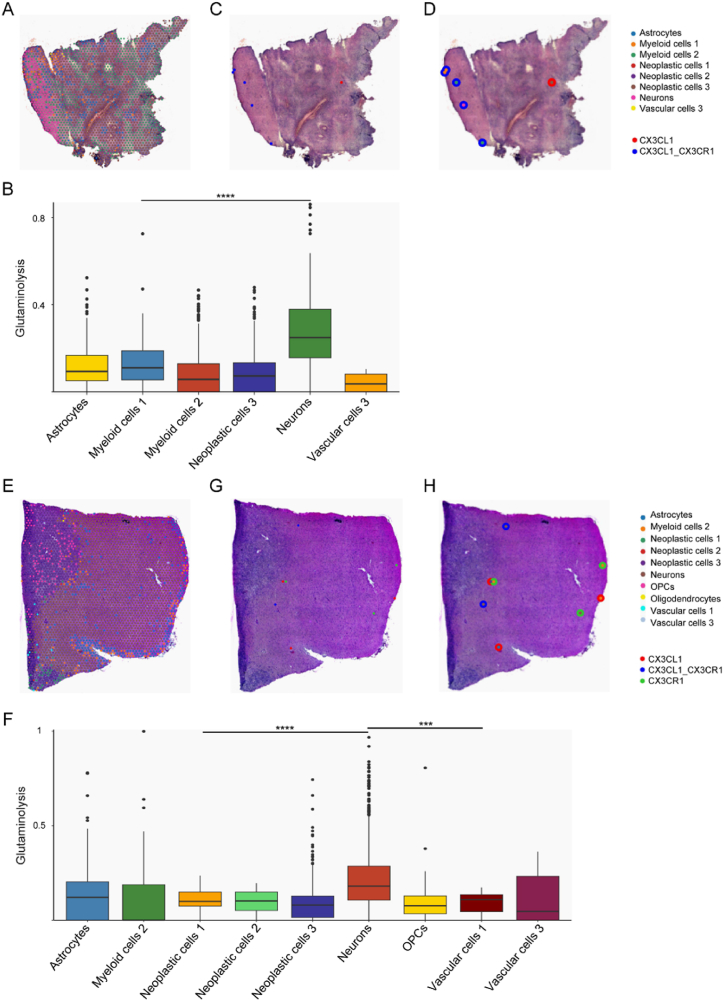
Then, we validated the CX3C pathway in single cell and spatial level using scRNA-seq and ST data. Firstly, CX3CL1 was uniquely expressed in peripheral neurons, and CX3CR1 expression was higher in glioma peripheral myeloid cells than these cells in the core region (Fig. 6E). Astrocytes, myeloid cells 1, myeloid cells 2, neoplastic cells 1, neoplastic cells 2, neoplastic cells 3, neurons and vascular cells 3 were annotated in sample UKF_243_RT (Fig. 7A). Neurons had the highest glutaminolysis pathway enrichment level (Fig. 7B), which was consistent with our former results (Fig. 4B–D). Furthermore, a significant CX3CL1/CX3CR1 pair was distributed in the area with more neurons, namely the periphery region (Fig. 7C and D). And these significant spots were annotated as myeloid cells 1, myeloid cells 2 and neurons, which was in harmony with the CX3CL1/CX3CR1 pair and was only expressed in myeloid cells and neurons (Fig. 6E). This finding was verified in the other glioma sample UKF_266_T (Fig. 7E ∼ H). However, no spot was mapped as a neuron, and the CX3CL1/CX3CR1 pair was negative in glioma sample UKF_313_RT. These findings confirmed that the CX3C interaction signal between neurons and myeloid cells was in the glioma periphery region.
Fig. 7.
Validation of CX3CL1/CX3CR1 pair by spatial transcriptomics data. A Cell2location-based cell type assignment in UKF_243 sample. B Enrichment score of glutaminolysis pathway among different cell-type spots in UKF_243 sample. C Binary CX3CL1/CX3CR1 pair co-expression plot for significant spots in UKF_243 sample. D Binary CX3CL1/CX3CR1 pair co-expression with spot annotations in UKF_243 sample. E Cell2location-based cell type assignment in UKF_266 sample. F Enrichment score of glutaminolysis pathway among different cell-type spots in UKF_266 sample. G Binary CX3CL1/CX3CR1 pair co-expression plot for significant spots in UKF_266 sample. H Binary CX3CL1/CX3CR1 pair co-expression with spot annotations in UKF_266 sample.
