Abstract
Recent publications have classified breast cancers on the basis of expression of cytokeratin-5 and -17 at the RNA and protein levels, and demonstrated the importance of these markers in defining sporadic tumours with bad prognosis and an association with BRCA1-related breast cancers. These important observations using different technology platforms produce a new functional classification of breast carcinoma. However, it is important in developing hypotheses about the pathogenesis of this tumour type to review the nomenclature that is being used to emphasize potential confusion between terminology that defines clinical subgroups and markers of cell lineage. This article reviews the lineages in the normal breast in relation to what have become known as the 'basal-like' carcinomas.
Introduction
Human breast cancers are heterogeneous in their morphology, response to therapy and clinical course. This heterogeneity may originate in differences in the underlying target cell population and/or it may be the result of different combinations of oncogene activation and loss of tumour suppressor gene function in a normal breast stem cell or committed progenitor. The concept of breast cancer stem cells and their relationship to kinetics in the normal breast was excellently reviewed by Behbod and Rosen [1]. The present review focuses on the characterization of the epithelium of the normal human breast and on what we know about the origin of breast cancer from morphological and cell biological viewpoints. Recent expression profiling data are considered within the context of this experimental classification of normal and neoplastic breast tissue, and of how these two valuable approaches are coming together to develop a new functional classification with predictive value for clinical behaviour and response to targeted therapies. In a recent commentary in this journal, Wilson and Dering [2] reviewed the current position in integrating available microarray data in relation to pathway signatures, with an emphasis on endocrine response. This review focuses on how these data relate to our limited knowledge of the cell-type origin of breast cancer.
Cell types in the normal breast: definition of a 'basal cell'
From 28 weeks of intrauterine life the normal human breast is composed of two cell layers, an inner luminal cell population and a distinct outer cell layer, juxtaposed to the basement membrane, termed the 'basal' layer [3]. Although the breast ductal system is comprised of domains with distinct morphology and function, this layered architecture is found throughout the mammary gland from the nipple to the terminal alveoli. This basal cell layer is morphologically heterogeneous in that cells appear either spindle-shaped or cuboidal, depending on their location in the branching structure of breast ducts and on the hormonal or menopausal status of the tissue. These cells can be distinguished from basal cells in stratified squamous epithelium because they exhibit many features of smooth muscle cells, including expression of smooth muscle actin (SMA), myosin [4] and neutral endopeptidase (CD10) [5] The term 'myoepithelial' cell was coined to describe cells that express both epithelial characteristics and these contractile proteins.
The use of the term 'basal' to refer to a cell population that expresses certain high-molecular-weight cytokeratins took its origin in the now classical papers of Moll and colleagues. In 1982 Moll, working in Werner Franke's laboratory, described by two-dimensional gel analysis the catalogue of human cytokeratins in cultured cells, normal epithelia and tumours [6]. That report also identified two main groups of breast cancers, based on their expression of simple or stratified epithelial cytokeratins. In 1983 Moll and coworkers [7] reported a more comprehensive comparison of cytokeratins expressed in tumours and their associated normal epithelia. This latter paper also confirmed that a small subgroup of breast cancers express stratified epithelial cytokeratins, including cytokeratin (CK)14 and CK17.
Over the next decade studies became more refined and antibodies became available that were specific for different cytokeratins, facilitating the mapping of individual cytokeratins at the cellular level. It was recognized by many groups that the high-molecular-weight cytokeratins CK5 and CK14, which form in vivo complexes, are expressed in the basal cells of stratified epithelium, and thus they became known as 'basal' keratins [8]. In many normal glandular epithelia, such as the salivary glands, CK5 and CK14 are found to be expressed in cells that sit adjacent to the basement membrane and in a 'basal position' from the ducts to the acini. Thus, in most tissues the nomenclature of a basal position and expression of 'basal' keratins defines the same population of cells. The term 'basal' then became synonymous with expression of basal keratins (CK5/CK14) rather than with a position adjacent to the basement membrane. However, confusion has arisen by applying this terminology in experimental model systems and in breast tumours to define the cell of origin because changes in intermediate filament expression can be modulated in tissue culture [9] and because, in the breast, expressions of CK5 and CK14 are apparently not restricted to myoepithelial cells.
