Abstract
Resveratrol, a natural compound found in many dietary plants and in red wine, plays an important role in the prevention of many human pathological processes, including inflammation, atherosclerosis and carcinogenesis. We have shown that the antiproliferative activity of resveratrol correlated with its ability to inhibit the replicative pols (DNA polymerases) α and δ in vitro [Stivala, Savio, Carafoli, Perucca, Bianchi, Maga, Forti, Pagnoni, Albini, Prosperi and Vannini (2001) J. Biol. Chem. 276, 22586–22594]. In this paper, we present the first detailed biochemical investigation on the mechanism of action of resveratrol towards mammalian pols. Our results suggest that specific structural determinants of the resveratrol molecule are responsible for selective inhibition of different mammalian pols, such as the family B pol α and the family X pol λ. Moreover, the resveratrol derivative trans-3,5-dimethoxy-4-hydroxystilbene, which is endowed with a strong antiproliferative activity (Stivala et al., 2001), can inhibit pols α and λ and also suppress the in vitro SV40 DNA replication. The potency of inhibition is similar to that of aphidicolin, an inhibitor of the three replicative pols α, δ and ε. Our findings establish the necessary background for the synthesis of resveratrol derivatives having more selective and potent antiproliferative activity.
Keywords: cell proliferation, DNA polymerase, DNA replication, enzyme kinetics, eukaryote, resveratrol
Abbreviations: BRCT, BRCA1 C-terminal domain; pol, DNA polymerase; ss, single-stranded; SV40, simian virus 40; t-DHMSB, trans-3,5-dihydroxy-4′-methoxystilbene; t-DHSB, trans-4,4′-dihydroxystilbene; t-DMHSB, trans-3,5-dimethoxy-4′-hydroxystilbene; DTT, dithiothreitol; TDT, terminal-deoxynucleotidyl transferase
INTRODUCTION
Resveratrol (3,4,5-trihydroxystilbene) is synthesized by several plants in response to adverse conditions such as environmental stress or pathogenic attack and it is classified as a phytoalexin [1–3]. Resveratrol has been found in a multitude of dietary plants and in grape skin, being present in relatively high concentrations in grape juice and, especially, in red wine [4–7]. Growing evidence suggests that resveratrol plays a role in the prevention of human pathological processes, such as inflammation [8–10], atherosclerosis [11–13] and carcinogenesis [14,15]. These protective effects have been attributed to its antioxidant and anticyclo-oxygenase activity [16–18], and to a modulating activity of lipid and lipoprotein metabolism [19,20]. Moreover, the proliferative activity of normal cells and various human malignant cell lines is slowed down by resveratrol [21–24], resulting in the accumulation of cells in S and G2 phases of the cell cycle [25].
By exploiting a series of resveratrol derivatives, we have shown that the 4′-hydroxy group in trans-conformation (hydroxystyryl moiety) was absolutely required for its antiproliferative activity and that there was a direct correlation, from a structural point of view, between the antiproliferative effect and the ability to inhibit the replicative pols (DNA polymerases) α and δ [26]. Thus one of the mechanisms underlying the inhibition of cell-cycle progression is probably the interaction between the 4′-hydroxystyryl moiety of trans-resveratrol and replicative pols. To expand further these results, we have undertaken an enzymological characterization of the inhibition of the DNA synthetic activity of other mammalian pols by selected resveratrol derivatives.
In mammalian cells, many different pols have been discovered so far, belonging to four different families: family A, comprising pol θ, pol ν and pol γ; family B, comprising pol α, pol δ, pol ε and pol ζ; family X, comprising pol β, pol λ, pol μ, pol σ and TDT (terminal-deoxynucleotidyl transferase); and family Y, comprising pol η, pol ι and pol κ [27].
In the present study, we focused our attention on the family X enzymes. Among family X members, pol β is the major DNA repair enzyme in mammalian cells and TDT is necessary for IgG gene diversity generation, whereas pol μ and pol λ probably play important roles in the non-homologous end-joining DNA repair pathway [28]. Interestingly, pol λ is also endowed with a terminal-transferase activity [29,30].
Besides providing a detailed investigation on the mechanism of action of resveratrol towards pols, our results show that specific structural determinants of the resveratrol molecule are responsible for selective inhibition of mammalian enzymes from different families, such as the family B pol α and the family X pol λ. Our findings provide the necessary background for the synthesis of resveratrol derivatives with more selective and potent antiproliferative activity.
MATERIALS AND METHODS
Chemicals
[3H]dTTP (30 Ci/mmol) was obtained from Amersham Biosciences and unlabelled dNTPs, poly(dA), and oligo(dT)12–18 were from Roche Molecular Biochemicals (Monza, Italy). The oligonucleotides were obtained from MWG Biotech (Florence, Italy). Whatman (Clifton, NJ, U.S.A.) was the supplier of the GF/C filters. Resveratrol was from Sigma. All other reagents were of analytical grade and were purchased from Merck or Fluka (Buchs, Switzerland).
Chemistry
The t-DHMSB (trans-3,5-dihydroxy-4′-methoxystilbene) and t-DMHSB (trans-3,5-dimethoxy-4′-hydroxystilbene) derivatives were synthesized as described in [26]. t-DHSB (trans-4,4′-dihydroxystilbene) was obtained by Perkin condensation by the following procedure: (a) 1.04 g of 4-hydroxybenzaldehyde (1 mol), 1.96 g of 4-hydroxyphenylacetic acid (1.5 mol), 15 ml of acetic anhydride and 3 ml of triethylamine were heated at 180 °C under N2 for 6 h, cooled at room temperature (25 °C) and diluted in 30 ml of 5% (v/v) HCl and 30 ml of ethyl acetate. The aqueous layer was extracted with ethyl acetate (3×30 ml). The organic layer was washed with 50 ml of brine, dried over MgSO4 and the solvent was removed under reduced pressure to obtain the diacetylated α,β-(p-hydroxyphenyl)-p-hydroxycinnamic acid, which was then deacetylated at room temperature in methanol with K2CO3. After removing the methanol under reduced pressure, the residue was diluted in 30 ml of 5% HCl and 30 ml of ethyl acetate. The aqueous layer was extracted with ethyl acetate (3×30 ml). The organic layer was washed with 50 ml of brine, dried over MgSO4 and the solvent was removed under reduced pressure to obtain the α,β-(p-hydroxyphenyl)-p-hydroxycinnamic acid. (b) To 2.5 g of α,β-(p-hydroxyphenyl)-p-hydroxycinnamic acid were added 12 ml of quinoline and 500 mg of copper chromite. The reaction flask was heated at 240 °C under N2 for 5 h. The mixture was then cooled, filtered over celite and the filtrate was then washed with 2×30 ml of ethyl acetate. The organic layer was washed with 2×30 ml of 5% HCl, dried over MgSO4 and the solvent was removed under reduced pressure. The black oil obtained was purified by chromatography on silica gel (petroleum ether/ethyl acetate in the ratios 8:2, 7:3, 6:4) to give t-DHSB as a pale yellow solid (0.43 g). Yield: 25%; 1H-NMR (Me2SO-d6, 200 MHz): δ (p.p.m.) 6.72 (AA′ multiplet, 4H, H-3,5, H-3′,5′); 6.88 (singlet, 2H, CH=CH); 7.33 (BB′ multiplet, 4H, H-2,6, H-2′,6′); 9.41 (singlet, 2H, OH).