OPC, oligodendrocyte precursor cell.
4. Discussion
Considering high heterogeneity in glioma, previous studies stratify glioma by unsupervised clustering of tumors based on genomic and transcriptomic profiles, which bring valuable discoveries of many group-specific differences [3,20,49,59,60]. Recently, metabolomic profiling has become an informative method to clarify tumor heterogeneity. Fan and colleagues used prognosis-related metabolic genes to stratify diffuse low-grade glioma into three distinct tumor subtypes [7]. To our knowledge, our work was the first to construct a novel metabolic classifier for several glioma cohorts based on 4 core metabolic pathways, namely glutaminolysis, glycolysis, PPP and FAO. While subtype 1 glioma preferred to utilize glutaminolysis for survival, subtype 2 was characterized by an increasing dependence on glycolysis, PPP and FAO. By assessing the variations between these subtypes, specific changes in clinical outcome, biological function and immunity were revealed, which indicated metabolic heterogeneity should be considered when developing personalized treatment. In addition, glioma metabolic heterogeneity was explored at single cell and spatial resolution, and it uncovered that TME profoundly contributed to metabolic heterogeneity, and that neurons mainly distributed in tumor periphery region and relied on glutaminolysis for life. These results indicated metabolic subtype 1 with a better prognosis was the sample with a high proportion of neurons and distributed near the tumor margin. Underlying clinical survival differences between glioma metabolic subtypes were further disclosed through cell-cell interactions. Our work would extend molecular subtyping of glioma, strengthen understanding of metabolic heterogeneity in glioma and provide novel comprehension for developing personalized treatment.
Glioma is a metabolically dynamic entity that rewires and adapts its metabolic properties [61]. Glucose and glutamine are two essential nutrients for cell growth, and the metabolic heterogeneity of GBM in terms of glucose and glutamine utilization is reported by Oizel and others [62]. By analyzing glucose, glutamine, alanine, and aspartate consumption and production, and determining mitochondrial oxidation and extracellular acidification as measures of the glycolytic rate of eight primary GBM cultures, they reveal two GBM subtypes, termed “glutaminolysis-high” and “glutaminolysis-low” group. The glutaminolysis-high type GBM is featured as a high glutamine consumption rate and the ability to convert glutamine (and other nutrients as well) to NADH. While the glutaminolysis-high subtype lacks CD133 expression, stem cell marker, glutaminolysis-low GBM cultures exhibit a strong dependency on glycolysis and express CD133. Similar observations were made by Tardito and others [63]. Furthermore, Marin-Valencia and others profiled metabolites of tumors formed by patient-derived genetically GBM cells implanted into mice without prior cell culture, and found these implanted tumors consumed more glucose but exhibited minimal glutaminolysis for survival [64]. These glycolysis-high cells expressed a significant amount of c-Myc, pyruvate carboxylase, and glutamine synthetase, but only traces of glutaminase. Our metabolic classifier was consistent with the above-mentioned studies, and it was significantly related to glioma patient clinical outcome and had prognostic value (Fig. 1). Metabolic subtype 1, an upregulated subtype of glutaminolysis, was associated with longer overall survival, whereas subtype 2 with a high level of glycolysis, PPP and FAO had a worse outcome in glioma (Fig. 1Ã G). Both enhanced glutaminolysis and glycolysis pathways drive the rapid growth of GBM [65]. While glutaminolysis is strengthened to provide biosynthetic precursors for cancer cells, glycolysis is enhanced to meet the increasing energy demand of cancer cells. Thus, inhibiting glutaminolysis and glycolysis pathways is an emerging drug discovery approach to combat cancers. Glutaminolysis in cancer can be activated by c-Myc for tumorigenesis and also by p53 for tumor suppression [13], which may explain to some extent why metabolic subtype 1 is less aggressive than subtype 2.