A major problem in the human breast literature is that there is no consensus view that can be derived on the cell types expressing CK5, CK14 and CK17. This may in part have arisen from variations in antibody specificity used and differences in staining and tissue processing schedules. There is general agreement that myoepithelial cells do express these proteins, but there are numerous publications from laboratories that have clearly demonstrated staining of luminal cells in large ducts [10] but particularly in the terminal duct lobular unit (TDLU) complex (Fig. 1) [8,11-14]. Figure 1 was taken from one of 20 reduction mammoplasties examined. It should be noted that there was considerable variation within the same breast and between breasts. Some lobules were totally negative, whereas others exhibited occasional cells that were positive. The majority of the staining was in luminal cells with very rare weak staining of myoepithelial cells. A limited study of in situ carcinomas, with the same reagents, demonstrated a clear positivity in the myoepithelial cells when associated with in situ malignancy. A similar switch from luminal to myoepithelial cell staining has been seen with another protein that forms part of the 'basal-like' gene signature, namely annexin VIII (Stein and coworkers, unpublished observation). In terms of comparative pathology, it is of interest that in the mouse CK14 has also been demonstrated in a defined subpopulation of luminal cells [15]. However, it is a consistent finding, using both immunocytochemistry and proteomic analysis of separated myoepithelial cells and luminal cells [16], that CK8 and CK18 are only expressed in luminal cells. This very careful proteomic study also demonstrated multiple keratin isoforms and the presence of significant levels of CK5, CK14 and CK17 in isolated luminal cells.
Figure 1.
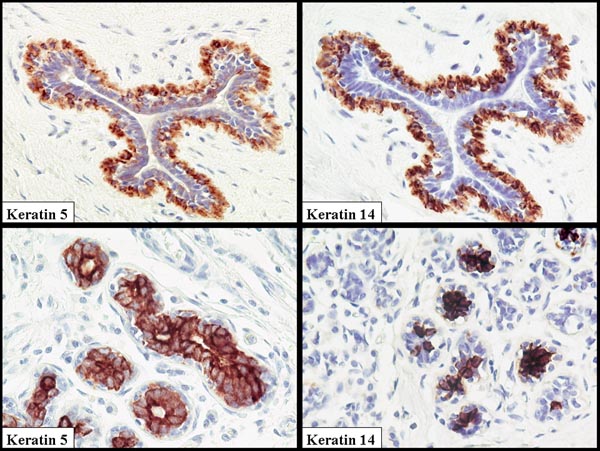
Normal breast duct and terminal duct lobular unit (TDLU) stained with antibodies to cytokeratin (CK)5 and CK14. Note the coexpression of these proteins that form a heterodimer. In this example the luminal cells are the dominantly stained population in the TDLU, but in the duct the myoepithelial cells are stained. Great variability can be seen both within the same breast and between specimens.
In the breast the term 'basal' thus has acquired two meanings. In one context it has become synonymous with breast myoepithelium and in the other it defines a specific subpopulation of 'basal' cytokeratin expressing cells that counter-intuitively may be found in either a luminal or basal location in normal glands.
Lineages in the normal breast?
The classic morphological work conducted by Wellings and coworkers [17-19], using whole mount preparations, clearly demonstrated that the majority of human breast cancers arise from the TDLU and not from the ductal system. Unfortunately, pathologists have consistently referred to two major subgroups of breast cancer as ductal and lobular carcinomas, both when invasive and in situ. The current evidence supports the contention that differences in morphology result from secondary genetic events [20,21] rather than a difference in the target cell, although the target cell for either major grouping has not been defined. The majority of breast cancers have a phenotype that supports origin from a cell(s) in the luminal compartment or from a cell that was committed to this lineage. This is based on the ability of the majority of breast carcinomas to form secretory acinar structures with polarized epithelial cells that express mucins on their luminal surface and have ultrastructural characteristics of secretory cells. It is generally accepted that breast cancers are clonal in their origin, but origin from a single cell is difficult to prove because X-linked inactivation studies have shown that the entire TDLU is clonal [22]. The cellular origin of breast cancers therefore requires knowledge of the normal lineages in the breast epithelium.