Enzymes and proteins
Recombinant human pol λ wild-type and the pol λΔN (amino acids 244–575) truncated form were cloned and expressed as described in [30]. After purification, the proteins were >90% homogenous, as judged by SDS/PAGE and Coomassie Blue staining (results not shown) and had specific activities of 200000 units/mg for wild-type pol λ and 180000 units/mg for the pol λΔN mutant; 1 unit of pol activity corresponds to the incorporation of 1 pmol of total dTMP into acid-precipitable material in 60 min at 37 °C in a standard assay containing 0.5 μg (as nucleotides) of poly(dA)/oligo(dT)10:1 and 10 μM dTTP. Saccaromyces cerevisiae pol IV was expressed and purified as described in [31] with the following modifications: the coding region of S. cerevisiae pol IV was amplified by PCR from the genomic DNA of S. cerevisiae. The primers used introduced XhoI and NcoI sites for cloning the pol IV gene into the pRSETb vector (Invitrogen). The primers used were: ScIV-F, 5′-CTGACTCGAGGTCTCTAAAGGGTAAATTTTTCG, and ScIV-R, 5′-CTGACCATGGCTTATGCAGTTTTTTTTTCCCATTC (restriction sites XhoI and NcoI are shown in boldface). Expression of pol IV was performed in Escherichia coli strain BL21(DE3) at an A600 of 0.4 by adding 0.3 mM isopropyl β-D-thiogalactoside at 28 °C for 3 h. Cells were harvested by centrifugation and the pellet was mixed with an equal volume of buffer A (30 mM phosphate buffer, pH 8.0, 10 mM Tris/HCl, pH 8.0, 500 mM NaCl, 10 mM imidazole, 1 mM PMSF, 1 μM benzamidine, 5 μg/ml leupeptin and 2 μg/ml pepstatin A). The cells were disrupted by a French press (twice) and sonicated on ice for 1 min. After centrifugation (20000 g for 30 min at 4 °C on a SS-34 rotor), the soluble fraction was loaded on to a 1 ml HiTrap Chelating (Ni+) column preequilibrated with buffer A. The column was washed with 50 ml of buffer A and 20 ml of buffer A containing 50 mM imidazole. The bound proteins were eluted by 300 mM imidazole in buffer A. After desalting to buffer B [40 mM Tris/HCl, pH 7.5, 50 mM NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, 15% (v/v) glycerol, 1 mM PMSF, 1 μM benzamidine, 5 μg/ml leupeptin and 2 μg/ml pepstatin] by using a HiTrap desalting column, the eluate was loaded on to a 1 ml HiTrap Heparin column pre-equilibrated with buffer B, the column was washed with 20 ml of buffer B and the proteins were eluted with a 20 ml linear NaCl gradient (50–1000 mM). Pol IV eluted at 250 mM NaCl as tested by pol activity, SDS/PAGE and Western blotting. The pool of pol IV was diluted to 50 mM NaCl and finally loaded on to a Mono S column pre-equilibrated with buffer B. Chromatography was performed exactly in the same way as for heparin–Sepharose. The yield from 1 litre of culture was approx. 30 mg of pol IV protein with purity above 95%. Human pol α was purified as described in [32]. Human pol β and calf thymus TDT were obtained from Trevigen (Gaithersburg, MD, U.S.A.).
Enzymatic assays
Pol assay
Human pol λ and S. cerevisiae pol IV activities on poly(dA)/oligo(dT)10:1 were determined in a final volume of 25 μl containing: 50 mM Tris/HCl (pH 7.0), 0.25 mg/ml BSA, 1 mM DTT (dithiothreitol), 0.5 mM MnCl2, 0.2 μM poly(dA)/oligo(dT)10:1 (3′-OH ends), 50 nM pol λ (or 0.1 unit of pol IV) and 5 μM [3H]dTTP (5 Ci/mmol), except where otherwise indicated in the Figure legends. All reactions were incubated for 15 min at 37 °C unless otherwise stated and the DNA was precipitated with 10% (w/v) trichloroacetic acid. Insoluble radioactive material was determined by scintillation counting as described in [32]. Pol β and pol α activities were assayed with poly(dA)/oligo(dT) as described in [32].
Terminal transferase assay
The terminal transferase activities of pol λ and TDT were assayed in a final volume of 25 μl containing: 50 mM Tris/HCl (pH 7.0), 0.25 mg/ml BSA, 1 mM DTT, 0.5 mM MnCl2 and 0.2 μM of ss (single-stranded) 27-mer DNA oligonucleotide, except where otherwise stated. The enzymes and [3H]dTTP (10 Ci/mmol) were added as indicated in the Figure legends. All reactions were incubated at 37 °C for 10 min, unless otherwise indicated in the Figures, and the DNA was precipitated with 10% trichloroacetic acid. Insoluble radioactive material was determined by scintillation counting as described in [32].
In vitro SV40 (simian virus 40) DNA replication assay
Reactions were performed in a final volume of 50 μl. S-100 extract (100 μg) from SV40-transformed African green monkey kidney cells (COS-7) were pre-incubated for 5 min at room temperature in the reaction buffer (50 mM Hepes/NaOH, pH 8.0, 0.5 mM DTT and 5 mM MgCl2), in the absence or presence of t-DMHSB or aphidicolin, as indicated in the Figure legends. Reactions were started by the addition of 30 μM [α-32P]-dCTP (300 Ci/mmol), 100 μM unlabelled dATP, dGTP, dTTP, UTP, CTP and GTP, 2 mM ATP and 200 ng of a DNA plasmid containing the SV40 origin of replication (pSVori). After 60 min at 37 °C, reactions were stopped by the addition of 2% (w/v) SDS and 50 mM EDTA and DNA was subjected to phenol extraction. Labelled DNA was precipitated with ethanol, resuspended in 10 mM Tris/HCl (pH 8.0) and loaded on to a 1% agarose gel. Products were resolved by electrophoresis at 100 V for 1 h and visualized by a phosphoimager.
Inhibition assays
Reactions were performed under the conditions described for the polymerase and terminal transferase activity assay. The incorporation of radioactive dTTP into the appropriate DNA substrates at different concentrations of DNA or dNTP was monitored in the presence of increasing amounts of inhibitor as indicated in the Figure legends. Dose–response curves were generated by computer fitting of the data to the equation:
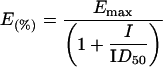 |
1 |
where E(%) is the fraction of enzyme activity measured in the presence of the inhibitor, Emax is the activity in the absence of the inhibitor, I is the inhibitor concentration and ID50 is the inhibitor concentration at which E(%)=0.5Emax.
Kinetic analysis
According to the ordered mechanism of the polymerization reaction, whereby the DNA primer (DNA) binds first, followed by the addition of dNTP, the enzyme can be present in three different catalytic forms: as a free enzyme, in a binary complex with the DNA or in a ternary complex with DNA and dNTP. The resulting rate equation for such a system is very complex and impractical to be used. For these reasons, the general steady-state kinetic analysis was simplified by varying one of the substrates (either DNA or dNTP) while the other was kept constant. When the DNA substrate was held constant at saturating concentrations (2–3-fold higher than its Michaelis constant Km) and the inhibition was analysed as a function of varying concentrations of dNTPs, at the steady state, all of the input enzyme was in the form of the enzyme–DNA binary complex, so that only the binary complex and the ternary complex with dNTP could react with the inhibitor. Similarly, when the dNTP concentration was kept constant at saturating levels (2–3-fold higher than its Km) and the inhibition was analysed as a function of varying DNA concentrations, the inhibitor could interact only with the free enzyme or with the ternary complex with DNA and dNTP.
The dependence of the initial velocities of the reaction was measured as a function of the DNA and dTTP substrate concentrations, by varying only one substrate and keeping the other saturating, as explained above, and analysed according to the Michaelis–Menten equation:
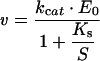 |
2 |
where kcat is the catalytic constant, E0 is the total input enzyme concentration and Vmax=kcatE0.
