Abstract
The introduction of chemically unique groups into proteins by means of non-natural amino acids has numerous applications in protein engineering and functional studies. One method to achieve this involves the utilization of a non-natural amino acid by the cell's native translational apparatus. Here we demonstrate that a methionine surrogate, azidohomoalanine, is activated by the methionyl-tRNA synthetase of Escherichia coli and replaces methionine in proteins expressed in methionine-depleted bacterial cultures. We further show that proteins containing azidohomoalanine can be selectively modified in the presence of other cellular proteins by means of Staudinger ligation with triarylphosphine reagents. Incorporation of azide-functionalized amino acids into proteins in vivo provides opportunities for protein modification under native conditions and selective labeling of proteins in the intracellular environment.
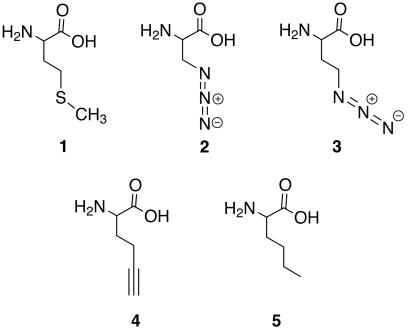
The 20 naturally occurring amino acids determine the structure and chemical reactivity of the proteins that they inhabit. Protein engineering by means of the introduction of non-natural amino acids is an important approach to the investigation of protein folding, structure, and function as well as the design of novel protein reactivity (1). Indeed, incorporation of non-natural amino acids into proteins by means of chemical methods such as solid-phase synthesis (2), native chemical ligation (3), and in vitro translation protocols (4, 5) has permitted characterization of protein folding pathways, enzymatic mechanisms, and ligand-receptor interactions. Recent advances in the adaptation of “unnatural amino acid mutagenesis” to cellular systems suggests a rich future for heterologous expression of novel proteins (6–9). At present, however, the most efficient means for introducing non-natural amino acids into recombinant proteins exploits the unnatural substrate tolerance of the native translational apparatus. For example, several methionine (1, Fig. 1) analogs are used in protein biosynthesis with high efficiency, particularly when competing methionine is eliminated by use of a methionine auxotrophic host (10).
Figure 1.
Structures of methionine and the analogs tested for utility as methionine surrogates. (1) Methionine, (2) azidoalanine, (3) azidohomoalanine, (4) 2-amino-5-hexynoic acid, and (5) norleucine.
Replacement of methionine by analogs that possess unique chemical reactivity offers the potential for posttranslational protein modification. Applications include selective tagging of proteins with probes and protein engineering for structure/function analysis (11, 12). Already the method has permitted the incorporation of a diverse set of methionine analogs with alkyl and unsaturated side chains (13, 14), the latter suggesting opportunities for subsequent chemical reaction (15–17).
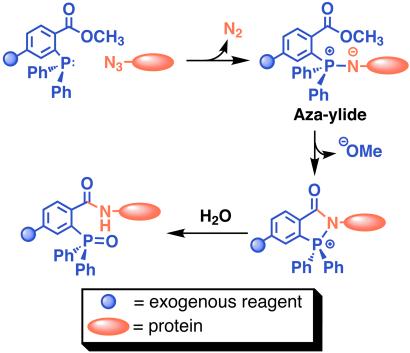
Toward a general method for chemical modification of proteins in vivo, we have co-opted the biosynthetic machinery of an Escherichia coli methionine auxotroph to introduce azides into proteins. The incorporation of azide-functionalized amino acids appeared especially attractive in light of the recent observation that azidosugars can be installed in cell-surface glycoproteins by means of the sialic acid biosynthetic machinery and subsequently modified by the Staudinger ligation (18). The ability of the azide group to survive cellular metabolism and react selectively with phosphine reagents under mild conditions suggested that proteins equipped with azide-functionalized amino acids could be modified according to Scheme S1. Incorporation of azide-functionalized amino acids as methionine surrogates may provide unique opportunities to manipulate protein–protein recognition, modify recombinant proteins, or selectively label proteins in the cell.
Scheme 1.
The Staudinger ligation between a protein containing azide functionalized amino acid side chains and a phosphine reagent.
The incorporation of methionine analogs into proteins is controlled most stringently by the methionyl-tRNA synthetase (MetRS) of the host. Accordingly, the rates of activation of methionine analogs by MetRS have been shown to correlate with their efficiencies of incorporation into target proteins (10, 14). In this article, two azide-functionalized amino acids, azidoalanine (2, Fig. 1) and azidohomoalanine (3, Fig. 1), were synthesized as potential methionine analogs. These two compounds were tested as in vitro substrates for MetRS and candidates for incorporation into proteins in vivo. The results of both in vitro and in vivo assays confirmed 3 as an excellent methionine surrogate. A target protein (murine dihydrofolate reductase, mDHFR) containing 3 was modified by Staudinger ligation with an appropriately engineered phosphine bearing an antigenic FLAG peptide. We have further demonstrated the selectivity of the Staudinger ligation by modifying the azide-labeled target protein within the context of all other cellular proteins.
