Abstract
We report here studies of tryptophan (Trp) solvation dynamics in water and in the Pyrococcus furiosus rubredoxin protein, including the native and its apo and denatured forms. We also report results on energy transfer from Trp to the iron-sulfur [Fe-S] cluster. Trp fluorescence decay with the onset of solvation dynamics of the chromophore in water was observed with femtosecond resolution (≈160 fs; 65% component), but the emission extended to the picosecond range (1.1 ps; 35% component). In contrast, the decay is much slower in the native rubredoxin; the Trp fluorescence decay extends to 10 ps and longer, reflecting the local rigidity imposed by residues and by the surface water layer. The dynamics of resonance energy transfer from the two Trps to the [Fe-S] cluster in the protein was observed to follow a temporal behavior characterized by a single exponential (15–20 ps) decay. This unusual observation in a protein indicates that the resonance transfer is to an acceptor of a well-defined orientation and separation. From studies of the mutant protein, we show that the two Trp residues have similar energy-transfer rates. The critical distance for transfer (R0) was determined, by using the known x-ray data, to be 19.5 Å for Trp-36 and 25.2 Å for Trp-3, respectively. The orientation factor (κ2) was deduced to be 0.13 for Trp-36, clearly indicating that molecular orientation of chromophores in the protein cannot be isotropic with κ2 value of 2/3. These studies of solvation and energy-transfer dynamics, and of the rotational anisotropy, of the wild-type protein, the (W3Y, I23V, L32I) mutant, and the fmetPfRd variant at various pH values reveal a dynamically rigid protein structure, which is probably related to the hyperthermophilicity of the protein.
Tryptophan (Trp) is the most important fluorophore among amino acid residues for optical probing of proteins. However, Trp fluorescence is complex because of different rotamers in the ground state and the two nearly degenerate electronic states (1La, 1Lb) with perpendicular transition moments. Accordingly, numerous studies (1–10) have focused on the lifetime, quantum yield, Stokes shift, and fluorescence anisotropy. Most of these studies were made with picosecond or nanosecond time resolution (3, 6–10). To probe the local protein dynamics, Trp solvation by neighboring soft/rigid water molecules, or by other polar amino acid residues, must be resolved on the femtosecond time scale. Moreover, such studies are important for examining the nature of resonance energy transfer (RET) that is used for deducing distances and orientations between the Trp and quenchers in the protein (e.g., see refs. 3 and 10, and references therein).
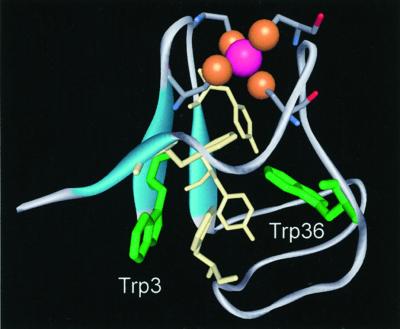
We choose the hyperthermophilic iron-sulfur protein, Pyrococcus furiosus rubredoxin (PfRd), as a prototype system (Fig. 1). The high-resolution x-ray crystallographic structures of PfRd at 0.95 Å and its formylmethionine variant (fmetPfRd) at 1.2 Å, have been recently reported (11). PfRd is a small protein of 53 amino acid residues with an iron atom coordinated by the sulfur atoms of four cysteine side chains and functions as an electron-transfer protein (12). It is approximately ellipsoidal in overall shape with a hydrophilic tail protruding into the solvent at the C terminus region. The structure consists of a three-stranded antiparallel β-sheet with a hydrophobic core containing six aromatic residues (Fig. 1); two of them are Trp-3 and Trp-36. The [Fe-S] cluster has a strong charge-transfer absorption band at 380 nm (13, 14), which overlaps with the Trp emission. Thus, RET between Trp and the cluster is expected and has been observed in other iron-sulfur proteins (15).
Figure 1.
Ribbon presentation of high-resolution x-ray structure of hyperthermophilic Pyrococcus furiosus rubredoxin (11). Six aromatic amino acid residues in the hydrophobic core are, along the primary sequence, Trp-3, Tyr-10, Tyr-12, Phe-29, Trp-36, and Phe-48. A tail at the C terminus protrudes into the solvent.
Experimental Methods
All experimental measurements were carried out by using the femtosecond-resolved fluorescence up-conversion technique (16). All protein samples were generously provided by the group of Michael W. W. Adams at the University of Georgia. l-Trp, N-acetyl-l-tryptophanamide (NATA) and N-acetyl-l-Trp ethyl ester (NATEE) were purchased from Sigma. Ultrapure guanidine hydrochloride (GdnHCl) was obtained from Baker, and trichloroacetic acid from Fisher. All chemicals were used as received.
The iron-sulfur protein rubredoxin we obtained was isolated and purified from Pyrococcus furiosus. The protein was concentrated and stored in 50 mM Tris buffer (pH 8) with 0.3 M NaCl at −20°C. Its purity was checked routinely by measuring the ratio of the extinction coefficient at 280 nm and 380 nm (17). For most experiments, the formylmethionine variant of PfRd (fmetPfRd) was used with a concentration of 0.3 mM in a 20 mM phosphate buffer at pH 7. Aside from the wild-type PfRd, we also examined the (W3Y, I23V, L32I) mutant of fmetPfRd in which the Trp-3 was replaced by Tyr, leaving only one Trp (Trp-36) in the protein.