For the non-competitive inhibition, the variation of the apparent maximal velocity of the reaction [Vmax(app)] as a function of the inhibitor concentration was analysed according to the linear relationship:
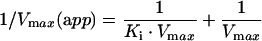 |
3 |
Similarly, for the competitive case, the variation of the apparent affinity of the enzyme for the competing substrate [Ks(app)] was analysed according to the linear relationship:
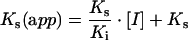 |
4 |
For the analysis of inhibitor-binding kinetics, the ratio between the enzyme activity at each pre-incubation time point (vt) and the enzyme activity without pre-incubation (v0), in the presence of different inhibitor concentrations (as indicated in the Figure legends), was plotted against the pre-incubation time.
Data were fitted to the simple exponential:
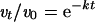 |
where t is time and k is the apparent exponential rate. Dose–response curves were generated at each pre-incubation time, and the corresponding apparent equilibrium dissociation constant [Ki(app)] values were calculated. The variation in Ki(app) values as a function of pre-incubation time was analysed according to the exponential equation:
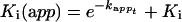 |
5 |
where t is time and kapp the apparent exponential rate. Since Ki(app) represents the equilibrium dissociation constant for the enzyme–inhibitor [E:I] complex at each pre-incubation time point, its rate of variation with time (kapp) is proportional to the apparent rate of [E:I] formation, and its value extrapolated at infinite time is equal to Ki. Thus the actual binding (kon) and dissociation (koff) rates at [I]=Ki can be calculated according to the equations:
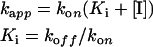 |
Each experiment was performed in triplicate and the mean values were used for interpolation. Curve fitting was performed with the computer program GraphPad Prism.
RESULTS
Effects of resveratrol derivatives on mammalian pols from the B and X families
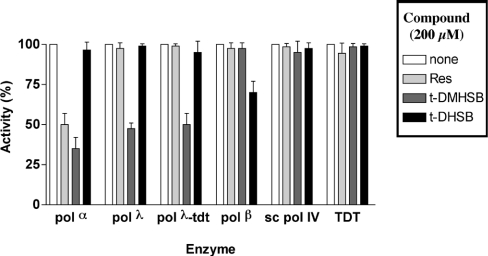
Resveratrol and its derivative t-DMHSB have been shown to inhibit mammalian replicative pols α and δ but not other noneukaryotic pols such as HSV-1 (herpes simplex virus) pol, HIV-1 reverse transcriptase or E. coli pol I [26]. We therefore compared the ability of resveratrol and its derivatives t-DMHSB, t-DHMSB and t-DHSB (Figure 1) to inhibit the mammalian family B pol α and the family X pol β, pol λ and TDT. As shown in Figure 2, whereas resveratrol itself inhibited only pol α but not the members of the family X pols, the t-DMHSB derivative inhibited both pol α and pol λ. Interestingly, t-DMHSB did inhibit both the template-dependent (i.e. pol) and the template-independent (i.e. terminal transferase, TDT) activities of pol λ. Despite their close similarity to pol λ, neither pol β nor TDT, two other family X pols, was inhibited by t-DMHSB. The t-DHSB derivative did not inhibit pol α, pol λ or TDT, and showed only weak inhibitory activity against pol β. The pol λ homologue from S. cerevisiae, pol IV, was not inhibited by any of the compounds tested, suggesting a specificity of the resveratrol derivatives towards mammalian X-family pols. The t-DHMSB analogue was inactive on all the enzymes tested (results not shown), confirming the absolute requirement of the 4′-hydroxy group for inhibition of pols.
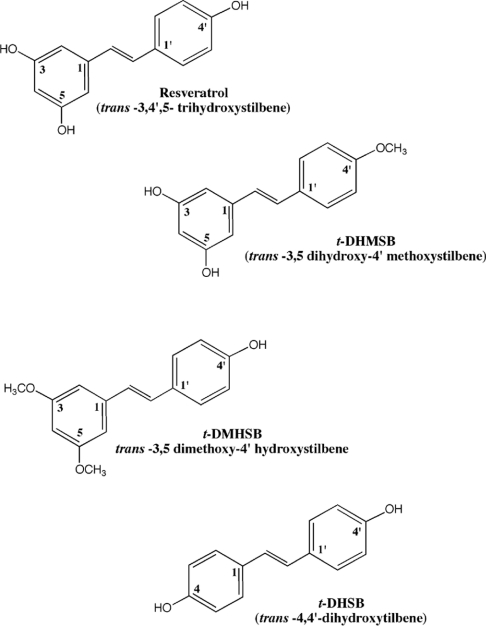
Figure 1. Resveratrol (trans-3,4′,5-trihydroxystilbene) and the three derivatives used in the present study.
Figure 2. Effects of resveratrol and its derivatives on the activity of human pols α, β, λ, TDT and S. cerevisiae pol IV.
Reactions were performed under the conditions described in the Materials and methods section either alone (white bars) or in the presence of 200 μM resveratrol (Res; light grey bars), 200 μM t-DMHSB (dark grey bars) or 200 μM t-DHSB (black bars) and in the presence of 0.1 unit of pol α, 0.1 unit of S. cerevisiae pol IV and 50 nM each of pol λ, pol β and TDT. Reaction mixtures were incubated for 15 min. Each experiment was performed in triplicate. Error bars represent ±S.D.
Resveratrol is a non-competitive, slow-binding, tight-binding inhibitor of pol α
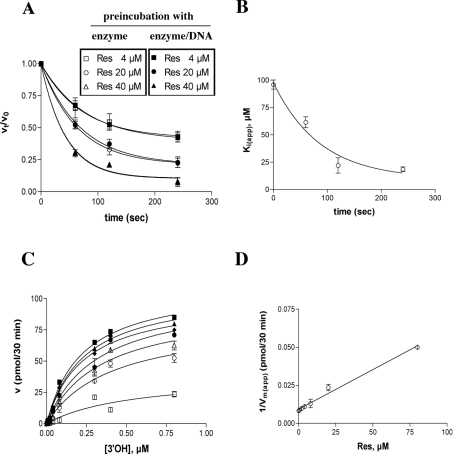
The effect of resveratrol on pol α activity was studied as a function of the pre-incubation time. As shown in Figure 3(A), inhibition by resveratrol increased as the time of pre-incubation was extended, regardless of whether the inhibitor was pre-incubated with the enzyme alone or in the presence of the DNA substrate. The variation in the apparent inhibition constant [Ki(app)] with time followed a simple exponential curve (Figure 3B), from which values of the equilibrium dissociation constant Ki=10 μM and the apparent rate kapp=0.008 s−1 were calculated for the enzyme–inhibitor complex formation. These values were then used to derive the true association (kon) and dissociation (koff) rates for resveratrol, which were 0.4×103 M−1·s−1 and 4×10−3 s−1 respectively. Thus resveratrol acted as a slow-binding, tight-binding inhibitor of pol α. Next, the dependence of the inhibition of pol α by resveratrol was studied as a function of the DNA and nucleotide substrate concentrations. As shown in Figure 3(C), resveratrol did not affect the affinity of the enzyme for the DNA substrate, but significantly decreased the apparent velocity of the reaction [Vmax(app)]. The reciprocal of the Vmax(app) values followed a linear relationship with the inhibitor concentrations (Figure 3D), allowing us to estimate the Ki value to be 9.5 μM. Similar results were obtained in the presence of different nucleotide substrate concentrations (results not shown), suggesting that resveratrol acts as a non-competitive inhibitor of pol α activity.
Figure 3. Resveratrol is a slow-binding, tight-binding non-competitive inhibitor of pol α.