In order to establish the relationship between the myoepithelial and luminal cells, O'Hare [23] developed methods to isolate pure populations of these two cell types using cell surface markers. These early studies indicated that luminal cells and myoepithelial cells would proliferate as isolated populations and that they would breed true. In 1999, Petersen and coworkers [24] extended these studies and demonstrated, with sorted luminal and myoepithelial cells, that approximately 4% of cells, when placed in culture conditions that supported myoepithelial cells, lost the luminal marker CK18. Approximately 2% of cells gained expression of the myoepithelial markers β4 integrin, CD10 and, later, SMA. The myoepithelial lineage showed no signs of conversion. In parallel with these studies, a number of workers demonstrated that antibody KA1, which was subsequently shown to recognize CK5 in association with CK14 [25], labelled a subpopulation of luminal cells in the TDLUs and basally located cells in the ducts [8,11]. Similar findings were reported by Otterbach and coworkers [13] and the group of Boecker and Buerger [14] using the anticytokeratin antibody no 5/6 (Boehringer, Mannheim, Germany). These observations have been extended by Boecker and Buerger [14] and by Böcker and coworkers [26] using in vivo multilabelling with CK8/18 and SMA as markers of the terminal luminal and myoepithelial lineage to generate a model in which the CK5+ cells are adult progenitor cells, which go through transitions of intermediary double labelling cells (CK5+/CK18/8+, CK+/SMA+) to produce fully differentiated secretary luminal cells (CK18/8+) and myoepithelial cells (SMA+). These data from the laboratories of Petersen and Boecker indicate that the putative stem cell is in the luminal/suprabasal compartment.
More sophisticated in vitro analyses followed on approaches in the haemopoietic system, where stem cell populations are defined by their ability to efflux the dye Hoechst 33342 [27]. Cells with this phenotype are called the 'side population' (SP) and can be readily purified using flow cytometry. Vivanco and coworkers [28] carried out the most comprehensive study to date on the characterization of human breast cells within the SP fraction. On isolation these cells do not express either the luminal cell marker epithelial membrane antigen or the myoepithelial marker common acute lymphoblastic leukaemia antigen (CD10), but as single cells give rise to four types of colonies, as assessed by their expression of CK18 and CK14 (CK18+, CK14+, CK18-/CK14- and CK18+/CK14+). These findings parallel those of Welm and coworkers [29] in the mouse, who also found that the SP fraction was originally null for differentiation markers. Unfortunately, the report from Vivanco and coworkers [28] does not allow for the fact that CK14 is a marker of a subpopulation of luminal cells, and so interpretation of these mixed phenotypes within the context of differentiation into myoepithelial cells is difficult.
CK19 has also been proposed to be a stem cell or progenitor marker. A recent report from Clarke and coworkers [30] identified a CK19-positive, oestrogen receptor (ER)-positive population of cells that have the capacity for self renewal. The suprabasal putative precursor cells described by Petersen and coworkers [31] were CK19-positive and may be the same as those described by Clarke and colleagues [30]. This may also be the ER-positive progenitor cell proposed by Dontu and coworkers [32] in their model of human breast progenitors. These observations are consistent with the earlier findings reported by Taylor-Papadimitriou and coworkers [33] on a CK19 subpopulation in the lobules of the normal breast. These cell biological approaches detail the complexities of the cell types in normal breast, but until these hierarchies are better defined it is difficult to interpret current knowledge in relation to immortalizing events and heterogeneity in breast cancer. It is clear, however, that breast cancers can be subclassified on the basis of their molecular signature, regardless of the significance, if any, to normal lineage phenotypes.
Carcinomas with 'basal' gene expression have a distinct molecular signature
Perou and colleagues originally demonstrated that the phenotypic diversity of breast carcinomas is reflected in corresponding systematic variation in gene expression patterns. Multiple independent studies have now demonstrated that a subset of tumours in sporadic breast carcinoma cohorts express a gene expression signature that includes relatively high level expression of stratified epithelial keratins (CK5 and CK17) [34-38]. As detailed below, although breast tumours with an aggressive phenotype that express these cytokeratins were previously characterized, this subtype of tumour gained considerable notoriety with their rediscovery by the use of cDNA microarrays. A consistent biological theme is apparent in the genes that distinguish these tumours, in that many encode proteins that are expressed by normal stratified epithelia. Included in this are genes that mediate cell–cell interactions (e.g. CDH3), matrix remodelling genes (e.g. MMP14), and genes that encode growth factor receptors (e.g. EGFR) and extracellular matrix proteins (e.g. LAMA3 and LAMC2). A comparison of the gene expression patterns between breast-derived tumours and cell lines in culture revealed that these tumours share signatures with breast tumours classified as 'luminal' or 'erbB2' by expression of common epithelial genes (e.g. CDH1 and JUP) but are distinguished by expression of the signature shared with cultured mammary epithelial cells (human mammary epithelial cells and immortalized normal breast epithelial cell lines) [39].