Materials and Methods
All chemical reagents were of analytical grade, obtained from commercial suppliers, and used without further purification unless otherwise noted. Reversed-phase HPLC was performed by using a Rainin Instruments Dynamax SD-200 HPLC system with 230-nm detection, on a Microsorb C-18 analytical column (4.6 × 250 mm) at a flow rate of 1 ml/min, a semipreparative column (10 × 250 mm) at a flow rate of 4 ml/min, or a preparative column (25 × 250 mm) at a flow rate of 20 ml/min. All runs used linear gradients of 30–100% buffer B in A (A = water containing 0.1% trifluoroacetic acid, B = acetonitrile containing 0.1% trifluoroacetic acid) over 60 min. 1H and 13C NMR spectra were measured on Bruker AMX-300, AMX-400, or DRX-500 MHz spectrometers as noted. Compounds 2 (19, 20) and 3 (21) were synthesized according to previously published procedures. Compounds 6 and 7 were synthesized according to procedures described in the supporting text, which is published on the PNAS web site, www.pnas.org.
FLAG (Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys) Peptide (8).
The peptide was synthesized by using established automated protocols on a Perkin–Elmer ABI 431 A peptide synthesizer (user-derived cycles) using fluorenylmethoxycarbonyl (Fmoc)-Lys(Boc)-Wang resin (Novabiochem; 0.83 mmol Lys/g resin), Nα-Fmoc-protected amino acids and 1,3-dicyclohexylcarbodiimide-mediated 1-hydroxybenzotriazole (HOBt) ester activation in 1-methyl-2-pyrrolidinone. Five equivalents of N-protected amino acid were activated for 30 min by using 10 equivalents each of 1,3-diisopropylcarbodiimide and HOBt in N,N-dimethylformamide (DMF). This solution was added to the resin and shaken for 30 min. The Fmoc-protecting group of the terminal Asp residue was removed by treatment with piperidine. The resin was washed with DMF followed by CH2Cl2 and transferred to a solid-phase reaction vessel for use in the following step without further purification.
Triarylphosphine-FLAG (9).
DMF (1 ml) followed by a solution of 7 (182 mg, 0.5 mmol), 1-hydroxybenzotriazole (68 mg, 0.5 mmol), O-benzotriazol-1-yl-N,N,N′,N′-tetramethyluronium hexafluorophosphate (190 mg, 0.5 mmol), diisopropylethylamine (87 μl, 0.5 mmol) in DMF (2 ml) was added to the solid-phase reaction vessel containing the resin-bound side-chain protected FLAG peptide (120 mg, 0.1 mmol). The reaction vessel was agitated for 4 h at room temperature, and then the resin was washed with DMF followed by CH2Cl2. The washed and dried resin was treated with 95% trifluoroacetic acid (3 ml) for 2 h at room temperature. Filtration afforded a solution of the crude product that was concentrated and then purified by reversed-phase HPLC. MS (electrospray ionization) confirmed the identity of the product with m/z 680.1 (MHH++).
Computation.
Single-point energy ab initio calculations (Hartree-Fock model, 6–31G* basis set) (22, 23) were performed for methionine and for analogs 2 and 3 with fully extended side chains. Electron density maps are shown as surfaces of electron density 0.08 electrons/au (3). Isopotential plots are represented as surfaces where the energy of interaction between the amino acid and a point positive charge is equal to −10 kcal/mole. Calculations were performed by using the program macspartan (Wavefunction, Irvine, CA).
Enzyme Purification and Activation Assays.
The fully active, truncated form of MetRS was purified from 24 h cultures of E. coli JM101 cells carrying the plasmid pGG3 (24) and was purified by size-exclusion chromotography as described (25). The activation of methionine analogs by MetRS was assayed by means of the amino acid-dependent ATP-32P-pyrophosphate exchange reaction, also as described (25–27). Specific experimental details are provided as supporting information on the PNAS web site.
Determination of Translational Activity.
The translational activity of the methionine analogs was assayed in two bacterial hosts, CAG18491/pQE15/pREP4 and CAG18491/pQE15-MRS/pREP4. Procedures for preparing these hosts and assaying translational activity are described in the supporting information on the PNAS web site.
Protein Expression and Purification.
mDHFR was expressed from 1-liter cultures of CAG18491/pQE15/pREP4, supplemented with either methionine or 3, using standard protocols. The mDHFR was purified from the cell pellet by using immobilized metal affinity chromatography. Incorporation of 3 was confirmed by amino acid analysis, N-terminal sequencing or matrix-assisted laser desorption ionization (MALDI) MS of the purified protein.