Apo-fmetPfRd was prepared (17) first by denaturing the holo-protein in 10% trichloroacetic acid. After a few hours, the precipitated apo-fmetPfRd was suspended twice in 6% trichloroacetic acid and then dissolved in 3 M GdnHCl. After renaturation of the protein by dialysis against 50 mM Tris buffer at pH 8, the apo-PfRd solution was clear. Circular dichroism showed that the apo protein has a similar secondary structure to the holo form.
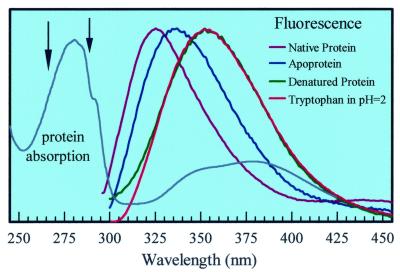
The steady–state absorption and fluorescence spectra (265-nm excitation) are shown in Fig. 2. The distinctive absorption spectrum has an intense band at 280 nm (Trp absorption). Other bands at 380 nm, 490 nm, and 570 nm (not shown) arise from charge transfer within the [Fe-S] cluster, from the cysteinyl thiolates to Fe(III) (13, 14). The mutant and the variant of PfRd show the same absorption as the wild-type PfRd, but their thermostabilities are slightly different (18). The fluorescence emission of apo-fmetPfRd peaks at 337 nm. For the native protein, the Trp emission at the red side strongly overlaps with the iron-sulfur charge-transfer absorption and thus it is quenched through RET by several orders of magnitude. Accordingly, the resulting spectrum is blue-shifted, peaking at 325 nm (Fig. 2) because of the comparable time scales of energy transfer and solvation (see below).
Figure 2.
Absorption of the PfRd protein and normalized fluorescence emission for different systems. Note that the spectral overlap between the Trp emission in apo-PfRd and the [Fe-S] cluster absorption in PfRd. The fluorescence intensity in PfRd is actually much weaker than that in apo-PfRd. The arrows mark two excitation wavelengths used in this study, 265 nm and 288 nm.
The fluorescence emission of Trp in buffer solution shifts to the red, 352 nm, because of solvation. Clearly, the Trp residues buried in apo-fmetPfRd (337 nm) are in a more hydrophobic environment than in the buffer solution. The denatured form of fmetPfRd has a similar emission spectrum to that of Trp in buffer solution, except for a tail at the blue side because of the tyrosine emission of the protein. To mimic the peptide bond in protein, we also studied the model systems: NATA in a phosphate buffer solution (30 mM) at pH 2 and NATEE in a solvent of p-dioxane. The emission spectrum of NATA is similar to that of Trp with a slight shift to the red side, by 5 nm; for NATEE it shifts to the blue side and peaks at 330 nm.
Results and Discussion
Excited States of Trp.
The two electronic excited states, 1La and 1Lb, are both involved in the absorption and fluorescence emission. Two transition dipoles are nearly perpendicular to each other, and the direction of the 1Lb transition dipole is along the side chain (19). The 1La state has a larger static dipole moment than its ground state so it is more sensitive to solvation. In polar solvents, the 1La state is red-shifted and becomes lower in energy than the 1Lb state. The observed steady–state fluorescence, especially at wavelengths longer than 340 nm, is mainly from the 1La state (8). For each electronic state, there are three rotamers of the alanyl side chain of Trp (1). The reported two principal lifetimes, ≈500 ps and ≈3 ns in bulk solution, were attributed to different conformers, and, as discussed below, our observed long-time component (>500 ps) at different wavelengths is an average value of the two lifetimes.
Solvation Dynamics of Trp.
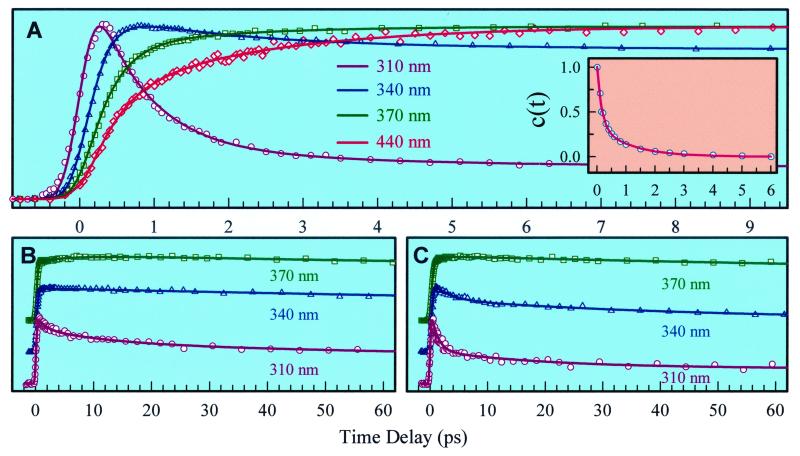
Fig. 3A shows femtosecond-resolved fluorescence transients of Trp in phosphate buffer at pH 2 with a systematic series of detection wavelengths. All transients have three distinct time scales. At the blue end of 310 nm, the signal decays with time constants of 700 fs (78%), 3.13 ps (8.6%), and ≈518 ps (13.4%); at 340 nm, it first rises in 200 fs and then decays in 1.8 ps (20%) and ≈865 ps (80%); at 370 nm, it rises in 330 fs (86%) and 1.9 ps (14%) and then decays in ≈1.12 ns; and at the red end of 440 nm, the transient rises in 410 fs (57%) and 2.21 ps (43%) and finally decays in ≈1.45 ns. We also studied NATA in the same buffer and similar transients were observed. For NATEE in p-dioxane, we observed slower temporal behaviors, e.g., at 310 nm the signal decays in 4.7 ps (58%) and ≈500 ps (42%) and at 340 nm it rises in 420 fs (72%) and 11 ps (28%) and then decays in ≈3.2 ns. These results are indicative of solvation.