Reactions were performed under the conditions described in the Materials and methods section. (A) Pol α (0.1 unit) was pre-incubated in the absence or presence of 4 μM (squares), 20 μM (circles) or 40 μM (triangles) resveratrol and in the absence (open symbols) or presence of 0.5 μM poly(dA)/oligo(dT) (closed symbols), for the indicated time periods. Reactions were started by adding the missing substrates and the mixtures were incubated for 15 min. The ratio between the enzyme activity at each pre-incubation time point (vt) and the activity without pre-incubation (v0), in the presence of each inhibitor concentration, was plotted against the pre-incubation time (see the Materials and methods section). Each experiment was performed in triplicate. (B) Dose–response curves were generated for the inhibition of pol α activity by resveratrol at different pre-incubation time points (see the Materials and methods section), and the Ki(app) values were plotted against the pre-incubation time. Each experiment was performed in triplicate. Error bars represent ±S.D. The calculated values were Ki=10±1 μM; kapp=0.008±0.001 s−1; kon=0.4×103 M−1·s−1 (±0.03); and koff=0.004 s−1 (±0.001). (C) The initial velocities of the reaction catalysed by 0.1 unit of pol α were measured as a function of increasing poly(dA)/oligo(dT) concentrations and in the absence (■) or in the presence of 0.5 μM (▲), 2 μM (◆), 4 μM (●), 8 μM (△), 20 μM (○) and 80 μM (□) resveratrol. The enzyme and inhibitor were pre-incubated for 5 min before starting the reactions. The DNA concentrations used (as 3′-OH ends) were 0.005, 0.01, 0.02, 0.04, 0.08, 0.3, 0.4 and 0.8 μM respectively. The TTP concentration was kept constant at 10 μM. Data were analysed as described in the Materials and methods section. Each experiment was performed in triplicate. (D) The variation in the apparent maximal velocity of the reaction [expressed as 1/Vmax(app)] was plotted as a function of the inhibitor concentration. The Ki value was calculated as described in the Materials and methods section and was 9.5 μM (±1). Error bars represent ±S.D.
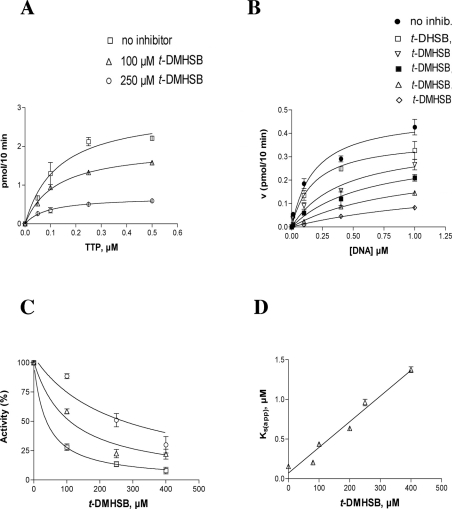
t-DMHSB is a non-competitive inhibitor of pol α
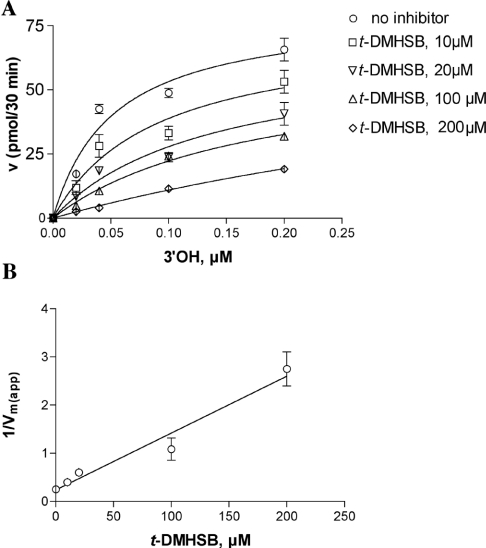
Similar experiments were performed to determine the mechanism of action of t-DMHSB towards pol α. Interestingly, pre-incubation experiments did not show any increase in the potency of inhibition, indicating a faster binding of t-DMHSB to pol α with respect to resveratrol (results not shown). As shown in Figure 4(A), addition of t-DMHSB to the reaction in the presence of increasing concentrations of the DNA substrate did not affect the apparent affinity (Ks) values, whereas the apparent velocity of the reaction Vmax(app) was decreased. The increase in the 1/Vmax(app) values followed a linear relationship with the inhibitor concentrations (Figure 4B), and a Ki value of 20 μM was calculated. Since no effect of t-DMHSB was found on the affinity of pol α for the nucleotide substrate (results not shown), it can be concluded that t-DMHSB inhibited pol α by a fully non-competitive mechanism.
Figure 4. The resveratrol derivative t-DMHSB is a non-competitive inhibitor of pol α.
Reactions were performed under the conditions described in the Materials and methods section. (A) The initial velocities of the reaction catalysed by 0.1 unit of pol α were measured as a function of increasing poly(dA)/oligo(dT) concentrations and in the absence (○) or in the presence of 10 μM (□), 20 μM (▽), 100 μM (△) and 200 μM (◇) t-DMHSB. The DNA concentrations used (as 3′-OH ends) were 0.02, 0.04, 0.1 and 0.2 μM respectively. The TTP concentration was kept constant at 10 μM. Data were analysed as described in the Materials and methods section. Each experiment was performed in triplicate. The Ks value in the absence of inhibitor was 0.05 μM and the corresponding values in the presence of different inhibitor concentrations were 0.05 (±0.01), 0.03 (±0.01), 0.07 (±0.01) and 0.05 (±0.01) μM respectively. The Vmax(app) value in the absence of inhibitor was 3.3 (±0.1) pmol/min and the corresponding values in the presence of different inhibitor concentrations were 2.2 (±0.1), 1.6 (±0.1), 1.1 (±0.1) and 0.4 (±0.07) pmol/min respectively. (B) The variation in the apparent maximal velocity of the reaction [expressed as 1/Vmax(app)] was plotted as a function of the inhibitor concentrations. The Ki value was calculated as described in the Materials and methods section and was 20 (±2) μM. Error bars represent ±S.D.
t-DMHSB is a non-competitive, slow-binding, tight-binding inhibitor of the pol but not of the terminal transferase activity of pol λ
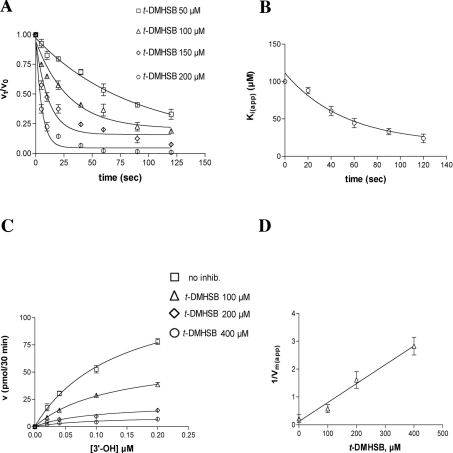
The inhibition of pol λ by t-DMHSB was also studied as a function of pre-incubation time. No differences in the potency of inhibition of the TDT activity of pol λ were observed on pre-incubation of t-DMHSB with the enzyme (results not shown). However, when similar experiments were performed to evaluate the inhibition of the pol activity of pol λ, a strong increase in the potency of t-DMHSB was observed as the pre-incubation time increased from 0 to 120 s (Figure 5A). This allowed us to estimate the decrease in Ki(app) as a function of the pre-incubation time. The variation of Ki(app) with time followed a simple exponential (Figure 5B), which allowed us to estimate Ki as 17.9 μM. The apparent rate at which the Ki varied (kapp) was used as an estimate of the apparent binding of the inhibitor to the enzyme. The calculated kapp value was then used to calculate the true association (kon) and dissociation (koff) rates, which were 0.4×103 M−1·s−1 and 9×10−3 s−1 respectively. Thus t-DMHSB showed slow-binding kinetics, but also a very low dissociation rate, acting as a slow-binding, tight-binding inhibitor, in a manner similar to resveratrol for pol α. Again, when the same experiments were repeated by pre-incubating the inhibitor in the presence of the enzyme and DNA substrate, no differences in the binding rate were detected, with respect to pre-incubation with the enzyme alone, indicating a slow inhibitor association also with the enzyme–DNA binary complex (results not shown). This is in agreement with a non-competitive mode of action (see below).