Chemical Modification of Purified Proteins by Staudinger Ligation.
Purified and lyophilized samples of mDHFR-Met and mDHFR-3 were dissolved at a concentration of ≈0.1 mg/ml in PBS at pH 7.4 containing 8 M urea. In a typical experiment, 10 μl of either solution was mixed with 10 μl of a solution of triarylphosphine-FLAG (500 μM in PBS, pH 7.4, 8 M urea) or 10 μl of the buffer without phosphine. The reactions were heated at 47°C for 6 h to obtain maximum ligation. Standard Western blotting procedures were then carried out to monitor the presence of all proteins bearing the N-terminal hexahistidine sequence as well as proteins that had reacted with the phosphine and were therefore FLAG-labeled. Reaction mixtures were divided in half and the two sets of reactions were subjected to SDS/PAGE in parallel. Proteins from the two gels were then transferred to nitrocellulose membranes. One nitrocellulose membrane was blocked with a solution of 5% dry nonfat milk in Tris-buffered saline (TBST, 0.05 M Tris, 0.15 M NaCl, 0.05% Tween, pH = 7.4) at room temperature for 2 h. The membrane was then washed with TBST (10 ml 2 × 1 min followed by 10 ml 2 × 30 min), and placed in a solution of anti-FLAG M2 mAb (Sigma, 1:10 000 in TBST) for 1 h at room temperature. After the membrane was washed as before, it was incubated with horseradish peroxidase (HRP)-rat anti-mouseIgG1 (Zymed, 1:20,000 in TBST) for 1 h at room temperature. Washing was repeated a third time and the membrane was developed by using Super Signal West Pico Chemiluminescent Substrate (Pierce).
The second nitrocellulose membrane was blocked with a solution of 2.5 mg/ml BSA in TBST at room temperature for 2 h. The membrane was then washed with TBST (10 ml 2 × 1 min followed by 10 ml 2 × 30 min), and placed in a solution of India HisProbe-HRP (Pierce, 1:5,000 in TBST) for 1 h at room temperature. After the membrane was washed as before, it was developed by using Super Signal West Pico Chemiluminescent Substrate (Pierce).
Chemical Modification of Proteins in Crude Cell Lysate Using the Staudinger Ligation.
Samples of CAG18491/pQE 15/pREP4 cultures (1 ml), supplemented either with methionine or with 3, were collected and sedimented 4 h after induction of protein synthesis. The samples were resuspended in PBS buffer, pH 7.4 to a normalized OD600 of 15, and then frozen immediately at −20°C. DNase 1, amplification grade (GIBCO, 5 μl) and RNase H (GIBCO, 2 μl) was added to the thawed cell lysate suspensions. The cell lysates were incubated at room temperature for 15 min followed by 55 min at 55°C. Aliquots of cell lysate (1 μl) were diluted with PBS, pH 7.4 (9 μl) and reacted with triarylphosphine-FLAG under conditions similar to those described above for the purified protein, without the addition of urea. Western blot analysis was performed as described above.
Results and Discussion
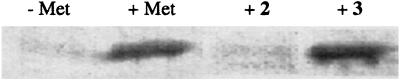
Our initial attempts to identify azide-functionalized methionine surrogates focused on azidoalanine (2) owing to the similarity of its side-chain length to that of methionine and the similarity of its side-chain geometry to that of the proven methionine surrogate 2-amino-5-hexynoic acid (4, Fig. 1) (27). We found, however, that 2 does not support protein synthesis in methionine-depleted cultures of E. coli (see Fig. 3), even under conditions where MetRS is overexpressed and the medium is supplemented with high concentrations of 2. These conditions have been shown previously to rescue the translational activity of methionine analogs that have kcat/Km values for activation by MetRS that are 340,000-fold lower than the kcat/Km value for methionine (14). Consistent with these observations, 2 does not support measurable levels of 32P-pyrophosphate exchange by MetRS in vitro under any assay conditions investigated.
Figure 3.
SDS/PAGE analysis of the translational activity of azide-functionalized amino acids. The SDS/PAGE analysis was conducted on whole-cell lysates of CAG18491/pQE15/pREP4 cultures, supplemented with nothing (−Met), methionine (+Met), 2, or 3.