Figure 3.
(A) Normalized, femtosecond-resolved fluorescence decay of Trp in buffer solution at pH 2 with a series of wavelength detection. (Inset) The constructed solvent response c(t). Normalized fluorescence decay of Trp in the apo-fmetPfRd protein (B) and in its denatured state (C). Note that the transient at 340-nm emission in B decays only with a long lifetime of ≈530 ps.
The initial femtosecond decay at the blue side and the rise at the red side dominantly result from solvation processes, and are not due to the electronic relaxation (1La and 1Lb coupling) and vibrational cooling. This conclusion was based on the following observations: First, the internal conversion between 1La and 1Lb occurs in <100 fs as measured by ultrafast anisotropy decay (see below). Recent studies deduced a time constant of 10–40 fs for the internal conversion of 5-methoxyindole in hexadecane (20). Second, Trp emission strongly depends on solvent polarity. From p-dioxane (or in the protein) to buffer solution, the emission peak shifts from 330 nm (337 nm) to 357 nm. Third, the initial Trp dynamics also show different temporal behaviors in different solvents. In p-dioxane it occurs in several picoseconds (21) whereas in water it is on the femtosecond time scale, as expected for water solvation (22, 23). Finally, all transients gated at various emission wavelengths are nearly independent of excitation wavelength (265 nm and 288 nm), ruling out a large contribution from vibrational cooling.
By following the time-resolved emission (Stokes shift with time), we constructed the correlation function (solvent response function) to obtain the solvation time: c(t) = [ν(t) − ν(∞)]/[ν(0) − ν(∞)], where ν(t), ν(0), and ν(∞) are time-resolved emission maxima in cm−1, respectively. The c(t) function, shown in Fig. 3A Inset gives an apparent biexponential behavior: 160 fs (65%) and 1.1 ps (35%). These two solvation times are close to the reported values (126 fs and 880 fs) in bulk water (23). The former reflects the librational motion of water molecules and the latter represents their diffusive motion.
After establishing the Trp solvation dynamics in bulk water, we studied Trp solvation in the apo-fmetPfRd and the denatured form of the protein. The results are given in Fig. 3 B and C, respectively. In apo-fmetPfRd, the transient at 310 nm decays in 1.2 ps (17%), 12 ps (26%), and ≈320 ps (57%); for 340 nm, it first rises in 200 fs and then decays ≈530 ps and for 370 nm, it rises in 200 fs (89%), 5.6 ps (11%) and then decays ≈640 ps. Clearly, Trp buried in apo-fmetPfRd shows multiexponential temporal behavior and the solvation processes become slower. According to the x-ray structure (11), both Trp-3 and Trp-36 face the hydrophobic core, but they are partially exposed to the protein surface and also interact with neighboring polar (Tyr) and negatively charged (Asp and Glu) residues. Thus, the observed multiple time scales represent dynamical motions of both water molecules in the rigid water layer around the protein surface and the interacting residues.
Very recently, Vivian and Callis (24) have carried out extensive theoretical studies of the interactions of Trp with water and other amino acid residues in proteins for interpretation of the fluorescence shifts. The simulation of a single Trp in proteins under the partial exposure to water gave a time scale of several picoseconds for the decay of the shifted spectral components because of the large-amplitude motions of the protein backbone and side chains and/or wholesale rearrangement of nearby hydrogen-bonded water clusters, consistent with our observation of solvation occurring up to 10 ps or longer. By placing an extrinsic dye probe in a protein pocket, solvation dynamics of polar amino acid residues or rigid water molecules on slower time scales also have been recently reported (25, 26). Ultrafast solvation dynamics in protein by using the intrinsic probe, Trp, were not reported before.