No ingredients or metabolism pathways can meet all the needs for cell survival, thus it is important for tumor cells to coordinate metabolic pathways to adapt to different TMEs, including hypoxic environments. While glycolysis and glutaminolysis are the major mechanisms of ATP production in tumors, PPP and FAO are essential to provide building blocks for cell growth. The well-known link among these 4 metabolism pathways is the Citrate cycle (TCA cycle) [30]. And functional links between glycolysis and glutaminolysis were described by Smith, Wang and others [66,67]: it is mediated through i) some intermediate metabolites, for example pyruvate; ii) serine synthesis pathway as glutamate provides an amine group to 3-phosphopyruvate, a product converted from glycolysis intermediate 3-phosphoglycerate, to form 3-phosphoserine, the precursor of serine; iii) synthesis of nucleotide hexosamine, a substrate for protein glycosylation, which requires input from both glucose and glutamine. However, how these pathways influence each other is still controversial and needs further studies to clarify.
Then, we looked into DEGs between metabolic group 1 and 2 to explore the different biological functions of them. Shared DEGs of metabolic subtypes among three independent glioma cohorts were used in enrichment analysis. Metabolic subtype 2 was related to migration, immunity and hypoxic tumoral microenvironment, but subtype 1 was associated with synapse and glutamate metabolism (Fig. 2). Tumor hypoxia is a condition characterized by insufficient oxygen (O2) supply. Because the O2 level can span from 45 to 50 mmHg in end-capillary blood to 0.02 mmHg partial pressure in cytochromes, the O2 level was lower in the tumor core than the periphery region [68]. Neurons can be induced death when they are co-cultured with glioma cells owing to glioma cell-derived xCT, which promotes cortical hyperexcitability, brain edema and neuronal cell death [69]. Therefore, metabolic subtype 1 was in the periphery region, whereas subtype 2 was in the tumor core. This was further verified by glioma scRNA-seq data and bulk RNA-seq data with anatomic information (Fig. 5).
Growing studies advocate that metabolic heterogeneity affects immune cell function and might lead to immunotherapy failure in cancer. For example, excessive consumption of nutrients (e.g., glucose) by cancer cells will suppress T cell activation and effector function [70]. Immune infiltration analyses were performed, which revealed distinct immune states among metabolic classifiers. Consistently, metabolic group 2 with higher levels of glycolysis, PPP and FAO had more M2 TAMs infiltration, and was associated with higher enrichment of tumor promotive immune infiltrates, such as TAMs, macrophage and DC traffic, Th2 signature and protumor cytokines (Fig. 3B–E). Conversely, metabolic group 1 had more glutaminolysis enrichment, which displayed anti-tumor microenvironment, for example, more infiltrations of activated mast cells and activated NK cells, and higher enrichment level of Th1 signature (Fig. 3C ∼ E). These results proved a close connection between tumor metabolism reprogramming and immune microenvironment.
Given the complexity of TME and lack of knowledge about cell-cell communication induced metabolic characteristics as well as mitochondria biological features, we studied cell-cell interaction in different tumor regions with distinct metabolic phenotypes. Certain pathways, for example FN1, ANGPT, PECAM1, ncWNT, GDF, LIFR, IL16, IL6, ESAM, CHEMERIN, VCAM, SEMA3, LAMININ, COLLAGEN, JAM and THBS, prominently increased their information flow in tumor core region and promoted tumor progression leading to bleak prognosis. Viswanadhapalli and others declared that LIFR takes part in glioma initiation and recurrence [71]. And cerebrospinal fluid cytokine, IL16, levels are associated with macrophage infiltration into glioma tissues [72]. FN1, ANGPT and IL6 pathways activate cell proliferation and tumor growth in glioma [[73], [74], [75]]. Wu and colleagues proclaimed CHEMERIN enhances mesenchymal features