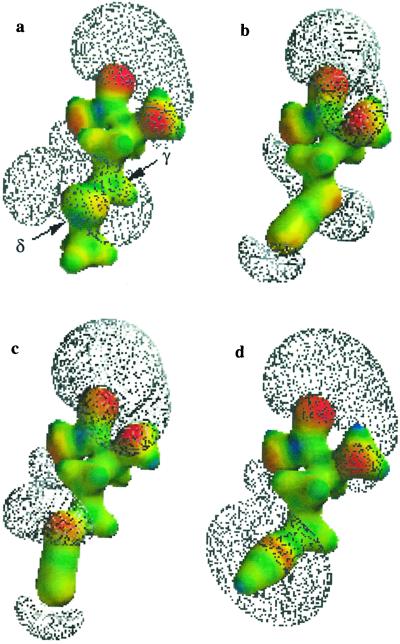
Electron density maps and isopotential surfaces of the extended side-chain structures of azidoalanine (2) and azidohomoalanine (3) were generated to determine whether 3 might better mimic methionine. The electron density maps (colored surfaces) and isopotential surfaces (meshes) for 2 and 3 are shown in Fig. 2; results for the known MetRS substrates, methionine and 2-amino-5-hexynoic acid (4), are shown for comparison. There is little similarity between the electronic structure of the side chain of 2 and that of either methionine or 4. The side-chain isopotential surface for 2 is most highly extended at the γ position, rather than at the δ position as observed for methionine and 4. Insertion of a γ-methylene group yields analog 3, in which the side-chain potential surface is most highly extended at the δ position. We proposed that this feature of the side-chain electronic structure might contribute to activation of 3 by MetRS. Indeed, the importance of electron density at the δ position has been supported by the observation that the thioether of the bound methionine substrate forms hydrogen bonds with both the side chain of Tyr-260 and the backbone NH of Leu-13 in the active site of the enzyme (28).
Figure 2.
Electron density maps (colored surfaces) and negative isopotential surfaces (meshes) for (a) methionine, (b) 2, (c) 3, and (d) 4. The electron density maps indicate electron-rich (red) and electron-poor (blue) regions of each molecule. For simplicity, the amino acid form is shown; this avoids representation of the highly extended isopotential surface of the carboxylate anion of the zwitterion and facilitates comparison of side-chain electronic structures.
The rate of activation of azidohomoalanine (3) by MetRS was determined in vitro by the ATP-32P-pyrophosphate exchange assay. Table 1 shows the kinetic parameters determined for 3; similar parameters measured for methionine, 2-amino-5-hexynoic acid (4), and norleucine (5), the three amino acids activated most efficiently by MetRS in our investigations, are given for comparison. A kcat/Km value of 1.42 × 10−3 s−1⋅μM−1 was obtained for activation of 3 by MetRS in vitro, indicating that 3 is a slightly better substrate than 4.
Table 1.
Comparison of kinetic parameters for 3 with those for methionine and methionine analogs 4 and 5
| Analog | kcat/Km (s−1⋅μM−1) | kcat/Km (rel)* | Relative protein yield (%)† |
|---|---|---|---|
| Met | 5.47 × 10−1 | 1 | 100 |
| 3 | 1.42 × 10−3 | 1/390 | 100 |
| 4 | 1.16 × 10−3 | 1/500 | 100 |
| 5 | 5.22 × 10−4 | 1/1,050 | 57 |
Relative to kcat/Km for Met.
Yield of mDHFR relative to that obtained from Met-supplemented cultures of E. coli strain CAG18491/pQE15/pREP4 (35 mg/liter).
The translational activity of 3 was assessed on the basis of its capacity to support synthesis of the target protein mDHFR in E. coli methionine auxotroph (CAG18491/pQE15/pREP4) cultures supplemented with the analog and depleted of methionine. Protein expression was monitored by SDS/PAGE analysis (Fig. 3). The target protein was not observed in the negative control culture, whereas mDHFR was clearly detected in positive control cultures supplemented with methionine and in cultures supplemented with 3. Consistent with the measured efficiency of activation of the analog by MetRS (Table 1), 3 supported protein synthesis in a conventional bacterial host; overexpression of MetRS was not required. Furthermore, CAG18491/pQE15/pREP4 cultures supplemented with 3 produced protein in yields nearly identical to those observed from cultures supplemented with methionine or 2-amino-5-hexynoic acid (4).
Azidohomoalanine-containing mDFHR (mDHFR-3) or methionine-containing mDHFR (mDHFR-Met) samples were purified and analyzed for amino acid content, identity of the N-terminal amino acid, and changes in mass based on replacement of methionine by 3. Amino acid analysis of mDHFR-3 yielded a methionine content of 0.2 mol% compared with 3.8 mol% found in mDHFR-Met. As 3 was unstable under the strongly acidic hydrolysis conditions required for amino acid analysis, this decrease in methionine content was taken as indirect evidence of replacement of methionine by 3. On this basis, amino acid analysis indicates 95 ± 2% replacement of methionine.