Several lines of evidence from molecular studies support the idea that these tumours are biologically distinct and may arise through distinct oncogenic pathways. Analysis of a breast cancer gene expression dataset that included both sporadic patients and carriers of the BRCA1 mutation demonstrated a strong association between BRCA1-related breast cancers and expression of the 'basal' gene expression signature [35]. This was supported by a study conducted by Foulkes and coworkers [40] that demonstrated an association between BRCA1-related breast cancers and immunoreactive basal CK5/6. Based on these observations, Foulkes [41] hypothesized that BRCA1 is a stem cell regulator. The model proposed by Behbod and Rosen [1] considers myoepithelial differentiation as being end-stage and 'basal-like' tumours arising from ER-negative long-term stem cells. This model would indicate that the stratified cytokeratins are markers of more than one cell population in the normal breast.
In addition, both cytogenetic and comparative genomic hybridization analyses suggest that breast tumours that express stratified epithelial keratins also have quantitative genomic abnormalities that distinguish them from tumours that express simple epithelial keratins [42,43]. Interestingly, Jones and coworkers [44] identified two subgroups of CK14-expressing tumours distinguished by cytogenetic differences. This may be related to the so-called 'normal' breast subtype identified in gene expression experiments that express gene patterns similar to the basal signature [34,35] and/or the subgroups of basal alluded to by Sortiriou and coworkers [37]. Moll's group and others [45] have pointed out that an explanation for stratified cytokeratin expression may be found in relation to squamous metaplasia. It is of interest in this context that the basal signature also distinguishes squamous cells from adenocarcinoma of the lung [46] and a subtype of head and neck tumours with poor prognosis [47]. This common signature in comparisons between tumours derived from different origins suggests that these tumours may arise in functionally related cell types that are unrelated to the tissue of origin. This suggests that, analogous to the classification of leukaemia subtypes, a novel classification of carcinomas may emerge that relates subtypes to differentiated cell types in normal epithelia that transcends tissue of origin.
What is the clinical significance of cytokeratin defined subtypes of breast cancer?
As it became clear that the various types of epithelial differentiation in the body and the tumours from which they were derived expressed different sets of cytokeratins, cytokeratin staining was rapidly applied in diagnostic pathology [48]. It was also noted at about the same time that the pattern of expression of CK5, CK14, and CK17 identified the myoepithelial cells of in situ breast lesions, and therefore was useful in distinguishing benign from malignant disease [13,25]. The potential poor survival or early recurrence associated with CK5/17 expression in tumour cells was first reported by Dairkee and coworkers [49] in 1987. This indicated that the tumours expressed phenotypes related to normal luminal cells as well as to stratified epithelial keratins, recapitulating the heterogeneity of the luminal cells in the normal TDLU. In 1991 Wetzels and coworkers [11] took the observations of Dairkee and colleagues forward with a detailed analysis of keratins in benign and malignant breast disease. In that study numerous antibodies were used against individual cytokeratins. Using three antibodies that recognize CK14 (i.e. LLO02, KA1 and EKH4), those investigators demonstrated that with minor variations all the reagents produced a similar pattern of immunoreactivity and that breast cancers fell into two main groupings: 38% of carcinomas expressed stratified epithelial cytokeratins (CK5, CK14 and/or CK17) and the rest expressed only simple epithelial keratins.
As pointed out by Moll and coworkers [12], the biological significance of the differential expression of cytokeratin polypeptides in breast carcinomas is unclear. Although we do not know the functional role that cytokeratins CK5, CK14 and CK17 play, it is clear that their expression is associated with poor prognosis. Moll and colleagues examined the keratin profile of 101 graded breast carcinomas. This is an important study because it clearly indicated that there was a correlation between grade and keratin profile, with grades 1 and 2 being associated with simple keratins and high-grade tumours being associated with the stratified epithelial keratins CK4, CK14 and/or CK17 [12]. This keratin phenotype was also associated with short overall and disease-free survival, and ER negativity, particularly in the node positive cohort. All tumours expressed simple epithelial cytokeratins (CK7, CK8, CK18, or CK19) and 62% of the 45 grade 3 carcinomas had a bimodal expression pattern, coexpressing at least one stratified cytokeratin (CK4 36%, CK5 18%, CK14 20% and CK17 38%). This important study, demonstrating the coexpression of simple and stratified cytokeratin expression in the same tumours, continued earlier work conducted by Nagle and coworkers [8] in 1986, and was confirmed recently by Abd El-Rehim and colleagues [50]. In this study 1944 carcinomas were examined, of which 98.8% were positive for simple epithelial (luminal) keratins. Combined luminal and basal cytokeratins were expressed in 27.4%, basal alone in 0.8%, and 0.4% expressed neither.