Fig. 3C shows the dynamics of Trp residues in the denatured fmetPfRd obtained by addition of 6 M GdnHCl into the protein solution at pH 2. At 310 nm, the transient decays in 1.2 ps (56%), 19 ps (22%), and ≈818 ps (22%); at 340 nm, it first rises in 280 fs and then decays 2.83 ps (25%), 40 ps (25%), and ≈1 ns (50%); and at 370 nm, it initially rises in 300 fs (91%), 1.52 ps (9%) and then decays with long components (≥1 ns). These results show a faster solvation process in the denatured fmetPfRd than in its apo form, consistent with the fact that both Trp-3 and Trp-36 are exposed to more water molecules in its denatured state. This is also evident from the emission spectra shifting from 337 nm in the apo form to 352 nm in the denatured form; see Fig. 2. However, the solvation process in the denatured state is much slower than that of Trp in water. Under 6 M GdnHCl, the ratio of water molecules to GdnHCl is ≈5:1. Thus, the observed slow dynamics results from the increased “viscosity” because of the high cationic and anionic concentrations (27) as well as the random-coiled polypeptide chain. But, the polarity of water molecules leads predominantly to the same steady–state emission spectra of Trp both in water and in the denatured protein.
Resonance Energy Transfer.
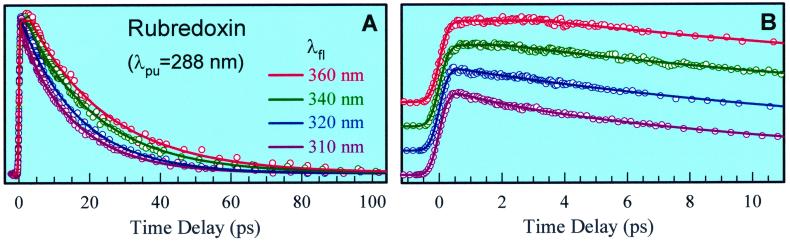
Fig. 4 shows the fluorescence transients of Trps in fmetPfRd for a series of fluorescence detection wavelengths after 288-nm excitation. Four striking results were observed: (i) The transient decay time systematically increases, from the blue side to the red side, and the decay because of RET follows a single-exponential behavior at all wavelengths (Fig. 4A) (ii); all transients decay to zero in 100 ps and no longer components are observed; (iii) an initial constant signal within 3 ps is observed at wavelengths longer than 340 nm (Fig. 4B); and (iv) transients obtained at 265-nm excitation (not shown) show similar temporal behavior. In the transients in which we observed the energy transfer, the effect of solvation is reflected on the shorter time scale: femtosecond rise at the red side and a small picosecond decay component (≈2 ps and 13%) at the blue side (310 nm) as observed in the apoprotein.
Figure 4.
Normalized, femtosecond-resolved fluorescence decay of Trp in the fmetPfRd protein at 288-nm excitation with a series of wavelength detection at long time scale (A) and for the initial part (B). Note that at wavelengths longer than 340 nm, a constant signal was observed up to 3 ps.
Specifically, at 310 nm the transient dominantly decays in 15.6 ps. At 320 nm, the transient follows a single-exponential decay in 16.4 ps. From 340 nm, we start to observe a constant signal in 2–3 ps and then it decays in 20.3 ps at 340 nm and 22.6 ps at 360 nm, respectively. The observed gradual increase of time constants (15–23 ps) toward longer wavelengths is from the influence of picosecond solvation of Trp in the protein. Thus, the decay time of ≈20 ps obtained at 340 nm best represents the dynamics of RET between Trps and the [Fe-S] cluster because we only observed a long component (>500 ps) of the solvated state in apo-fmetPfRd at this wavelength (Fig. 3B).
According to the theory of Förster energy transfer (28), the RET rate from Trp to the [Fe-S] cluster depends on the relative position (r) and orientations of donor (Trp) and acceptor ([Fe-S] cluster), and the rate of transfer kRET can be expressed as follows: kRET = (R0/r)6/τD, R0 = 9.78 × 102(κ2⋅n−4⋅QD⋅J)1/6. R0 (in nm), the critical transfer distance, is defined as the donor-acceptor distance at which the transfer efficiency is 50%. κ2 is the orientation factor, n is the refractive index of the medium (≈1.4), τD and QD are the donor excited-state lifetime and quantum yield in the absence of the acceptor (here in apoprotein), respectively, and J is the spectral overlap integral (in unit of cm3/M) between donor-emission and acceptor-absorption.
It is striking that for RET each transient decays to zero with only a single exponential temporal behavior although the fmetPfRd protein has two Trps (Trp-3 and Trp-36). According to the x-ray structure, the distance between Trp-36 (the middle point of the C–C bridge in the indole ring) and the center of [Fe-S] cluster is ≈9.6 Å and it is ≈13.2 Å for Trp-3. Both Trps in apo-fmetPfRd have a similar lifetime τD and quantum yield QD, and if they have the same R0 we should observe two distinct RET rates differing by a factor of 6.8, obviously inconsistent with our result of a single exponential decay. Thus, our observations indicate that either the two Trps have similar energy-transfer rates, but with different R0 (or κ2), or one of Trps has κ2 = 0; i.e., no energy transfer.
During RET, Trps in the protein do not undergo significant tumbling motions (see below); the anisotropy studies indicate that they are actually rigid. In such a short time of 20 ps, each Trp has a certain value of the orientation factor κ2. Here, we must also consider Trp–Trp RET. The critical distance for Trp–Trp is in the range of 5–12 Å (29, 30), and the distance between Trp-3 and Trp-36 is ≈10.6 Å. The energy transfer between Trp-3 and Trp-36 takes >100 ps. If one Trp doesn't transfer energy to the [Fe-S] cluster (κ2 = 0), but it transfers energy to the other Trp, we would observe a longer component (>100 ps) in the transients, again inconsistent with our observation. Thus, the case for κ2 = 0 is excluded.