Figure 5. The resveratrol derivative t-DMHSB is a slow-binding, tight-binding non-competitive inhibitor of the template-dependent activity of pol λ.
Reactions were performed under the conditions described in the Materials and methods section. (A) Pol λ (50 nM) was pre-incubated in the absence or presence of 100 μM (△), 150 μM (◇) or 200 μM (○) t-DMHSB for the indicated time periods. Reactions were started by adding poly(dA)/oligo(dT) and labelled dTTP, and the mixtures were incubated for 15 min. The ratio between the enzyme activity at each pre-incubation time point (vt) and the activity without pre-incubation (v0), in the presence of each inhibitor concentration was plotted against the pre-incubation time (see also the Materials and methods section). Each experiment was performed in triplicate. (B) Dose–response curves were generated for the inhibition of pol λ activity by t-DMHSB at different pre-incubation timepoints (see the Materials and methods section), and the Ki(app) values were plotted against pre-incubation time. The binding (kon) and dissociation (koff) rates were calculated as described in the Mateirals and methods section. Each experiment was performed in triplicate. Error bars represent ±S.D. The calculated values were: Ki=17.9 μM (±2), kapp=0.019 s−1 (±0.005), kon=0.4×103 M−1·s−1 (±0.05) and koff=0.009 s−1 (±0.002). (C) The initial velocities of the reaction catalysed by 50 nM pol λ were measured as a function of increasing poly(dA)/oligo(dT) concentrations and in the absence (□) or in the presence of 100 μM (△), 200 μM (◇) and 400 μM (○) t-DMHSB. The enzyme and inhibitor were pre-incubated for 5 min before starting the reactions. DNA concentrations used (as 3′-OH ends) were 0.02, 0.04, 0.1 and 0.2 μM respectively. The TTP concentration was kept constant at 10 μM. Data were analysed as described in the Materials and methods section. Each experiment was performed in triplicate. Error bars represent ±S.D. The Ks value in the absence of inhibitor was 0.15 (±0.03) μM and the corresponding values in the presence of different inhibitor concentrations were: 0.13 (±0.03), 0.12 (±0.01) and 0.12 (±0.01) μM respectively. The Vmax(app) value in the absence of inhibitor was 4.5 (±0.5) pmol/min and the corresponding values in the presence of different inhibitor concentrations were: 2.06 (±0.5), 0.7 (±0.1) and 0.35 (±0.1) pmol/min respectively. (D) The variation in the apparent maximal velocity of the reaction [expressed as 1/Vmax(app)] is plotted as a function of the inhibitor concentration. The Ki value was calculated as described in the Materials and methods section and was 17.5 (±1) μM. Error bars represent ±S.D.
Next, the inhibition of pol λ DNA polymerase activity by t-DMHSB was studied as a function of the DNA substrate concentration. As shown in Figure 5(C), t-DMHSB did not affect the apparent affinity of pol λ for the DNA substrate (expressed as available concentration of 3′-OH ends), decreasing only the Vmax of the reaction. Similar results were observed in the presence of increasing concentrations of the nucleotide substrate (results not shown). The variation of the apparent velocity of the reaction [Vmax(app)] as a function of the t-DMHSB concentration was analysed by plotting the reciprocal of Vmax(app) values as a function of inhibitor concentrations. As shown in Figure 5(C), the 1/Vmax(app) values followed a linear relationship with respect to the inhibitor, as expected for a purely non-competitive inhibitor. From the curve, a Ki value of 17.5 μM was calculated for t-DMHSB inhibition.
t-DMHSB is a competitive inhibitor of the terminal transferase activity of pol λ with respect to the DNA substrate
To determine the mechanism of inhibition of pol λ–TDT by t-DMHSB, increasing concentrations of either the nucleotide or the DNA substrates were tested in the absence or presence of different amounts of the inhibitor. As shown in Figure 6(A), addition of t-DMHSB did not influence the apparent affinity (Ks) of the enzyme for the nucleotide substrate, affecting only the velocity of the reaction (Vmax). On the other hand, when the effects of t-DMHSB were studied as a function of the DNA substrate concentrations, the inhibitor caused an increase in the Ks values, whereas the Vmax of the reaction was unaffected (Figure 6B). When the inhibition of pol λ–TDT by t-DMHSB was measured at different DNA substrate concentrations, the potency of inhibition of t-DMHSB decreased as the DNA concentration increased, as shown in Figure 6(C). Taken together, these results indicated that t-DMHSB was a competitive inhibitor of the pol λ–TDT reaction with respect to the DNA substrate. Accordingly, the decrease in the apparent affinity for the DNA substrate [as indicated by an increase in the Ks(app) values] followed a linear relationship with the inhibitor concentrations, as shown in Figure 6(D). From these data, the inhibition constant Ki=21 μM was calculated.
Figure 6. The resveratrol derivative t-DMHSB is a competitive inhibitor of the terminal transferase activity of pol λ with respect to the DNA substrate.
Reactions were performed under the conditions described in the Materials and methods section. (A) The initial velocities of the reaction catalysed by 50 nM pol λ were measured as a function of increasing dTTP concentrations and in the absence (□) or in the presence of 100 μM (△) or 250 μM (○) t-DMHSB. The dTTP concentrations used were 0.05, 0.1, 0.25 and 0.5 μM respectively. The polyd(A)/oligo(dT) concentration was kept constant at 1 μM. Data were analysed as described in the Materials and methods section. The Ks value in the absence of inhibitor was 0.13 (±0.02) μM and the corresponding values in the presence of different inhibitor concentrations were: 0.11 (±0.01) and 0.1 (±0.01) μM respectively. The Vmax(app) value in the absence of inhibitor was 0.29 (±0.1) pmol/min and the corresponding values in the presence of different inhibitor concentrations were: 0.19 (±0.1) and 0.06 (±0.01) pmol/min respectively. (B) The initial velocities of the reaction catalysed by 50 nM pol λ were measured as a function of increasing concentrations of ss 27-mer oligodeoxynucleotide substrate (see the Materials and methods section) and in the absence (●) or in the presence of 80 μM (□), 100 μM (▽), 200 μM (■), 250 μM (△) and 400 μM (◇) t-DMHSB. The DNA concentrations used (as 3′-OH ends) were 0.025, 0.1, 0.4 and 1 μM respectively. The TTP concentration was kept constant at 5 μM. Data were analysed as described in the Materials and methods section. The Ks value in the absence of inhibitor was 0.4 (±0.02) μM and the corresponding values in the presence of different inhibitor concentrations were 0.6 (±0.02), 0.9 (±0.05) and 1.5 (±0.09) μM respectively. The Vmax(app) value in the absence of inhibitor was 0.38 (±0.08) pmol/min and the corresponding values in the presence of different inhibitor concentrations were 0.37 (±0.09), 0.29 (±0.08) and 0.28 (±0.07) pmol/min respectively. (C) Dose–response curves were generated for the inhibition of pol λ activity by t-DMHSB at DNA substrate concentrations (see the Materials and methods section) of 0.01 μM (□), 0.4 μM (△) and 1 μM (○) and the corresponding 50% inhibitory constant (ID50) values were derived. ID50 values at the different DNA concentrations tested were: 33 (±2), 141 (±10) and 248 (±20) μM. (D) The variation in the affinity of pol λ for the DNA substrate [Ks(app)] was plotted as a function of the inhibitor concentrations. The Ki value was calculated as described in the Materials and methods section and was 21 (±2) μM. (A–D) Each experiment was performed in triplicate. Error bars represent ±S.D.