of glioblastoma [76]. PECAM1 and SEMA3 are considered vital regulators involved in angiogenesis [77,78]. As reported, ncWNT, LAMININ, COLLAGEN, JAM, THBS, VCAM and ESAM pathways mediate migration and invasion in glioma and other cancers [[79], [80], [81], [82], [83], [84]]. Zahra and others validated that GDF affects cancer growth and progression, drug resistance, and metastasis [85]. However, specific pathways, for example CD45, SOMATOSTATIN, VIP, RELN, CX3C, CALCR, NRXN, NCAM and SEMA5, predominantly functioned in tumor periphery region and inhibited tumor prognosis, which resulted in better clinical outcome. TAM expressing high levels of CD45 are suppressive as well as pro-inflammatory characteristics in glioma [86]. SOMATOSTATIN inhibits production of vascular endothelial growth factor in human glioma cells and suppresses glioma angiogenesis [87]. Cochaud and colleagues stated neuropeptides of the VIP family inhibit glioblastoma cell invasion [88]. As compared to non-neoplastic tissue, both RELN and its main downstream effector DAB1 are silenced in glioblastoma and their RNA expressions are inversely correlated with pathological grade; RELN RNA expression is positively related to glioma patient survival and stimulation of RELN signaling can reduce proliferation and glioblastoma growth [89]. GBM patients with mutated CALCR or reduced transcript level predict poor prognosis [90]. In thyroid cancer cells, the NRXN pathway possesses a tumor suppressor potential through suppressing tumor growth [91]. NCAM and SEMA5 can inhibit human glioma cell motility [92,93]. The CX3C pathway was further verified in glioma using ST data (Fig. 7).
This study highlights the potential for targeting distinct metabolic pathway in the glioma core and periphery region. Although current evidence is limited, future research should explore this approach to develop more effective therapeutic strategies for glioma treatment.
5. Conclusions
In summary, this work provided a new perspective for elucidating metabolic heterogeneity of glioma. Our robustly new metabolic classifier proved that reprogrammed metabolism was associated with patient prognosis and distinct tumor region in glioma. The advent of single-cell RNA sequence and spatial transcriptomic had opened the door to explore glioma metabolic heterogeneity. Emerging technologies, including metabolomic-based single-cell or spatial profiling, would further improve the understanding of tumor cell state and TME features.
CRediT authorship contribution statement
Yong Xiao: Writing – original draft, Methodology, Formal analysis, Conceptualization. Mengjie Zhao: Writing – original draft. Ran Wang: Methodology, Formal analysis. Liang Liu: Data curation. Chong Xiang: Methodology. Taiping Li: Data curation. Chunfa Qian: Writing – review & editing. Hong Xiao: Writing – review & editing. Hongyi Liu: Writing – review & editing. Yuanjie Zou: Methodology, Conceptualization. Xianglong Tang: Supervision, Methodology, Conceptualization. Kun Yang: Supervision, Data curation, Conceptualization.
Ethical statement
Not applicable.
Ethical approval statement
Not applicable.
Data availability statement
Publicly available datasets were analyzed in this study. These data can be found at: the CGGA database (http://www.cgga.org.cn/), TCGA database (https://www.cancer.gov/ccg/research/genome-sequencing/tcga), the IGAP database (http://glioblastoma.alleninstitute.org/), and the GEO database (https://www.ncbi.nlm.nih.gov/geo) with accession number GSE108474 and GSE84465.
Funding
This research was funded by the National Natural Science Foundation of China (No. 82203637), Healthy Science and Technology Development Foundation of Nanjing (No. YKK22139), and Science and Technology Development Foundation of Nanjing Medical University (Nos. NMUB20210220 and NMUB20220131).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dr. Liangtao Zheng at Peking University for preparing the manuscript.
Contributor Information
Yuanjie Zou, Email: zouyuanjie0115@126.com.
Xianglong Tang, Email: xltang0326@163.com.
Kun Yang, Email: yk_nj@hotmail.com.