The observed single-exponential decay in 20 ps indicates that the two Trps have similar RET rates but with different κ2 values. We further carried out site-directed mutagenesis studies by replacing Trp-3 with tyrosine. At 265-nm excitation, we observed for RET a single exponential decay time of 13 ps for the mutant with only one energy donor Trp-36, and 17.5 ps for fmetPfRd with two energy donors Trp-36 and Trp-3 at 340-nm detection (Fig. 5). Therefore, the decay time for Trp-3 because of RET is estimated to be ≈20 ps by simulations of our transients.
Figure 5.
(A) Normalized, femtosecond-resolved fluorescence transients of Trp in various systems and under different conditions for 340-nm detection at 265-nm excitation. The corresponding initial parts are shown in B. Note that only the wild-type PfRd and the variant fmetPfRd at pH 7 have an initial constant signal for 2–3 ps; see text for detail.
The lifetimes of Trps in apo-fmetPfRd were measured to be 290 ps (53%), 1.44 ns (37%), and 4.03 ns (10%). The average lifetime (τD) is ≈1.1 ns. Thus, the deduced average critical distance R0 for RET between Trp-36 and the [Fe-S] cluster is ≈19.5 Å and ≈25.2 Å for Trp-3. The overall quantum yield (QD) was estimated to be 0.15, and the spectral overlap integral was evaluated as 1.2 × 10−14 cm3/M. We obtained the orientation factor κ2 to be 0.13 for Trp-36 and 0.62 for Trp-3. The orientation factor for Trp-36 former is far away from 0.67, the value for assumption of isotropic orientation distributions for both donor and acceptor molecules.
On the other hand, if we use the isotropic κ2 value in our case of PfRd, we would obtain a critical distance (R0) of 25.5 Å for Trp. The resulting distance, r, from Trp-36 to the cluster center is deduced to be 12.5 Å. Compared to the x-ray structural distance of 9.6 Å for Trp-36, a 30% error in this case is introduced for use of the isotropic value of 0.67 for κ2. Thus, care must be taken when the isotropic value is used to calculate the distance in protein. The consequence of the new κ2 values of Trp for more theoretical studies of electron transfer in the [Fe-S] cluster of the rubredoxin is clear.
The distinct initial flatness for a time of 2–3 ps, observed at wavelengths longer than 340 nm before the decay by RET, may indicate the presence of an early build-up process. One possibility is RET between Tyr and Trp and this process must take place in several picoseconds. In the hydrophobic core of the protein, there are another four aromatic residues, Tyr-10 and Tyr-12, Phe-29, and Phe-48. At 265-nm excitation, the absorption is distributed by ≈75% for Trp, ≈22% for Tyr and ≈3% for Phe, and the distribution becomes ≈90% for Trp and ≈10% for Tyr at 288-nm excitation (31). RET may occur between these aromatic residues, as observed in other proteins (32, 33), especially from Tyr to Trp. The calculated κ2 values for different energy-transfer pairs based on the x-ray structure are given in Table 1. The estimated RET time constants are in the range of 2–6 ps for Tyr-12–Trp-36 (1Lb) and 12–27 ps for Tyr-12–Trp-36 (1La) with a separation of 5.9 Å by using a critical distance of 14–17 Å (29, 34) and a lifetime (τD) of 1.7 ns for Tyr residues (35). The energy transfer of all other pairs takes much longer time. Further studies can be made by tuning the excitation wavelength to the red side at 295–300 nm to excite Trp only and eliminate the Tyr contribution.
Table 1.
Orientation factors based on the high-resolution x-ray crystallographic structure
| Trp-3–Trp-36 | Tyr-12–Trp-36 | Tyr-10–Trp-36 | Tyr-12–Trp-3 | Tyr-10–Trp-3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| La–La | La–Lb | Lb–La | Lb–Lb | Lb–La | Lb–Lb | Lb–La | Lb–Lb | Lb–La | Lb–Lb | Lb–La | Lb–Lb | |
| κ2 | 0.50 | 0.22 | 0.66 | 0.61 | 0.35 | 1.62 | 0.32 | 1.23 | 0.14 | 0.17 | 0.25 | 2.41 |
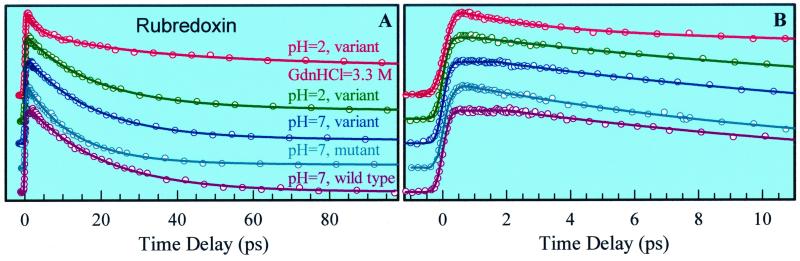
To examine the influence of the structure on the observed rates of RET, we compared the transients for all different forms. Fig. 5 shows fluorescence transients gated at 340-nm emission, under 265-nm excitation, for the different systems and conditions reported here. Except for the partial denatured protein at 3.3 M GdnHCl, all transients show for RET a single exponential decay time: 20 ps for the wild-type PfRd, 13 ps for the mutant, 17.5 ps for the variant (fmetPfRd) at pH 7, and 18 ps for the variant at pH 2. We also observed a long component (>250 ps; 15%) in the case of the variant PfRd under the condition of pH 2, indicating that Trp residues in some protein conformers with unique orientations escape the quenching by RET. This observation shows a more flexible structure at pH 2 than those at pH 7 and in the wild-type form.