Observations from Moll's laboratory were recently extended by numerous other investigators who have shown similar correlations between poor prognosis and expression of CK5, CK14 and CK17 [39,50,51]. Korsching and coworkers [43], using immunohistochemical expression analysis of 15 proteins with hierarchical cluster analysis, extended these findings to identify clinical subgroups. They confirmed that CK5/6 immunoreactive tumours are generally negative for ER and progesterone receptor, and form a clinical subgroup distinct from c-erbB-2 expressing tumours. They also found a strong correlation between this subgroup with increased expression of p53, epidermal growth factor receptor (EGFR) and proliferative index. Other groups have described a correlation between basal cytokeratin expression and atypical and typical medullary carcinomas [52].
More recently, Nielsen and colleagues [53] took 21 tumours that were defined as 'basal' by microarray, and performed an immunohistochemical analysis with six distinguishing markers (ER, CK5/6, CK17, EGFR, HER2, c-Kit). They concluded that an immunohistochemical surrogate for gene array experiments to identify 'basal-like' breast cancers is ER-negative, HER2-negative/low, and CK5/6-positive and/or HER1-positive. Thus, 16 out of 21 tumours expressing the basal gene expression signature would have been identified using these criteria, providing a sensitivity of 76% and a specificity of 100%. It is clear from the discussion in this paper that the term 'basal-like tumour' is still to be clearly defined at the immunohistochemical level, because the presence of basal cytokeratin positivity was not a requirement. There is a tendency to move toward a definition of a basal-like tumour by exclusion on the basis that these tumours are ER negative and c-erbB-2 negative. Because there is no absolute correlation between HER1, c-Kit, CK5/14/17, ER negativity and c-erbB-2 negativity, it may be best to await standardization of methodologies, protocols and agreements on reagents for immunocytochemistry before new classifications are developed. For clinical purposes, it would seem appropriate to reconsider the terminology and that, instead of rigidly defining subgroups, a pragmatic marker-driven approach is required.
The possibility that detection of basal-like tumours might be relevant to development of tailored treatment approaches is raised by their high level expression of EGFR and c-Kit, both targets of recently approved therapies. An interesting study conducted by Troester and coworkers [54] identified chemotherapy-induced gene expression signatures in cell line models of luminal epithelial tumours (MCF-7 and ZR-75-1) and compared them with induced genes in telomerase-immortalized cell line models of basal epithelial tumours. They then identified gene expression signatures in comparison between two samples from patients taken before and after 16 weeks of chemotherapy, including both luminal and basal classified cases. It was intriguing in these very different experimental approaches that they identified some genes induced in common between basal cell lines and tumours that were different from the individual genes in common between 'luminal' cell lines and tumours. This suggests that the physiological differences between luminal and basal breast cancers may have implications for their response to standard chemotherapy.
Conclusion
In this article we have tried to produce an overview of current knowledge relating to cytokeratin expression and the basal cell phenotype. Biochemical, immunohistochemical and gene expression profiling technologies are inherently descriptive and therefore do not directly address hypotheses on the lineage of different cancer types. A definitive statement cannot be made on the normal localization of cytokeratins, and thus the use of cytokeratin expression to define relationships between cells and the origins of cancer is premature and might be misleading. Whatever the histogenic relationships, if any, between the cells expressing CK5 and CK14 and the group of tumours defined as 'basal-like', it is clear that the combination of morphology and expression profiling has defined a potentially important tumour subset that can be identified as a poor prognostic group and should be considered for individual management. The clustering data have revealed a number of known and new potential targets for diagnosis, predictive testing and therapy.
Abbreviations
CK = cytokeratin; EGFR = epidermal growth factor receptor; ER = oestrogen receptor; SMA = smooth muscle actin; SP = side population; TDLU = terminal duct lobular unit.
Competing interests
The author(s) declare that they have no competing interests.
Acknowledgments
Acknowledgements
The laboratory of BAG is supported by a programme grant from Breakthrough Breast Cancer UK.
Contributor Information
Barry A Gusterson, Email: bag5f@clinmed.gla.ac.uk.
Douglas T Ross, Email: dross@applied-genomics.com.
Victoria J Heath, Email: v.heath@clinmed.gla.ac.uk.
Torsten Stein, Email: ts33s@udcf.gla.ac.uk.