References
- 1.Lapointe S., Perry A., Butowski N.A. Primary brain tumours in adults. Lancet. 2018;392(10145):432–446. doi: 10.1016/S0140-6736(18)30990-5. [DOI] [PubMed] [Google Scholar]
- 2.Stringer B.W., et al. Human cerebrospinal fluid affects chemoradiotherapy sensitivities in tumor cells from patients with glioblastoma. Sci. Adv. 2023;9(43):eadf1332. doi: 10.1126/sciadv.adf1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee E., et al. Comparison of glioblastoma (GBM) molecular classification methods. Semin. Cancer Biol. 2018;53:201–211. doi: 10.1016/j.semcancer.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Brennan C.W., et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neftel C., et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849. doi: 10.1016/j.cell.2019.06.024. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L.B., et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell. 2021;39(4):509–528. doi: 10.1016/j.ccell.2021.01.006. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu F., et al. Metabolic expression profiling stratifies diffuse lower-grade glioma into three distinct tumour subtypes. Br. J. Cancer. 2021;125(2):255–264. doi: 10.1038/s41416-021-01418-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshmukh R., Allega M.F., Tardito S. A map of the altered glioma metabolism. Trends Mol. Med. 2021;27(11):1045–1059. doi: 10.1016/j.molmed.2021.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Lin G.L., et al. Therapeutic strategies for diffuse midline glioma from high-throughput combination drug screening. Sci. Transl. Med. 2019;11(519) doi: 10.1126/scitranslmed.aaw0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jothi J., Janardhanam V.A., Krishnaswamy R. Metabolic variations between low-grade and high-grade gliomas-profiling by (1)H NMR spectroscopy. J. Proteome Res. 2020;19(6):2483–2490. doi: 10.1021/acs.jproteome.0c00243. [DOI] [PubMed] [Google Scholar]
- 11.Yu D., et al. Metabolic alterations related to glioma grading based on metabolomics and lipidomics analyses. Metabolites. 2020;10(12) doi: 10.3390/metabo10120478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saurty-Seerunghen M.S., et al. Capture at the single cell level of metabolic modules distinguishing aggressive and indolent glioblastoma cells. Acta Neuropathol Commun. 2019;7(1):155. doi: 10.1186/s40478-019-0819-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Márquez J., et al. Glutamine addiction in gliomas. Neurochem. Res. 2017;42(6):1735–1746. doi: 10.1007/s11064-017-2212-1. [DOI] [PubMed] [Google Scholar]
- 14.Maus A., Peters G.J. Glutamate and alpha-ketoglutarate: key players in glioma metabolism. Amino Acids. 2017;49(1):21–32. doi: 10.1007/s00726-016-2342-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang M., Vousden K.H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer. 2016;16(10):650–662. doi: 10.1038/nrc.2016.81. [DOI] [PubMed] [Google Scholar]
- 16.Jiang P., Du W., Wu M. Regulation of the pentose phosphate pathway in cancer. Protein & Cell. 2014;5(8):592–602. doi: 10.1007/s13238-014-0082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye L., et al. Exosomes-regulated lipid metabolism in tumorigenesis and cancer progression. Cytokine Growth Factor Rev. 2023;73:27–39. doi: 10.1016/j.cytogfr.2023.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Comandatore A., et al. Lactate Dehydrogenase and its clinical significance in pancreatic and thoracic cancers. Semin. Cancer Biol. 2022;86:93–100. doi: 10.1016/j.semcancer.2022.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Z., et al. Comprehensive RNA-seq transcriptomic profiling in the malignant progression of gliomas. Sci. Data. 2017;4 doi: 10.1038/sdata.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verhaak R.G., et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gusev Y., et al. The REMBRANDT study, a large collection of genomic data from brain cancer patients. Sci. Data. 2018;5 doi: 10.1038/sdata.2018.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puchalski R.B., et al. An anatomic transcriptional atlas of human glioblastoma. Science. 2018;360(6389):660–663. doi: 10.1126/science.aaf2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louis D.N., et al. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 24.Zakharova G., et al. Reclassification of TCGA diffuse glioma profiles linked to transcriptomic, epigenetic, genomic and clinical data, according to the 2021 WHO CNS tumor classification. Int. J. Mol. Sci. 2022;24(1) doi: 10.3390/ijms24010157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liberzon A., et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prusinkiewicz M.A., et al. Survival-associated metabolic genes in human papillomavirus-positive head and neck cancers. Cancers. 2020;12(1) doi: 10.3390/cancers12010253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karasinska J.M., et al. Altered gene expression along the glycolysis-cholesterol synthesis Axis is associated with outcome in pancreatic cancer. Clin. Cancer Res. 2020;26(1):135–146. doi: 10.1158/1078-0432.CCR-19-1543. [DOI] [PubMed] [Google Scholar]