PfRd is a hyperthermophilic protein and its melting temperature is as high as ≈200°C (36). It is not fully denatured at pH 7, but it easily unfolds at pH 2 when using denaturants to disrupt all of the salt bridges (37, 38). Experimental results (38) at pH 2 have shown two different intermediates occurring in the unfolding pathway at 2.4 M and 3.3 M GdnHCl. Our studies of fmetPfRd at the latter concentration show a triple-exponential temporal behavior: 2.6 ps (25%), 25 ps (32%) and ≈432 ps (43%); the initial decay reflects solvation processes as observed in the denatured protein (Fig. 3C). The decay time of 25 ps is for RET between Trp residues and the [Fe-S] cluster. The last long component is the lifetime of unquenched Trp residues of the denatured protein. Thus, the ratio of the folded to the unfolded species is about 1:1.3.
The initial temporal behaviors are shown in Fig. 5B. Only the wild-type and the variant proteins at pH 7 show a constant signal for ≈3 ps. This observation indicates that both the wild-type and the variant (at pH 7) have a more rigid structure and favorable RET between Trp-36 and Tyr-12. On the other hand, the variant at pH 2 and the mutant must have a more flexible structure, consistent with their lower thermostability (11).
Anisotropy and Internal Conversion.
The initial anisotropy of Trp, “r0”, has been extensively studied and discussed (3, 4, 8, 39, 40). In brief, it shows strong dependence on excitation wavelength. Even under subpicosecond resolution, the measured value is still far from the ideal value of 0.4. For example, it is ≈0.2 at 330-nm emission under 295-nm excitation (4). The observed smaller value r0 is due to ultrafast internal conversion between two nearly degenerate electronic states of 1La and 1Lb with mutually perpendicular transition dipole moments, and is the average one after the internal mixing. The dependence of r0 on excitation wavelength is attributed to the different contributions of the two states at different wavelength.
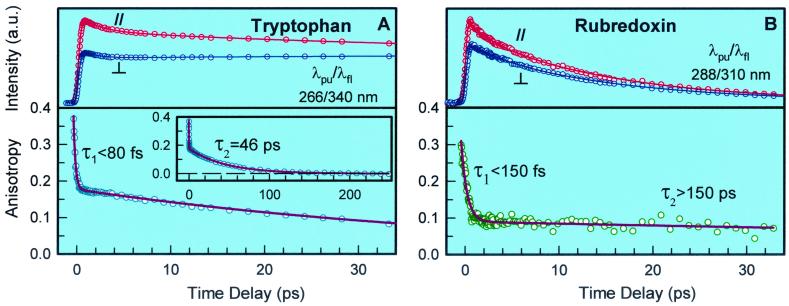
Fig. 6 shows our measured anisotropy of Trp in buffer solution and in the fmetPfRd protein. At 265-nm excitation (Fig. 6A), the anisotropy gated at 340 nm promptly drops to 0.19 in <200 fs and then decays with a time constant of 46 ps because of the orientation relaxation. Thus, internal conversion is ultrafast and the time constant is estimated to be <80 fs. Here, we observed high initial anisotropy (≈0.37) at negative time (t < 0) because of the broad experimental response function (≈300 fs) (41, 42). The observed apparent r0 (0.19) here is consistent with the reported value of ≈0.2 (4).
Figure 6.
(A) Femtosecond-resolved fluorescence transients of Trp in water at parallel and perpendicular conditions (Upper) for 340-nm emission at 265-nm excitation and the corresponding anisotropy decay (Lower). (Inset) The anisotropy decay for the long time scale. (B) Femtosecond-resolved fluorescence transients of Trps in the fmetPfRd protein at parallel and perpendicular conditions (Upper) for 310-nm emission at 288-nm excitation and the corresponding anisotropy decay. The anisotropy at 340-nm emission shows an identical temporal behavior (not shown). Note that the anisotropy of Trps in the protein stays constant on the picosecond time scale.
The anisotropy of Trp in the protein at 288-nm excitation shows a similar behavior. It initially drops to 0.1 in <450 fs but then stays at this value during RET. The measured anisotropy of r0 (0.1) is also close to the reported value of ≈0.12 (4). The deduced time constant for internal conversion is <150 fs. In contrast with Trp in water, the Trp residue is rigid in the protein (43, 44). Thus, during RET from Trp to the [Fe-S] cluster, their orientations are relatively frozen and the orientation factor κ2 is uniquely determined, consistent with the single-exponential decay observed in RET. This result also reveals a rigid structure of the hydrophobic core in PfRd.