References
- Behbod F, Rosen JM. Will cancer stem cells provide new therapeutic targets? Carcinogenesis. 2004;26:703–711. doi: 10.1093/carcin/bgh293. [DOI] [PubMed] [Google Scholar]
- Wilson CA, Dering J. Recent translational research: microarray expression profiling of breast cancer – beyond classification and prognostic markers? Breast Cancer Res. 2004;6:192–200. doi: 10.1186/bcr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbazhagan R, Osin PP, Bartkova J, Nathan B, Lane B, Gusterson BA. The development of epithelial phenotypes in the human fetal and infant breast. J Pathol. 1998;184:197–206. doi: 10.1002/(SICI)1096-9896(199802)184:2<197::AID-PATH992>3.3.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Gusterson BA, Warburton MJ, Mitchell D, Ellison M, Neville AM, Rudland PS. Distribution of myoepithelial cells and basement membrane proteins in the normal breast and in benign and malignant breast disease. Cancer Res. 1982;42:4763–4770. [PubMed] [Google Scholar]
- Gusterson BA, Monaghan P, Mahendran R, Ellis J, O'Hare MJ. Identification of myoepithelial cells in human and rat breasts by anti-common acute lymphoblastic leukaemia antigen antibody A12. J Natl Cancer Inst. 1986;77:343–349. [PubMed] [Google Scholar]
- Moll R, Franke WW, Schiller DL. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- Moll R, Krepler R, Frank WW. Complex cytokeratin polypeptide patterns observed in certain human carcinomas. Differentiation. 1983;23:256–269. doi: 10.1111/j.1432-0436.1982.tb01291.x. [DOI] [PubMed] [Google Scholar]
- Nagle RB, Bocker W, Davis JR, Heid HW, Kaufmann M, Lucas DO, Jarasch ED. Characterization of breast carcinomas by two monoclonal antibodies distinguishing myoepithelial from luminal epithelial cells. J Histochem Cytochem. 1986;34:869–881. doi: 10.1177/34.7.2423579. [DOI] [PubMed] [Google Scholar]
- Ben-Ze'ev A. Cell density and cell shape-related regulation of vimentin and cytokeratin synthesis. Exp Cell Res. 1985;157:520–532. doi: 10.1016/0014-4827(85)90137-5. [DOI] [PubMed] [Google Scholar]
- Su L, Morgan PR, Lane EB. Expression of cytokeratin messenger RNA versus protein in the normal mammary gland and in breast cancer. Hum Pathol. 1996;27:800–806. doi: 10.1016/S0046-8177(96)90452-9. [DOI] [PubMed] [Google Scholar]
- Wetzels RHW, Kuijpers HJH, Lane EB, Leigh IM, Troyanovsky SM, Holland R, van Haelst UJGM, Ramaekers PCS. Basal cell specific and hyperproliferation-related keratins in human breast cancer. Am J Pathol. 1991;138:751–763. [PMC free article] [PubMed] [Google Scholar]
- Malzahn K, Mitze M, Thoenes M, Moll R. Biological and prognostic significance of stratified epithelial cytokeratins in infiltrating ductal breast carcinomas. Virchows Arch. 1998;433:119–129. doi: 10.1007/s004280050226. [DOI] [PubMed] [Google Scholar]
- Otterbach F, Bànkfalvi À, Bergner S, Decker T, Krech R, Boecker W. Cytokeratin 5/6 immunohistochemistry assists the differential diagnosis of atypical proliferations of the breast. Histopathology. 2000;37:232–240. doi: 10.1046/j.1365-2559.2000.00882.x. [DOI] [PubMed] [Google Scholar]
- Boecker W, Buerger H. Evidence of progenitor cells of glandular and myoepithelial cell lineages in the human adult female breast epithelium: a new progenitor (adult stem) cell concept. Cell Prolif. 2003;36:73–84. doi: 10.1046/j.1365-2184.36.s.1.7.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GH, Mehrel T, Roop DR. Differential keratin gene expression in developing, differentiating, preneoplastic, and neoplastic mouse mammary epithelium. Cell Growth Differ. 1990;1:161–170. [PubMed] [Google Scholar]
- Page MJ, Amess B, Townsend RR, Parekh R, Herath A, Brusten L, Zvelebil MJ, Stein RC, Waterfield MD, Davies SC, et al. Proteomic definition of normal human luminal and myoepithelial breast cells purified from reduction mammoplasties. Proc Natl Acad Sci USA. 1999;96:12589–12594. doi: 10.1073/pnas.96.22.12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellings SR. A hypothesis of the origin of human breast cancer from the terminal ductal lobular unit. Pathol Res Pract. 1980;166:515–535. doi: 10.1016/S0344-0338(80)80248-2. [DOI] [PubMed] [Google Scholar]