- 28.Possemato R., et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476(7360):346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016;2(5) doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu T.J., et al. Bulk and single-cell transcriptome profiling reveal the metabolic heterogeneity in human breast cancers. Mol. Ther. 2021;29(7):2350–2365. doi: 10.1016/j.ymthe.2021.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao Y., et al. Single-cell transcriptomics revealed subtype-specific tumor immune microenvironments in human glioblastomas. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.914236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ritchie M.E., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malik S., et al. Antitumor efficacy of a sequence-specific DNA-targeted γPNA-based c-Myc inhibitor. Cell Reports Medicine. 2024;5(1) doi: 10.1016/j.xcrm.2023.101354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M., et al. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshihara K., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newman A.M., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37(7):773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bagaev A., et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell. 2021;39(6):845–865. doi: 10.1016/j.ccell.2021.04.014. e7. [DOI] [PubMed] [Google Scholar]
- 41.Darmanis S., et al. Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017;21(5):1399–1410. doi: 10.1016/j.celrep.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao Y., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587. doi: 10.1016/j.cell.2021.04.048. e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao Z., Dai Z., Locasale J.W. Metabolic landscape of the tumor microenvironment at single cell resolution. Nat. Commun. 2019;10(1):3763. doi: 10.1038/s41467-019-11738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin S., et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravi V.M., et al. Spatially resolved multi-omics deciphers bidirectional tumor-host interdependence in glioblastoma. Cancer Cell. 2022;40(6):639–655. doi: 10.1016/j.ccell.2022.05.009. e13. [DOI] [PubMed] [Google Scholar]
- 46.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(1):15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kleshchevnikov V., et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat. Biotechnol. 2022;40(5):661–671. doi: 10.1038/s41587-021-01139-4. [DOI] [PubMed] [Google Scholar]
- 48.Pham D., et al. Cell-Cell Interactions and Spatial Trajectories within Undissociated Tissues. 2020. stLearn: integrating spatial location, tissue morphology and gene expression to find cell types. [Google Scholar]
- 49.Ceccarelli M., et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao Y., et al. CD44-Mediated poor prognosis in glioma is associated with M2-polarization of tumor-associated macrophages and immunosuppression. Frontiers in Surgery. 2022;8 doi: 10.3389/fsurg.2021.775194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martins F., et al. Take advantage of glutamine anaplerosis, the kernel of the metabolic rewiring in malignant gliomas. Biomolecules. 2020;10(10) doi: 10.3390/biom10101370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quail D.F., Joyce J.A. The microenvironmental landscape of brain tumors. Cancer Cell. 2017;31(3):326–341. doi: 10.1016/j.ccell.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Q., et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2018;33(1):152. doi: 10.1016/j.ccell.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rong Y., et al. ‘Pseudopalisading’ necrosis in glioblastoma: a familiar morphologic feature that links vascular pathology, hypoxia, and angiogenesis. J. Neuropathol. Exp. Neurol. 2006;65(6):529–539. doi: 10.1097/00005072-200606000-00001. [DOI] [PubMed] [Google Scholar]