Conclusions
The reported studies with femtosecond time resolution of Trp elucidate its solvation dynamics in different environments and its resonance energy transfer in Pyrococcus furiosus rubredoxin. The solvation process of Trp in water was observed to be ultrafast, 160 fs and 1.1 ps, but in the protein it covers a wide range of a much longer time scale. This slow solvation process, which is evident in the time-dependent spectral relaxation, is the origin for nonexponential subnanosecond decays of Trp fluorescence in proteins. The internal conversion between 1La and 1Lb states occurs in <100 fs, and the frequently reported r0 value is actually the average anisotropy after the internal state mixing. These results are significant for future studies of local protein structures and dynamics by using Trp as an intrinsic probe. Resonance energy transfer between Trps and the [Fe-S] cluster in the protein was observed to follow a single-exponential temporal behavior on the picosecond time scale. The two Trp residues have similar rates. The critical distance and the corresponding orientation factor for each Trp are uniquely determined. Studies involving measurements of both the population decay and the anisotropy for the wild-type, the mutant and the variant at different pH values reveal a dynamically rigid protein structure. This inflexible structure is probably related to its thermostability (45).
The reported studies indicate that energy transfer occurs on a much faster time scale than the local orientation relaxation (τRET≪τorien) and that the transfer efficiency is as high as 100%. This finding contrasts the other limit where τorien≪τRET; in this limit, a mobile energy donor results in multiple energy-transfer rates with relatively low efficiency, contrary to our observation. Solvation in the protein occurs on a similar time scale to that of energy transfer (τsolv≈τRET), resulting in wavelength-dependent transfer rates, as observed in this study. With τorien≫τsolv≈τRET, energy transfer is separated from solvation by femtosecond-resolved fluorescence gating of the relaxed state as observed here. If τorien≈τsolv≈τRET, energy transfer is convoluted with both orientational relaxation and solvation. Thus, the elucidation of the time scales, in this case τorien, τsolv, and τRET, is crucial to the understanding of protein dynamics.
Acknowledgments
We like to thank Prof. Michael W. W. Adams and Dr. Francis E. Jenney, Jr. (University of Georgia) for the generous gift of all proteins reported here. We acknowledge the assistance of Dr. Spencer Baskin for the measuring of the lifetime of the apoprotein and for helpful discussion. This work was supported by the National Science Foundation.
Abbreviations
- Trp
tryptophan
- fmetPfRd
formylmethionine variant of Pyrococcus furiosus rubredoxin
- RET
resonance energy transfer
- NATA
N-acetyl-l-tryptophanamide
- NATEE
N-acetyl-l-Trp ethyl ester
Note
In the process of writing this work, we learned of a study of Trp solvation in water (46). The ultrafast solvation in 160 fs was not resolved and the reported ≈1.2 ps solvation time is consistent with our observed long-time component (1.1 ps) reported here.
References
- 1.Szabo A G, Rayner D M. J Am Chem Soc. 1980;102:554–563. [Google Scholar]
- 2.Petrich J W, Chang M C, McDonald D B, Fleming G R. J Am Chem Soc. 1983;105:3824–3832. [Google Scholar]
- 3.Beechem J M, Brand L. Annu Rev Biochem. 1985;54:43–71. doi: 10.1146/annurev.bi.54.070185.000355. [DOI] [PubMed] [Google Scholar]
- 4.Ruggiero A J, Todd D C, Fleming G R. J Am Chem Soc. 1990;112:1003–1014. [Google Scholar]
- 5.Callis R R. Methods Enzymol. 1997;278:113–150. doi: 10.1016/s0076-6879(97)78009-1. [DOI] [PubMed] [Google Scholar]
- 6.Hochstrasser R M, Negus D K. Proc Natl Acad Sci USA. 1984;81:4399–4403. doi: 10.1073/pnas.81.14.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eftink M R. In: Methods of Biochemical Analysis: Protein Structure Determination. Suelter C H, editor. Vol. 35. New York: Wiley; 1991. [DOI] [PubMed] [Google Scholar]
- 8.Hansen J E, Rosenthal S J, Fleming G R. J Phys Chem. 1992;96:3034–3040. [Google Scholar]
- 9.Lakowicz J R. Photochem Photobiol. 2000;72:421–437. doi: 10.1562/0031-8655(2000)072<0421:OSRIP>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lakowicz J R, editor. Topics in Fluorescence Spectroscopy: Protein Fluorescence. New York: Kluwer Academic/Plenum; 2000. [Google Scholar]