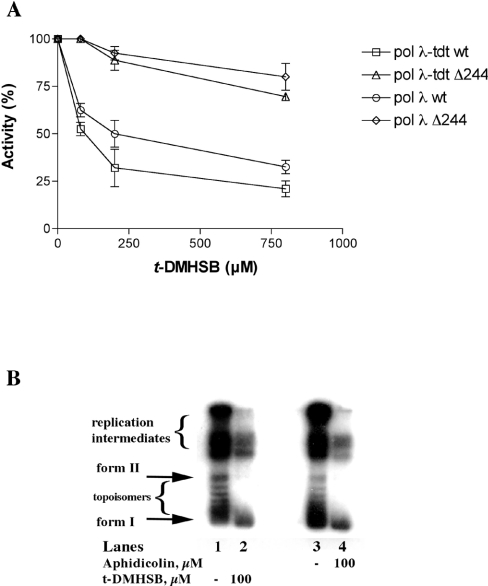
t-DMHSB is sensitive to the absence of the N-terminal BRCT (BRCA1 C-terminal domain) and proline-rich domains of pol λ
Pol λ has a BRCT domain (amino acids 36–132) and a proline-rich domain (amino acids 133–244) at its N-terminus [28]. These domains have been shown to be entirely dispensable for all the catalytic activities of pol λ (pol, lyase and TDT), which reside in its C-terminal part (amino acids 244–575) [29,30]. However, the phenolic compound petasiphenol, an inhibitor of pol λ DNA polymerase activity, was shown to bind to a pocket in the BRCT domain (amino acids 76–104). Accordingly, petasiphenol did not inhibit a truncated form of pol λ, lacking the first 132 amino acids [33]. To test whether the t-DMHSB derivative also shared the same binding site, its inhibition potency was determined against the truncated form of pol λ, the Δ244λ mutant, which lacked the first 244 amino acids, comprising both the BRCT and the prolinerich domains. As shown in Figure 7(A), the t-DMHSB derivative inhibited neither the template-dependent nor the TDT incorporation activities of the truncated enzyme. This result suggested that t-DMHSB might bind to the N-terminal part of pol λ, similar to petasiphenol.
Figure 7. The resveratrol derivative t-DMHSB is sensitive to absence of the N-terminal BRCT and proline-rich domains of pol λ and inhibits the in vitro SV40 DNA replication reaction.
Reactions were performed under the conditions described in the Materials and methods section. (A) Increasing concentrations of t-DMHSB were titrated in the presence of 50 nM pol λ wild-type or the truncated form pol λΔ244, under both template-dependent and template-independent (TDT) conditions. □, TDT activity of pol λ wild-type; ○, template-dependent activity of pol λ wild-type; △, TDT activity of pol λΔ244; ◇, template-dependent activity of pol λΔ244. Each experiment was performed in triplicate. Error bars represent ±S.D. (B) In vitro SV40 DNA replication reactions were performed as described in the Materials and methods section. Reaction products were analysed on a 1% agarose gel. Lanes 1 and 3, control reaction without inhibitors; lane 2, reaction performed in the presence of 100 μM t-DMHSB; lane 4, reaction performed in the presence of 100 μM t-DMHSB. Form I, fully replicated circular supercoiled plasmid DNA; form II, fully replicated circular relaxed plasmid DNA.
t-DMHSB can inhibit in vitro SV40 DNA replication
Next, we examined whether t-DMHSB is capable of suppressing replicative pols activity in a more physiological context. For this purpose, we exploited the in vitro SV40 DNA replication system. S-100 extracts from COS-7 cells, constitutively expressing the SV40 tag (see the Materials and methods section), were pre-incubated for 5 min in the absence or in the presence of either t-DMHSB or aphidicolin, a well-known inhibitor of mammalian family B pols α, δ and ε. Reactions were started with the addition of labelled dNTPs and a plasmid containing the SV40 origin of replication. The products of the reaction were resolved on a 1% agarose gel and revealed by densitometric scanning with a phosphoimager. As shown in Figure 7(B), in the control reactions (lanes 1 and 3), high-molecular-mass reaction intermediates as well as form II (relaxed) and form I (supercoiled) DNA products were detected. Addition of t-DMHSB greatly decreased all the reaction products in a manner similar to aphidicolin (compare lane 2 with lane 4). Thus, t-DMHSB could also inhibit the in vitro SV40 DNA replication, further supporting its observed effect on DNA replication in vivo [26].
DISCUSSION
In agreement with other studies [21–25], we have described that trans-resveratrol inhibited cell growth, in a dose- and time-dependent manner, both in normal fibroblasts as well as in fibrosarcoma cells [26]. Among different resveratrol analogues, an antiproliferative activity comparable with that of trans-resveratrol has been observed only for the t-DMHSB derivative [26]. Our previous results demonstrated that both trans-resveratrol and the t-DMHSB derivative significantly inhibited pols α and δ. Interestingly, there was an increase in specificity for the inhibition of pol α with respect to pol δ, from trans-resveratrol to t-DMHSB, suggesting a possible role of the substituent groups at positions 3 and 5 in the binding to different pols [26].
In the present study, we expanded our area of research by (i) characterizing the mechanism of action of resveratrol and t-DMHSB against the family A pol α and (ii) evaluating the ability of selected resveratrol derivatives to inhibit other mammalian pols from the X family, including pol λ, pol β and TDT. To this aim, we compared trans-resveratrol with three derivatives (Figure 1): first, t-DMHSB, carrying methoxy groups at positions 3 and 5; secondly, t-DHMSB, carrying a methoxy group at position 4′; and thirdly, t-DHSB, lacking the substituents at the 3,5 positions, but having a hydroxy group at position 4 and 4′. The results of the in vitro assays confirmed that the 4′-hydroxy group was essential for pol inhibition, since the compound t-DHMSB, having a methoxy group at 4′, lacked any activity. In addition, positions 3, 4 and 5 were found to be important for the selectivity of pol inhibition. In fact, replacing the 3,5-hydroxy groups of resveratrol with methoxy substituents, as in the case of t-DMHSB, maintained the activity towards pol α, but also conferred the ability to inhibit pol λ. This effect was, however, very specific, since the closely related pol β and TDT (from the same family) were not affected. On the other hand, eliminating the substituents at positions 3 and 5, but introducing a hydroxy group at position 4, completely changed the selectivity of inhibition, since the resulting t-DHSB derivative lost all its activity towards pol α and pol λ and showed only a very weak activity against pol β. Both t-DMHSB and t-DHSB were inactive against the only known family X homologue of budding yeast, the S. cerevisiae pol IV, indicating a strict preference for mammalian enzymes.
Resveratrol and its active derivative t-DMHSB were non-competitive inhibitors with respect to the substrates of the pol reaction, i.e. DNA and nucleotides. However, interesting differences in their binding mode were found. Resveratrol acted as a slow-binding, tight-binding inhibitor of pol α, displaying not only a low association rate but also a slow dissociation from the enzyme. As a consequence, its potency of inhibition was found to be strongly enhanced by pre-incubation with the enzyme. The t-DMHSB derivative, on the other hand, showed no increase in its potency of inhibition on pre-incubation with pol α, but retained a slow-binding, tight-binding mechanism towards pol λ. This might indicate that the 3,5-methoxy groups of t-DMHSB (i) increased binding to pol α (thus overcoming the requirement for a pre-incubation step) and (ii) made the drug able to bind to pol λ, even though with slow kinetics. It should be pointed out that the same effect could be observed in the case of fast initial binding of the inhibitor, after a slow (i.e. rate-limiting) conformational change of the enzyme–substrate complex into a stable equilibrium state. Under the steady-state conditions used, we were not able to discriminate between these two possibilities. However, regardless of the precise identity of the rate-limiting step, it is clear that formation of a stable enzyme–inhibitor complex between resveratrol or its analogue t-DMHSB and their target enzymes proceeds with slow kinetics. Exploiting additional substitutions at the 3, 4 and 5 positions of the resveratrol pharmacophore is warranted to improve its potency and selectivity.