- 55.Brat D.J., Van Meir E.G. Vaso-occlusive and prothrombotic mechanisms associated with tumor hypoxia, necrosis, and accelerated growth in glioblastoma. Lab. Invest. 2004;84(4):397–405. doi: 10.1038/labinvest.3700070. [DOI] [PubMed] [Google Scholar]
- 56.Robin E.D., Murphy B.J., Theodore J. Coordinate regulation of glycolysis by hypoxia in mammalian cells. J. Cell. Physiol. 2005;118(3):287–290. doi: 10.1002/jcp.1041180311. [DOI] [PubMed] [Google Scholar]
- 57.Sciume G., et al. CX3CR1/CX3CL1 axis negatively controls glioma cell invasion and is modulated by transforming growth factor-beta1. Neuro Oncol. 2010;12(7):701–710. doi: 10.1093/neuonc/nop076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng X., et al. Loss of CX3CR1 increases accumulation of inflammatory monocytes and promotes gliomagenesis. Oncotarget. 2015;6(17):15077–15094. doi: 10.18632/oncotarget.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun Y., et al. A glioma classification scheme based on coexpression modules of EGFR and PDGFRA. Proc. Natl. Acad. Sci. U. S. A. 2014;111(9):3538–3543. doi: 10.1073/pnas.1313814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan W., et al. Molecular classification of gliomas based on whole genome gene expression: a systematic report of 225 samples from the Chinese Glioma Cooperative Group. Neuro Oncol. 2012;14(12):1432–1440. doi: 10.1093/neuonc/nos263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shibao S., et al. Metabolic heterogeneity and plasticity of glioma stem cells in a mouse glioblastoma model. Neuro Oncol. 2018;20(3):343–354. doi: 10.1093/neuonc/nox170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oizel K., et al. Efficient mitochondrial glutamine targeting prevails over glioblastoma metabolic plasticity. Clin. Cancer Res. 2017;23(20):6292–6304. doi: 10.1158/1078-0432.CCR-16-3102. [DOI] [PubMed] [Google Scholar]
- 63.Tardito S., et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat. Cell Biol. 2015;17(12):1556–1568. doi: 10.1038/ncb3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marin-Valencia I., et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metabol. 2012;15(6):827–837. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Akins N.S., Nielson T.C., Le H.V. Inhibition of glycolysis and glutaminolysis: an emerging drug discovery approach to combat cancer. Curr. Top. Med. Chem. 2018;18(6):494–504. doi: 10.2174/1568026618666180523111351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang L., et al. Molecular link between glucose and glutamine consumption in cancer cells mediated by CtBP and SIRT4. Oncogenesis. 2018;7(3) doi: 10.1038/s41389-018-0036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith H., et al. The effects of severe hypoxia on glycolytic flux and enzyme activity in a model of solid tumors. J. Cell. Biochem. 2016;117(8):1890–1901. doi: 10.1002/jcb.25488. [DOI] [PubMed] [Google Scholar]
- 68.Amberger-Murphy V. Hypoxia helps glioma to fight therapy. Curr. Cancer Drug Targets. 2009;9(3):381–390. doi: 10.2174/156800909788166637. [DOI] [PubMed] [Google Scholar]
- 69.Radin D.P., Tsirka S.E. Interactions between tumor cells, neurons, and microglia in the glioma microenvironment. Int. J. Mol. Sci. 2020;21(22) doi: 10.3390/ijms21228476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Macintyre A.N., et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metabol. 2014;20(1):61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Viswanadhapalli S., et al. Targeting LIF/LIFR signaling in cancer. Genes Dis. 2022;9(4):973–980. doi: 10.1016/j.gendis.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kemmerer C.L., et al. Cerebrospinal fluid cytokine levels are associated with macrophage infiltration into tumor tissues of glioma patients. BMC Cancer. 2021;21(1):1108. doi: 10.1186/s12885-021-08825-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen J., et al. Identification of new therapeutic targets and natural compounds against diffuse intrinsic pontine glioma (DIPG) Bioorg. Chem. 2020;99 doi: 10.1016/j.bioorg.2020.103847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reiss Y., Machein M.R., Plate K.H. The role of angiopoietins during angiogenesis in gliomas. Brain Pathol. 2005;15(4):311–317. doi: 10.1111/j.1750-3639.2005.tb00116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang H., et al. Targeting interleukin 6 signaling suppresses glioma stem cell survival and tumor growth. Stem Cell. 2009;27(10):2393–2404. doi: 10.1002/stem.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu J., et al. Chemerin enhances mesenchymal features of glioblastoma by establishing autocrine and paracrine networks in a CMKLR1-dependent manner. Oncogene. 2022;41(21):3024–3036. doi: 10.1038/s41388-022-02295-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiao B., et al. Class-3 semaphorins: potent multifunctional modulators for angiogenesis-associated diseases. Biomed. Pharmacother. 2021;137 doi: 10.1016/j.biopha.2021.111329. [DOI] [PubMed] [Google Scholar]