- 11.Bau R, Rees D C, Kurtz D M, Jr, Scott R A, Huang H, Adams M W W, Eidsness M K. J Biol Inorg Chem. 1998;3:484–493. [Google Scholar]
- 12.Sieker L C, Stenkamp R E, Legall J. Methods Enzymol. 1994;243:203–216. doi: 10.1016/0076-6879(94)43016-0. [DOI] [PubMed] [Google Scholar]
- 13.Lovenberg W, editor. Iron Sulfur Proteins. New York: Academic; 1973. [Google Scholar]
- 14.Lowery M D, Guckert J A, Gebhard M S, Solomon E I. J Am Chem Soc. 1993;115:3012–3013. [Google Scholar]
- 15.Dorovska-Taran V, van Hoek A, Link T A, Visser A J W G, Hagen W R. FEBS Lett. 1994;348:305–310. doi: 10.1016/0014-5793(94)00606-7. [DOI] [PubMed] [Google Scholar]
- 16.Fiebig T, Chachisvilis M, Manger M, Zewail A H, Douhal A, Garcia-Ochoa I, Ayuso A D H. J Phys Chem A. 1999;103:7408–7418. [Google Scholar]
- 17.Blake P R, Park J B, Bryant F O, Aono S, Magnuson J K, Eccleston E, Howard J B, Sunmers M F, Adams M W W. Biochemistry. 1991;30:10885–10895. doi: 10.1021/bi00109a012. [DOI] [PubMed] [Google Scholar]
- 18.Eidsness M K, Richie K A, Burden A E, Kurtz D M, Jr, Scott R A. Biochemistry. 1997;36:10406–10413. doi: 10.1021/bi970110r. [DOI] [PubMed] [Google Scholar]
- 19.Albinsson B, Kubista M, Norden B, Thulstrup E W. J Phys Chem. 1989;93:6646–6654. [Google Scholar]
- 20.Shen X, Knutson J R. Chem Phys Lett. 2001;339:191–196. [Google Scholar]
- 21.Reynolds L, Gardecki J A, Frankland S J V, Horng M L, Maroncelli M. J Phys Chem. 1996;100:10337–10354. [Google Scholar]
- 22.Jarzeba W, Walker G C, Johnson A E, Kahlow M, Barbara P F. J Phys Chem. 1988;92:7039–7041. [Google Scholar]
- 23.Jimenez R, Fleming G R, Kumar P V, Maroncelli M. Nature (London) 1994;369:471–473. [Google Scholar]
- 24.Vivian J T, Callis P R. Biophys J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jordanides X J, Lang M J, Song X, Fleming G R. J Phys Chem B. 1999;102:3044–3052. [Google Scholar]
- 26.Changenet-Barret P, Choma C T, Gooding E F, DeGrado W F, Hochstrasser R M. J Phys Chem B. 2000;104:9322–9329. [Google Scholar]
- 27.Nandi N, Bhattacharyya K, Bagchi B. Chem Rev. 2000;100:2013–2045. doi: 10.1021/cr980127v. [DOI] [PubMed] [Google Scholar]
- 28.Förster Th. In: Modern Quantum Chemistry. Sinanoglu O, editor. Vol. 3. New York: Academic; 1965. pp. 93–137. [Google Scholar]
- 29.Steinberg I Z. Annu Rev Biochem. 1971;40:83–114. doi: 10.1146/annurev.bi.40.070171.000503. [DOI] [PubMed] [Google Scholar]
- 30.Griep M A, McHenry C S. J Biol Chem. 1990;265:20356–20363. [PubMed] [Google Scholar]
- 31.Rava R P, Spiro T G. J Phys Chem. 1985;89:1856–1861. [Google Scholar]
- 32.Wu P G, James E, Brand L. Biophys Chem. 1993;48:123–133. doi: 10.1016/0301-4622(93)85004-2. [DOI] [PubMed] [Google Scholar]
- 33.Andrews D L, Demidov A A, editors. Resonance Energy Transfer. New York: Wiley; 1999. [Google Scholar]
- 34.Weber G. Biochem J. 1960;75:335–345. doi: 10.1042/bj0750335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferreira S T, Stella L, Gratton E. Biophys J. 1994;66:1185–1196. doi: 10.1016/S0006-3495(94)80901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hiller R, Zhou Z H, Adams M W W, Englander S W. Proc Natl Acad Sci USA. 1997;94:11329–11332. doi: 10.1073/pnas.94.21.11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavagnero S, Zhou Z H, Adams M W W, Chan S I. Biochemistry. 1998;37:3377–3385. doi: 10.1021/bi9721804. [DOI] [PubMed] [Google Scholar]
- 38.Zhang D, Ramakrishnan V, Chan S I. J Inorg Biochem. 1999;74:348–348. [Google Scholar]
- 39.Valeur B, Weber G. Photochem Photobiol. 1977;25:441–444. doi: 10.1111/j.1751-1097.1977.tb09168.x. [DOI] [PubMed] [Google Scholar]
- 40.Hu Y, Fleming G R. J Chem Phys. 1991;94:3857–3866. [Google Scholar]
- 41.Myers A B, Holt P L, Pereira M A, Hochstrasser R M. Chem Phys Lett. 1986;132:585–590. [Google Scholar]
- 42.Baskin J S, Zewail A H. J Phys Chem. 1994;98:3337–3351. [Google Scholar]
- 43.Munro I, Pecht I, Stryer L. Proc Natl Acad Sci USA. 1979;76:56–60. doi: 10.1073/pnas.76.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ichiye T, Karplus M. Biochemistry. 1983;22:2884–2893. doi: 10.1021/bi00281a017. [DOI] [PubMed] [Google Scholar]
- 45.Lazaridis T, Lee I, Karplus M. Protein Sci. 1997;6:2589–2605. doi: 10.1002/pro.5560061211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen X, Knutson J R. J Phys Chem B. 2001;105:6260–6265. [Google Scholar]