- Jensen HM, Rice JR, Wellings SR. Preneoplastic lesions in the human breast. Science. 1976;191:295–297. doi: 10.1126/science.1246614. [DOI] [PubMed] [Google Scholar]
- Wellings SR, Jensen HM, Marcum RG. An atlas of subgross pathology of the human breast with special reference to possible precancerous lesions. J Natl Cancer Inst. 1975;55:231–273. [PubMed] [Google Scholar]
- Osin P, Shipley J, Lu YJ, Crook T, Gusterson BA. Experimental pathology and breast cancer genetics: new technologies. Recent Results Cancer Res. 1998;152:35–48. doi: 10.1007/978-3-642-45769-2_4. [DOI] [PubMed] [Google Scholar]
- Simpson PT, Reis-Filho JS, Gale T, Lakhani SR. Molecular evolution of breast cancer. J Pathol. 2005;205:248–254. doi: 10.1002/path.1691. [DOI] [PubMed] [Google Scholar]
- Diallo R, Schaefer K-L, Poremba C, Shivazi N, Willman V, Buerger H, Dockhorn-Dworniczak B, Boecker W. Monoclonality in normal epithelium and in hyperplastic and neoplastic lesions of the breast. J Pathol. 2001;193:27–32. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH747>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- O'Hare MJ, Ormerod MG, Monaghan P, Lane EB, Gusterson BA. Characterization in vitro of luminal and myoepithelial cells isolated from the human mammary gland by cell sorting. Differentiation. 1991;46:209–221. doi: 10.1111/j.1432-0436.1991.tb00883.x. [DOI] [PubMed] [Google Scholar]
- Péchoux C, Gudjonsson T, Rønnov-Jessen L, Bissell MJ, Petersen OW. Human mammary luminal epithelial cells contain progenitors to myoepithelial cells. Dev Biol. 1999;206:88–99. doi: 10.1006/dbio.1998.9133. [DOI] [PubMed] [Google Scholar]
- Jarasch ED, Nagle RB, Kaufmann M, Maurer C, Bocker WJ. Differential diagnosis of benign epithelial proliferations and carcinomas of the breast using antibodies to cytokeratins. Hum Pathol. 1988;19:276–289. doi: 10.1016/s0046-8177(88)80520-3. [DOI] [PubMed] [Google Scholar]
- Böcker W, Moll R, Poremba C, Holland R, van Diest PJ, Dervan P, Bürger J, Wai D, Diallo RI, Brandt B, et al. Common adult stem cells in the human breast give rise to glandular and myoepithelial cell lineages: a new cell biological concept. Lab Invest. 2002;82:737–745. doi: 10.1097/01.lab.0000017371.72714.c5. [DOI] [PubMed] [Google Scholar]
- Alvi AJ, Clayton H, Joshi C, Enver T, Ashworth A, Vivanco M dM, Dale TC, Smalley MJ. Functional and molecular characerisation of mammary side population cells. Breast Cancer Res. 2002;5:R1–R8. doi: 10.1186/bcr547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton H, Titley I, Vivanco M. Growth and differentiation of progenitor/stem cells derived from the human mammary gland. Exp Cell Res. 2004;297:444–460. doi: 10.1016/j.yexcr.2004.03.029. [DOI] [PubMed] [Google Scholar]
- Welm BE, Tepera SB, Venezia T, Graubert TA, Rosen JM, Goodell MA. Sca-1pos cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- Clarke RB, Spence K, Anderson E, Howell A, Okano H, Potten CS. A putative breast stem cell population is enriched for steroid receptor-positive cells. Dev Biol. 2005;277:443–456. doi: 10.1016/j.ydbio.2004.07.044. [DOI] [PubMed] [Google Scholar]
- Petersen OW, Gudjonsson T, Villadsen R, Bissell MJ, Rønnov-Jessen L. Epithelial progenitor cell lines as models of normal breast morphogenesis and neoplasia. Cell Prolif. 2003;36:33–44. doi: 10.1046/j.1365-2184.36.s.1.4.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, El-Ashry D, Wicha MS. Breast cancer, stem/progenitor cells and the estrogen receptor. Trends Endocrinol Metab. 2004;15:193–197. doi: 10.1016/j.tem.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Taylor-Papadimitriou J, Stampfer M, Bartek J, Lewis A, Boshell M, Lane EB, Leigh IM. Keratin expression in human mammary epithelial cells cultured from normal and malignant tissue: relation of in vivo phenotypes and influence of medium. J Cell Sci. 1989;94:403–413. doi: 10.1242/jcs.94.3.403. [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Langerod A, Ji Y, Nowels KW, Nesland JM, Tibshirani R, Bukholm IK, Karesen R, Botstein D, Borresen-Dale AL, et al. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15:2523–2536. doi: 10.1091/mbc.E03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross DT, Perou CM. A comparison of gene expression signatures from breast tumors and breast tissue derived cell lines. Dis Markers. 2001;17:99–109. doi: 10.1155/2001/850531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Stefansson IM, Chappuis PO, Bégin LR, Goffin JR, Wong N, Trudel M, Akslen LA. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95:1482–1485. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- Foulkes WD. BRCA1 functions as a breast stem cell regulator. J Med Genet. 2004;41:1–5. doi: 10.1136/jmg.2003.013805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, Nonni AV, Fulford L, Merrett S, Chaggar R, Eusebi V, Lakhani SR. CGH analysis of ductal carcinoma of the breast with basaloid/myoepithelial cell differentiation. Br J Cancer. 2001;85:422–427. doi: 10.1054/bjoc.2001.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsching E, Packeisen J, Agelopoulos K, Eisenacher M, Voss R, Isola J, van Diest PJ, Brandt B, Boecker W, Buerger H. Cytogenetic alterations and cytokeratin expression patterns in breast cancer: integrating a new model of breast differentiation into cytogenetic pathways of breast carcinogenesis. Lab Invest. 2002;82:1525–1533. doi: 10.1097/01.lab.0000038508.86221.b3. [DOI] [PubMed] [Google Scholar]
- Jones C, Ford E, Gillett C, Ryder K, Merrett S, Reis-Filho JS, Fulford LG, Hanby A, Lakhani SR. Molecular cytogenetic identification of subgroups of grade III invasive ductal breast carcinomas with different clinical outcomes. Clin Cancer Res. 2004;10:5988–5997. doi: 10.1158/1078-0432.CCR-03-0731. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Milanezi F, Paredes J, Silva P, Pereira EM, Maeda SA, de Carvalho LV, Schmitt FC. Novel and classic myoepithelial/stem cell markers in metaplastic carcinomas of the breast. Appl Immunohistochem Mol Morphol. 2003;11:1–8. doi: 10.1097/00022744-200303000-00001. [DOI] [PubMed] [Google Scholar]
- Chung CH, Bernard PS, Perou CM. Molecular portraits and the family tree of cancer. Nat Genet. 2002;(Suppl):533–540. doi: 10.1038/ng1038. [DOI] [PubMed] [Google Scholar]
- Chung CH, Parker JS, Karaca G, Wu J, Funkhouser WK, Moore D, Butterfoss D, Xiang D, Zanation A, Yin X, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489–500. doi: 10.1016/S1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- Nagle RB. Intermediate filaments: efficacy in surgical pathologic diagnosis. Am J Clin Pathol. 1989;91:14–18. [PubMed] [Google Scholar]
- Dairkee SH, Mayall BH, Smith HS, Hackett AJ. Monoclonal marker that predicts early recurrence of breast cancer. Lancet. 1987;1:514. doi: 10.1016/s0140-6736(87)92129-5. [DOI] [PubMed] [Google Scholar]
- Abd El-Rehim DM, Pinder SE, Paish CE, Bell J, Blamey RW, Robertson JFR, Nicholson RI, Ellis IO. Expression of luminal and basal cytokeratins in human breast carcinoma. J Pathol. 2004;203:661–671. doi: 10.1002/path.1559. [DOI] [PubMed] [Google Scholar]
- Van de Rijn M, Perou CM, Tibshirani R, Haas P, Kallioniemi O, Kononen J, Torhorst J, Sauter G, Zuber M, Köchli OR, et al. Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol. 2002;161:1991–1996. doi: 10.1016/S0002-9440(10)64476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tot T. The cytokeratin profile of medullary carcinoma of the breast. Histopathology. 2000;37:175–181. doi: 10.1046/j.1365-2559.2000.00889.x. [DOI] [PubMed] [Google Scholar]
- Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandex-Boussard T, Livasy C, Cowan D, Dressler L, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- Troester MA, Hoadley KA, Sorlie T, Herbert BS, Borresen-Dale AL, Lonning PE, Shay JW, Kaufmann WK, Perou CM. Cell-type-specific responses to chemotherapeutics in breast cancer. Cancer Res. 2004;64:4218–4226. doi: 10.1158/0008-5472.CAN-04-0107. [DOI] [PubMed] [Google Scholar]