- 78.Feng Y.M., Chen X.H., Zhang X. Roles of PECAM-1 in cell function and disease progression. Eur. Rev. Med. Pharmacol. Sci. 2016;20(19):4082–4088. [PubMed] [Google Scholar]
- 79.Gulaia V., et al. Molecular mechanisms governing the stem cell's fate in brain cancer: factors of stemness and quiescence. Front. Cell. Neurosci. 2018;12:388. doi: 10.3389/fncel.2018.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kawataki T., et al. Laminin isoforms and their integrin receptors in glioma cell migration and invasiveness: evidence for a role of alpha5-laminin(s) and alpha3beta1 integrin. Exp. Cell Res. 2007;313(18):3819–3831. doi: 10.1016/j.yexcr.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 81.Tenan M., et al. Cooperative expression of junctional adhesion molecule-C and -B supports growth and invasion of glioma. Glia. 2010;58(5):524–537. doi: 10.1002/glia.20941. [DOI] [PubMed] [Google Scholar]
- 82.Seliger C., et al. Lactate-modulated induction of THBS-1 activates transforming growth factor (TGF)-beta2 and migration of glioma cells in vitro. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0078935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li M., et al. miR-7 reduces breast cancer stem cell metastasis via inhibiting RELA to decrease ESAM expression. Mol Ther Oncolytics. 2020;18:70–82. doi: 10.1016/j.omto.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen Q., Massague J. Molecular pathways: VCAM-1 as a potential therapeutic target in metastasis. Clin. Cancer Res. 2012;18(20):5520–5525. doi: 10.1158/1078-0432.CCR-11-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hasanpour Segherlou Z., et al. GDF-15: diagnostic, prognostic, and therapeutic significance in glioblastoma multiforme. J. Cell. Physiol. 2021;236(8):5564–5581. doi: 10.1002/jcp.30289. [DOI] [PubMed] [Google Scholar]
- 86.Brandenburg S., et al. Myeloid cells expressing high level of CD45 are associated with a distinct activated phenotype in glioma. Immunol. Res. 2017;65(3):757–768. doi: 10.1007/s12026-017-8915-1. [DOI] [PubMed] [Google Scholar]
- 87.Mentlein R., et al. Somatostatin inhibits the production of vascular endothelial growth factor in human glioma cells. Int. J. Cancer. 2001;92(4):545–550. doi: 10.1002/ijc.1223. [DOI] [PubMed] [Google Scholar]
- 88.Cochaud S., et al. Neuropeptides of the VIP family inhibit glioblastoma cell invasion. J. Neuro Oncol. 2015;122(1):63–73. doi: 10.1007/s11060-014-1697-6. [DOI] [PubMed] [Google Scholar]
- 89.Schulze M., et al. RELN signaling modulates glioblastoma growth and substrate-dependent migration. Brain Pathol. 2018;28(5):695–709. doi: 10.1111/bpa.12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pal J., et al. Loss-of-Function mutations in calcitonin receptor (CALCR) identify highly aggressive glioblastoma with poor outcome. Clin. Cancer Res. 2018;24(6):1448–1458. doi: 10.1158/1078-0432.CCR-17-1901. [DOI] [PubMed] [Google Scholar]
- 91.Ma C., Zhang Y. NRXN2 possesses a tumor suppressor potential via inhibiting the growth of thyroid cancer cells. Comput. Math. Methods Med. 2021;2021 doi: 10.1155/2021/7993622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Z., et al. MiR-30a-5p is induced by Wnt/beta-catenin pathway and promotes glioma cell invasion by repressing NCAM. Biochem. Biophys. Res. Commun. 2015;465(3):374–380. doi: 10.1016/j.bbrc.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 93.Li X., Lee A.Y. Semaphorin 5A and plexin-B3 inhibit human glioma cell motility through RhoGDIalpha-mediated inactivation of Rac1 GTPase. J. Biol. Chem. 2010;285(42):32436–32445. doi: 10.1074/jbc.M110.120451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found at: the CGGA database (http://www.cgga.org.cn/), TCGA database (https://www.cancer.gov/ccg/research/genome-sequencing/tcga), the IGAP database (http://glioblastoma.alleninstitute.org/), and the GEO database (https://www.ncbi.nlm.nih.gov/geo) with accession number GSE108474 and GSE84465.