Besides being a DNA polymerase, thus synthesizing DNA in a strictly template-dependent manner, pol λ can also act as a TDT, adding nucleotides to the 3′-OH end of an ss DNA substrate in a template-independent manner [29,30]. The t-DMHSB derivative was capable of inhibiting this activity also, but with a competitive mechanism towards the DNA substrate. The difference in the mechanism of inhibition of t-DMHSB against the templatedependent (non-competitive) and template-independent (competitive) activities of pol λ suggested that its binding induced a conformational change in the enzyme, which did not affect the binding of partially double-stranded template-primer DNA (the substrate for the pol activity) but prevented the interaction with an ss DNA (the substrate for the TDT activity). The idea that t-DMHSB bound to the DNA-binding site of pol λ could be ruled out based on the following considerations: (i) the DNA polymerase and TDT activities of pol λ have been shown to share the same active site, which is located in the C-terminal part of the enzyme [34]. Thus, if t-DMHSB bound to the DNA-binding site, it should display the same competitive mechanism of inhibition towards both activities. (ii) The N-terminal domain of pol λ (amino acids 1–244) has been shown to be dispensable for both the polymerase and TDT activities of pol λ, as well as for the interaction with the DNA and nucleotide substrates [35]. However, t-DMHSB was unable to inhibit a pol λ mutant lacking the first 244 amino acids (pol λΔ244), clearly suggesting that its binding site was located in the N-terminal region of the enzyme.
The inhibition of pol λ-TDT activity by t-DMHSB was no longer dependent on its pre-incubation with the target enzyme, in contrast with what was observed in the case of the template-dependent activity of pol λ. Due to the ordered mechanism of the reaction catalysed by pols, whereby DNA binds first, followed by the nucleotide, binding of t-DMHSB in the case of the TDT reaction is restricted to the free enzyme since it is a competitor of the DNA substrate. On the other hand, acting as a non-competitive inhibitor of the template-dependent reaction, t-DMHSB could, in principle, target either the binary enzyme–DNA or the ternary enzyme–DNA–nucleotide complexes. Thus the binding kinetics observed for the two reactions probably reflect these differences.
In the present study, pol λ activity was measured in the presence of Mn2+, because, as we have already shown, Mn2+ is the metal ion preferred by pol λ; Mg2+ can substitute for Mn2+, but with lower efficiency, even resulting in strong inhibition of the enzyme at concentrations above 4–5 mM [36]. Thus, we cannot rule out that the differences observed between pol α and pol λ for t-DMHSB inhibition might be, at least partially, due to the different metal ion cofactor used in the assays. Although the Mn2+ concentrations used in the present study (0.5–1 mM) cannot be considered physiological in absolute terms (the physiological range of Mn2+ is estimated to be <0.1 mM), it should be noted that free metal ion concentrations are tightly regulated in vivo by special metal ion-binding proteins and their concentrations may vary considerably, even within the nucleus of a cell. Thus it is possible that Mn2+ is the physiological cofactor for pol λ.
The fact that t-DMHSB was able to inhibit both pol λ and pol α suggests the existence of a similar binding pocket in these two enzymes. Indeed, the existence of similar binding sites for the interaction of natural compounds with the pols from families B and X has already been suggested by the observation that some terpeno-benzoic acids and triterpenoids can inhibit pol α as well as pol β and pol λ [37]. Unfortunately, no detailed information about their binding site(s) are available so far.
The experiments with the pol λΔ244 mutant seemed to indicate that t-DMHSB binds to the N-terminal region of pol λ. However, multiple alignment analysis of the entire sequence of the human pol α catalytic subunit, as well as of the two primase subunits, with the first 244 amino acids of pol λ failed to reveal significant similarity, at least at the primary amino acid-sequence level. However, the best characterized natural pol λ inhibitor so far, petasiphenol, has been shown to bind to a pocket located between amino acids 74 and 107 of pol λ [33]. When the alignment was repeated using the sequence of the petasiphenol-binding site only, significant identity was found with a region at the very C-terminus (amino acids 1285–1327) of the human pol α catalytic subunit (Figure 8). Most of the amino acid residues that were predicted to make essential contacts with petasiphenol in the pol λ sequence are also conserved in human pol α (Figure 8, shown shaded). The main difference was the fact that there were short amino acid insertions (three, one, one and ten amino acids respectively from the N- to C-terminus) in the pol α sequence with respect to pol λ. Interestingly, these insertions were located corresponding to a putative loop predicted by homology modelling to occur in the pol λ structure (amino acids 84–107), suggesting that this loop might be larger in pol α. It is then tempting to speculate that t-DMHSB might bind to both pol λ and pol α pockets, whereas petasiphenol could only fit to the binding site in pol λ, but failed to bind to the slightly larger loop of pol α. Site-directed mutagenesis studies will be required to test directly this intriguing but highly speculative hypothesis.
Figure 8. A putative common resveratrol-binding site of human pols α and λ.
Sequence alignment of the C-terminal sequence (amino acids 1286–1327) of the catalytic subunit of human pol α (hupolalpha, SwissProt accession no. P09884) and of the predicted inhibitor-binding site [33] in the N-terminal domain (amino acids 77–103) of human pol λ (hulam, SwissProt accession no. Q9UGP5). Amino acids in the pol λ sequence predicted to make direct contacts with the phenolic compound petasiphenol [33] are indicated by asterisks. Amino acids that are conserved in the pol α sequence are shaded. Multiple alignment has been performed with the CLUSTAL W program (http://www.ebi.ac.uk/clustalw/).
In summary, we presented the first biochemical characterization of the mechanism of action of trans-resveratrol against mammalian pols. Our results indicated that specific structural determinants of resveratrol might confer selectivity towards different families of mammalian pols. Moreover, in the present study, we have further substantiated the hypothesis that the antiproliferative activity of resveratrol and its derivatives on cultured cells was due to inhibition of replicative pols [26], by showing that t-DMHSB was capable of suppressing the in vitro SV40 DNA replication with a potency similar to aphidicolin. Our results lay the background for the rational design of resveratrol analogues with more potent and very specific antiproliferative and antitumoral properties.
Acknowledgments
This work was supported by the Swiss National Science Foundation (grant 3100.061361.00) to K.R.; the EU (project no. QLK3-CT-2002-02071 REBIOTECH) to U.H., K.R., I.S. and G.M.; the BBW 02.2286 to U.H., K.R. and I.S.; the Kanton of Zürich to G.M., I.S. and U.H.; and the CARIPLO Foundation Project ‘Oncogenetica e Proteomica della Replicazione’ (2003.1663/10.8441) to G.M.