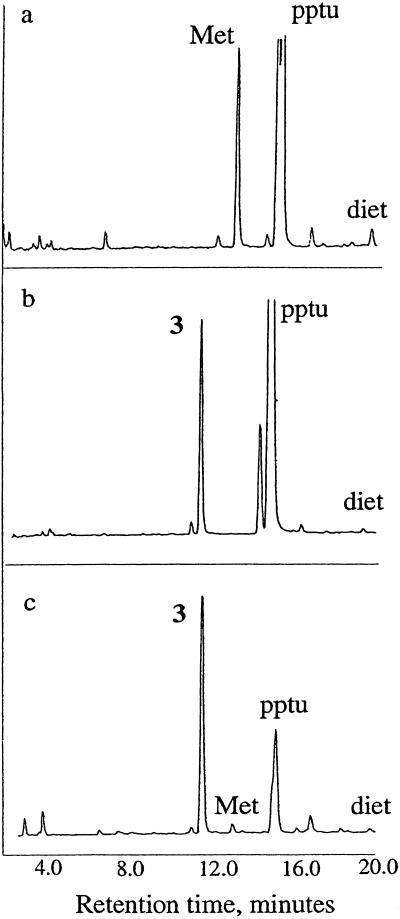
A more direct measure of the incorporation of 3 could be obtained from N-terminal sequencing of mDHFR-3 (Fig. 4). Unlike the conditions required for total amino acid analysis, sequencing does not cause degradation of 3. mDHFR expressed in pQE15 is expected to retain its N-terminal methionine residue (29). As shown in Fig. 4a, the methionine residue cleaved from the N terminus of mDHFR-Met eluted at ≈13.0 min. The large peaks at ≈14.7 min correspond to piperidylphenylthiourea (pptu), a product of the analysis resulting from the buffer, and the small peak at ≈18.5 min corresponds to diethylphthalate (diet), an internal standard. When a known standard of 3 was analyzed under the same conditions, it eluted at 11.5 min (Fig. 4b). N-terminal sequencing of mDHFR-3 (Fig. 4c) revealed that the first amino acid site was predominantly occupied by 3, with only a small amount of methionine remaining. Integration of the two peak areas indicates 97 ± 2% replacement of methionine. The reduction product of 3, 2,4-diaminobutyric acid, elutes at 15.1 min; however, no such peak was observed in the analysis of the N-terminal residue of mDHFR-3 (data not shown).
Figure 4.
N-terminal sequencing results indicating occupancy of the initiator site in mDHFR produced in bacterial cultures supplemented with 3. N-terminal residues are shown for (a) DHFR-Met, (b) the free amino acid 3, and (c) DHFR-3, as determined by Edman degradation. pptu, piperidylphenylthiourea; diet, diethylphthalate.
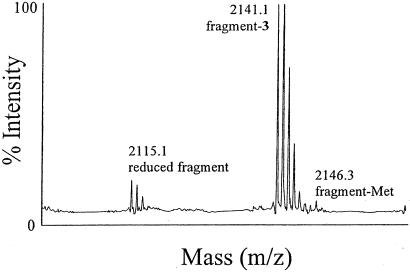
The replacement of methionine by 3 was further confirmed by mass analysis of tryptic fragments of mDHFR-3. A mass spectrum for a representative fragment comprising residues 153–170, with the encoded sequence IMQEFESDTFFPEIDLGK, is given in Fig. 5. The expected mass of the fragment containing methionine is 2,146.4 Da, whereas substitution of the single methionine by 3 yields a fragment with a mass of 2,141.3 Da. As shown in Fig. 5, a mass of 2,141.1 Da was observed for this fragment in the MALDI-MS analysis of the tryptic digest of mDHFR-3, with no evidence of a peak at 2,146.4 Da. In contrast, a peak at 2,146.4 Da was reliably observed for the corresponding fragment obtained from tryptic digest/MALDI-MS analysis of mDHFR-Met (data not shown). Similar results were observed for the other methionine-containing fragments. These results strongly suggest replacement of methionine by 3. However, in Fig. 5, there is clear evidence of a peak at 2,115.1 Da. This molecular mass may correspond to a peptide fragment in which the azide had been converted to an amine, resulting in a mass loss of 26 Da. Similar mass losses are observed in the additional tryptic fragments of mDHFR-3 (data not shown). This finding suggests that the intracellular reducing potential of E. coli may be sufficient to cause partial reduction of 3 to 2,4-diaminobutyric acid (DABA), either before or after it is incorporated into mDHFR-3. Because DABA was not observed at the N-terminal residue, it is possible that this position is uniquely shielded from reduction relative to the rest of the protein. Future experiments using strains of E. coli that have been engineered to maintain a more oxidizing environment within the cytoplasm could circumvent this problem (30).
Figure 5.
Mass spectrometric analysis of a tryptic fragment of mDHFR-3. The tryptic digest of mDHFR-3 was analyzed by MALDI-MS; results are shown for a fragment with an expected mass of 2,146.4 Da. The observed peak at a mass of 2,141.1 Da is consistent with that expected for the fragment in which 3 replaces methionine. The peak at 2,115.1 Da may correspond to a fragment in which 3 has been reduced to 2,4-diaminobutyric acid.
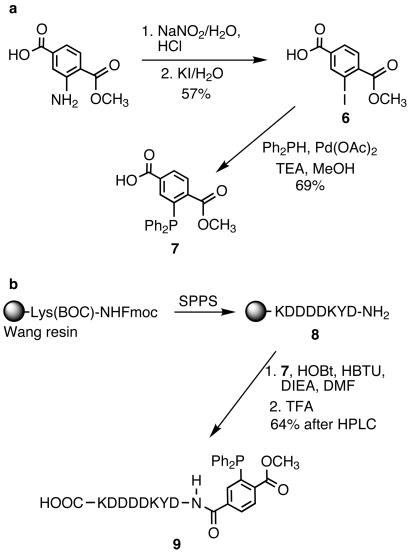
Having expressed an azide-modified form of mDHFR we next investigated the reactivity of this protein with a phosphine in the Staudinger ligation. We designed triarylphosphine-FLAG conjugate 9 for selective reaction with mDHFR at sites substituted with 3 (Scheme S2). The phosphine moiety (7) possesses three aryl substituents to limit air oxidation. One of the aryl rings is derivatized with a methyl ester ortho to the phosphorus atom, providing an electrophilic trap for the nucleophilic nitrogen in the aza-ylide formed on reaction with azides. This type of phosphine has been successfully used in several model Staudinger ligations as well as those described for labeling cell surface azide-bearing sialic acids with a biotinylated phosphine (18). The addition of a FLAG peptide to 7 enhanced water solubility (crucial for biological applications) and provided an epitope for detection with anti-FLAG antibodies.
Scheme 2.
Synthesis of triarylphosphine-FLAG conjugate 9.
The synthesis depicted in Scheme S2a began with the Sandmeyer reaction of commercially available 1-methyl-2-aminoterephthalate to form the aryl-iodide 6. This compound was reacted with diphenylphosphine by means of a palladium-mediated cross-coupling reaction to provide phosphine 7. Standard methods of solid-phase peptide synthesis provided the side-chain-protected FLAG peptide bound to Wang resin at the C terminus and a free amine at the N terminus (Scheme S2b). The resin-bound peptide was coupled to 7 and the crude product was simultaneously deprotected and cleaved from the resin with trifluoroacetic acid. Purification by reversed-phase HPLC provided the final product 9.
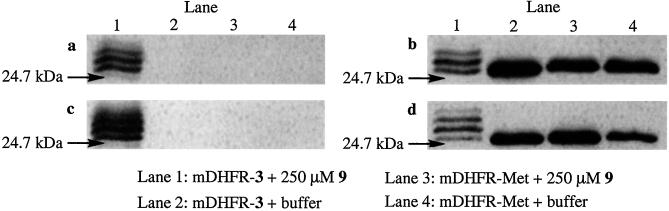
Purified mDHFR-Met and mDHFR-3 each were reacted with 250 μM 9 or with buffer alone as a negative control. The products of those four reactions were separated by SDS/PAGE and examined for the presence of the FLAG epitope (Fig. 6a). In a parallel experiment, the products of the four reactions were examined for the presence of the hexahistidine sequence of mDHFR, to ensure that the FLAG peptide was ligated to mDHFR and that protein degradation was not taking place under the reaction conditions (Fig. 6b). It is evident from Fig. 6a that Staudinger ligation proceeded when both mDHFR-3 and 9 were present in the reaction (lane 1). Untreated mDHFR-3 did not show any anti-FLAG immunoreactivity (Fig. 6a, lane 2). Likewise, when mDHFR-Met was treated with 9 or with buffer alone, no FLAG peptide was associated with the protein (Fig. 6a, lanes 3 and 4). A similar Western blot using the anti-His6 antibody confirmed that FLAG immunoreactivity was coincident with mDHFR (Fig. 6b, lane 1). The results shown in Fig. 6b also demonstrate that possible side reactions such as proteolysis are not occurring, because lanes 2–4 each show a single immunoreactive protein at the appropriate molecular mass for mDHFR.
Figure 6.
Western blot analysis of the products of Staudinger ligation. (a) Purified protein (mDHFR-3 or mDHFR-Met) was used in the ligation, and the blot was labeled with anti-FLAG M2 mAb followed by HRP-rat anti-mouse IgG1. (b) Similar to a but labeled with India HisProbe-HRP. (c) Crude cell lysate (containing either mDHFR-3 or mDHFR-Met) was used in the ligation, and the blot was labeled with anti-FLAG M2 mAb followed by HRP-rat anti-mouse IgG1. (d) Similar to c but labeled with India HisProbe-HRP.
Complete ligation of triarylphosphine-FLAG (1.3 kDa) at eight possible positions in mDHFR-3 is expected to produce a shift in molar mass of ≈10.4 kDa relative to the unligated protein. As shown in lanes 1 of Fig. 6, however, a distribution of molecular masses was observed, indicating modification of mDHFR-3 by triarylphosphine-FLAG (9) at fewer than the eight possible sites. The resolution of SDS/PAGE analysis is insufficient to assign an exact number of triarylphosphine-FLAG conjugates to each band, and each band may represent more than one ligation product. Given these limitations, it appears that at least five different ligation products are formed based on the Western blot analysis.
To optimize the conditions for Staudinger ligation of mDHFR-3, a range of temperatures, triarylphosphine-FLAG (9) concentrations, and reaction times was studied. Reaction for 6 h with 250 μM 9 at 47°C was found to be optimal. Longer reaction times, higher concentrations of 9, and higher temperatures did not produce additional ligation products, nor a more uniform product distribution.
The incomplete modification of all eight sites in mDHFR-3 may be the result of several factors. Reduction of the azide within 3 to the amine would preclude ligation at any residue where this has taken place. As described previously, some reduction was suggested from tryptic digest/MALDI-MS analysis of mDHFR-3. It is also possible that the quantitative yields obtained from reaction of azides with triarylphosphine reagents demonstrated in model reactions (18) do not apply within the context of the protein.
Despite the product heterogeneity observed upon ligation of mDHFR-3 with 9, these results confirmed the high selectivity of the Staudinger ligation. To place even higher demands on selectivity, we performed the reaction on crude cell lysates containing either mDHFR-Met or mDHFR-3. Samples of each cell lysate mixture were reacted with or without 9 and examined for the presence of both FLAG (Fig. 6c) and hexahistidine immunoreactivity (Fig. 6d). Similar to results using purified protein, FLAG immunoreactivity depended on the presence of both mDHFR-3 and 9 (Fig. 6c, lane 1). Neither ligation nor degradation occurred in the control reactions, as shown by the consistent appearance of hexahistidine immunoreactivity (Fig. 6d). The impressive selectivity of the reaction underscores its potential to label target azide-bearing proteins among a multitude of cellular components.
Conclusions
Our results demonstrate that azidohomoalanine (3) is recognized as a methionine surrogate by the translational apparatus of E. coli. Incorporation of other azidoamino acids into proteins in vivo may also be possible. Arylazides, for example, would provide opportunities for photoactivated modification of proteins as well as Staudinger ligations. Furthermore, these studies indicate that translationally active amino acid analogs can be rationally designed based on side-chain features important in analog recognition by the appropriate aminoacyl-tRNA synthetase (aaRS). Computational methods that can determine energies for the binding of various non-natural amino acids to an aaRS may facilitate the rational design of new translationally active analogs.
The chemoselective modification of mDHFR-3 demonstrated herein is an example of a route for selective modification of recombinant proteins. The selectivity of the Staudinger ligation even within a complex cell lysate mixture suggests the exciting prospect of intracellular applications. If the reaction were to proceed similarly within an intact cell, functionalities such as spin labels, fluorophores, and radiolabels may be appended to a phosphine and delivered to an azide-labeled protein of interest installed in a cell. Given the concise structure of the azide, it is possible that azide-substituted versions of other amino acids will be tolerated by the translational machinery. In this case, the Staudinger ligation may prove to be a highly versatile tool for posttranslational protein engineering.
Supplementary Material
Acknowledgments
We acknowledge the generous donation of plasmids encoding MetRS from H. Jakubowski and Y. Mechulam and the assistance of J. Kua in modeling the azide-functionalized amino acids. This research was supported by the Office of Naval Research, Grant N00014–98-1–0605 and Order N00014–98-F-0402 through the U.S. Department of Energy under Contract DE-AC03–76SF00098, the National Institutes of Health (GM58867–01), the Polymers and Genetics Programs of the U.S. National Science Foundation, and the U.S. Army Research Office. K.L.K. thanks the U.S. Department of Defense for a National Defense Science and Engineering Graduate Fellowship. E.S. was supported by a Howard Hughes Medical Institute Predoctoral Fellowship. The Center for New Directions in Organic Synthesis is supported by Bristol-Myers Squibb as a supporting member.
Abbreviations
- MetRS
methionyl-tRNA synthetase
- mDHFR
murine dihydrofolate reductase
- DMF
N,N-dimethylformamide
- MALDI
matrix-assisted laser desorption ionization
- TBST
Tris-buffered saline
- HRP
horseradish peroxidase
References
- 1.Dougherty D A. Curr Opin Chem Biol. 2000;4:645–652. doi: 10.1016/s1367-5931(00)00148-4. [DOI] [PubMed] [Google Scholar]
- 2.Marcaurelle L A, Bertozzi C R. Chem Eur J. 1999;5:1384–1390. [Google Scholar]
- 3.Cotton G J, Muir T W. Chem Biol. 1999;6:R247–R256. doi: 10.1016/s1074-5521(99)80109-4. [DOI] [PubMed] [Google Scholar]
- 4.Cornish V W, Mendel D, Schultz P G. Angew Chem Int Ed Engl. 1995;34:621–633. [Google Scholar]
- 5.Mendel D, Ellman J A, Chang Z, Veenstra D L, Kollman P A, Schultz P G. Science. 1992;256:1798–1802. doi: 10.1126/science.1615324. [DOI] [PubMed] [Google Scholar]
- 6.Furter R. Protein Sci. 1998;7:419–426. doi: 10.1002/pro.5560070223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Brock A, Herberich B, Schultz P G. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 8.Doring V, Mootz H D, Nangle L A, Hendrickson T L, de Crecy-Lagard V, Schimmel P, Marliere P. Science. 2001;292:501–504. doi: 10.1126/science.1057718. [DOI] [PubMed] [Google Scholar]
- 9.Li L T, Zhong W G, Zacharias N, Gibbs C, Lester H A, Dougherty D A. Chem Biol. 2001;8:47–58. doi: 10.1016/s1074-5521(00)00055-7. [DOI] [PubMed] [Google Scholar]
- 10.Kiick K L, Tirrell D A. Tetrahedron. 2000;56:9487–9493. [Google Scholar]
- 11.Cornish V W, Hahn K M, Schultz P G. J Am Chem Soc. 1996;118:8150–8151. [Google Scholar]
- 12.Griffin B A, Adams S R, Tsien R Y. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 13.Kiick K L, van Hest J C M, Tirrell D A. Angew Chem Int Ed Engl. 2000;39:2148–2152. [PubMed] [Google Scholar]
- 14.Kiick K L, Weberskirch R, Tirrell D A. FEBS Lett. 2001;502:25–30. doi: 10.1016/s0014-5793(01)02657-6. [DOI] [PubMed] [Google Scholar]
- 15.Rutjes F P J T, Wolf L B, Schoemaker H E. J Chem Soc Perkin Trans. 2000;1:4197–4212. [Google Scholar]
- 16.Furstner A, Guth O, Rumbo A, Seidel G. J Am Chem Soc. 1999;121:11108–11113. [Google Scholar]
- 17.Trnka T M, Grubbs R H. Acc Chem Res. 2001;34:18–29. doi: 10.1021/ar000114f. [DOI] [PubMed] [Google Scholar]
- 18.Saxon E, Bertozzi C R. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 19.Arnold L D, May R G, Vederas J C. J Am Chem Soc. 1988;110:2237–2241. [Google Scholar]
- 20.Arnold L D, Kalantar T H, Vederas J C. J Am Chem Soc. 1985;107:7105–7109. [Google Scholar]
- 21.Mangold J B, Mischke M R, LaVelle J M. Mutat Res. 1989;216:27–33. doi: 10.1016/0165-1161(89)90020-4. [DOI] [PubMed] [Google Scholar]
- 22.Hehre W J, Ditchfield R, Pople J A. J Chem Phys. 1972;56:2257–2261. [Google Scholar]
- 23.Francl M M, Pietro W J, Hehre W J, Binkley J S, Gordon M S, DeFrees D J, Pople J A. J Chem Phys. 1982;77:3654–3665. [Google Scholar]
- 24.Ghosh G, Brunie S, Schulman L H. J Biol Chem. 1991;266:17136–17141. [PubMed] [Google Scholar]
- 25.Mellot P, Mechulam Y, LeCorre D, Blanquet S, Fayat G. J Mol Biol. 1989;208:429–443. doi: 10.1016/0022-2836(89)90507-x. [DOI] [PubMed] [Google Scholar]
- 26.Blanquet S, Fayat G, Waller J-P. Eur J Biochem. 1974;44:343–351. doi: 10.1111/j.1432-1033.1974.tb03491.x. [DOI] [PubMed] [Google Scholar]
- 27.van Hest J C M, Kiick K L, Tirrell D A. J Am Chem Soc. 2000;122:1282–1288. [Google Scholar]
- 28.Serre L, Verdon G, Choinowski T, Hervouet N, Risler J-L, Zelwer C. J Mol Biol. 2001;306:863–876. doi: 10.1006/jmbi.2001.4408. [DOI] [PubMed] [Google Scholar]
- 29.Hirel P H, Schmitter J M, Dessen P, Fayat G, Blanquet S. Proc Natl Acad Sci USA. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bessette P H, Aslund F, Beckwith J, Georgiou G. Proc Natl Acad Sci USA. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.