References
- 1.Langcake P., Pryce R. J. A new class of phytoalexins from grapevines. Experientia. 1977;33:151–152. doi: 10.1007/BF02124034. [DOI] [PubMed] [Google Scholar]
- 2.Hain R., Bieseler B., Kindl H., Schroder G., Stocker R. Expression of a stilbene synthase gene in Nicotiana tabacum results in synthesis of the phytoalexin resveratrol. Plant Mol. Biol. 1990;15:325–335. doi: 10.1007/BF00036918. [DOI] [PubMed] [Google Scholar]
- 3.Soleas G. J., Diamandis E. P., Goldberg D. M. Resveratrol: a molecule whose time has come? And gone? Clin. Biochem. 1997;30:91–113. doi: 10.1016/s0009-9120(96)00155-5. [DOI] [PubMed] [Google Scholar]
- 4.Jang M., Cai L., Udeani G. O., Slowing K. V., Thomas C. F., Beecher C. W. W., Fong H. H. S., Farnsworth N. R., Kinghorn A. D., Menta R. G., et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 5.Adrian M., Jeandet P., Douillet-Breuil A. C., Tesson L., Bessis R. Stilbene content of mature Vitis vinifera berries in response to UV-C elicitation. J. Agric. Food Chem. 2000;48:6103–6105. doi: 10.1021/jf0009910. [DOI] [PubMed] [Google Scholar]
- 6.Goldberg D., Tsang E., Karumanchiri A., Diamandis E. P., Soleas G., Ng E. Method to assay the concentrations of phenolic constituents of biological interest in wines. Anal. Chem. 1996;68:1688–1694. doi: 10.1021/ac951083i. [DOI] [PubMed] [Google Scholar]
- 7.Soleas G. J., Diamandis E. P., Goldberg D. M. Wine as a biological fluid: history, production, and role in disease prevention. J. Clin. Lab. Anal. 1997;11:287–313. doi: 10.1002/(SICI)1098-2825(1997)11:5<287::AID-JCLA6>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura Y., Okuda H., Arichi S. Effects of stilbenes on arachidonate metabolism in leukocytes. Biochim. Biophys. Acta. 1985;834:275–278. [PubMed] [Google Scholar]
- 9.Rotondo S., Rajtar G., Manarini S., Celardo A., Rotilio D., de Gaetano G., Evangelista V., Cerletti C. Effect of trans-resveratrol, a natural polyphenolic compound, on human polymorphonuclear leukocyte function. Br. J. Pharmacol. 1998;123:1691–1699. doi: 10.1038/sj.bjp.0701784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang D. S., Kang B. S., Ryu S. Y., Chang I. M., Min K. R., Kim Y. Inhibitory effects of resveratrol analogs on unopsonized zymosan-induced oxygen radical production. Biochem. Pharmacol. 1999;57:705–712. doi: 10.1016/s0006-2952(98)00350-5. [DOI] [PubMed] [Google Scholar]
- 11.Pace-Asciak C. R., Hahn S., Diamandis E. P., Soleas G., Goldberg D. M. The red wine phenolics trans-resveratrol and quercetin block human platelet aggregation and eicosanoid synthesis: implications for protection against coronary heart disease. Clin. Chim. Acta. 1995;235:207–219. doi: 10.1016/0009-8981(95)06045-1. [DOI] [PubMed] [Google Scholar]
- 12.Pace-Asciak C. R., Rounova O., Hahn S., Diamandis E. P., Goldberg D. M. Wines and grape juices as modulators of platelet aggregation in healthy human subjects. Clin. Chim. Acta. 1996;246:163–182. doi: 10.1016/0009-8981(96)06236-5. [DOI] [PubMed] [Google Scholar]
- 13.Fauconneau B., Waffo-Teguo P., Huguet F., Barrier L., Decendit A., Merillon J. M. Comparative study of radical scavenger and antioxidant properties of phenolic compounds from Vitis vinifera cell cultures using in vitro tests. Life Sci. 1997;61:2103–2110. doi: 10.1016/s0024-3205(97)00883-7. [DOI] [PubMed] [Google Scholar]
- 14.Mgbonyebi O. P., Russo J., Russo I. H. Antiproliferative effect of synthetic resveratrol on human breast epithelial cells. Int. J. Oncol. 1998;12:865–869. [PubMed] [Google Scholar]
- 15.Hsieh T. C., Burfeind P., Laud K., Bacher J. M., Traganos F., Darzynkiewicz Z., Wu J. M. Cell cycle effects and control of gene expression by resveratrol in human breast carcinoma cell lines with different metastatic potentials. Int. J. Oncol. 1999;15:245–252. doi: 10.3892/ijo.15.2.245. [DOI] [PubMed] [Google Scholar]
- 16.Frankel E. N., Waterhouse A. L., Kinsella J. E. Inhibition of human LDL oxidation by resveratrol. Lancet. 1993;341:1103–1104. doi: 10.1016/0140-6736(93)92472-6. [DOI] [PubMed] [Google Scholar]
- 17.Kerry N. L., Abbey M. Red wine and fractionated phenolic compounds prepared from red wine inhibit low density lipoprotein oxidation in vitro. Atherosclerosis. 1997;135:93–102. doi: 10.1016/s0021-9150(97)00156-1. [DOI] [PubMed] [Google Scholar]
- 18.Maccarrone M., Lorenzon T., Guerrieri P., Finazzi Agrò A. Resveratrol prevents apoptosis in K562 cells by inhibiting lipoxygenase and cyclooxygenase activity. Eur. J. Biochem. 1999;265:27–34. doi: 10.1046/j.1432-1327.1999.00630.x. [DOI] [PubMed] [Google Scholar]
- 19.Arichi H., Kimura Y., Okuda H., Baba K., Kozawa M., Arichi S. Effects of stilbene components of the roots of Polygonum cuspidatum Sieb. et Zucc. on lipid metabolism. Chem. Pharm. Bull. 1982;30:1766–1770. doi: 10.1248/cpb.30.1766. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg D. M., Hahn S. E., Parkes J. G. Beyond alcohol: beverage consumption and cardiovascular mortality. Clin. Chem. Acta. 1995;237:155–187. doi: 10.1016/0009-8981(95)06069-p. [DOI] [PubMed] [Google Scholar]
- 21.Della Ragione F., Cucciolla V., Borriello A., Della Pietra V., Racioppi L., Soldati G., Manna C., Galletti P., Zappia V. Resveratrol arrests the cell division cycle at S/G2 phase transition. Biochem. Biophys. Res. Commun. 1998;250:53–58. doi: 10.1006/bbrc.1998.9263. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh T., Juan G., Darzynkiewicz Z., Wu J. M. Resveratrol increases nitric oxide synthase, induces accumulation of p53 and p21(WAF1/CIP1), and suppresses cultured bovine pulmonary artery endothelial cell proliferation by perturbing progression through S and G2. Cancer Res. 1999;59:2596–2601. [PubMed] [Google Scholar]
- 23.Zou J., Huang Y., Chen Q., Wang N., Cao K., Hsieh T. C., Wu J. M. Suppression of mitogenesis and regulation of cell cycle traverse by resveratrol in cultured smooth muscle cells. Int. J. Oncol. 1999;15:647–651. doi: 10.3892/ijo.15.4.647. [DOI] [PubMed] [Google Scholar]
- 24.Babich H., Reisbaum A. G., Zuckerbraun H. L. In vitro response of human gingival epithelial S-G cells to resveratrol. Toxicol. Lett. 2000;114:143–153. doi: 10.1016/s0378-4274(99)00288-x. [DOI] [PubMed] [Google Scholar]
- 25.Hsieh T., Wu J. M. Differential effects on growth, cell cycle arrest, and induction of apoptosis by resveratrol in human prostate cancer cell lines. Exp. Cell Res. 1999;249:109–115. doi: 10.1006/excr.1999.4471. [DOI] [PubMed] [Google Scholar]
- 26.Stivala L. A., Savio M., Carafoli F., Perucca P., Bianchi L., Maga G., Forti L., Pagnoni U. M., Albini A., Prosperi E., Vannini V. Specific structural determinants are responsible for the antioxidant activity and the cell cycle effects of resveratrol. J. Biol. Chem. 2001;276:22586–22594. doi: 10.1074/jbc.M101846200. [DOI] [PubMed] [Google Scholar]
- 27.Hübscher U., Maga G., Spadari S. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 28.Ramadan K., Shevelev I., Hübscher U. The DNA-polymerase-X family: controllers of DNA quality? Nat. Rev. Mol. Cell Biol. 2004;5:1038–1043. doi: 10.1038/nrm1530. [DOI] [PubMed] [Google Scholar]
- 29.Ramadan K., Maga G., Shevelev I. V., Villani G., Blanco L., Hübscher U. Human DNA polymerase lambda possesses terminal deoxyribonucleotidyl transferase activity and can elongate RNA primers: implications for novel functions. J. Mol. Biol. 2003;328:63–72. doi: 10.1016/s0022-2836(03)00265-1. [DOI] [PubMed] [Google Scholar]
- 30.Maga G., Ramadan K., Locatelli G. A., Shevelev I., Spadari S., Hübscher U. DNA elongation by human DNA polymerase lambda polymerase and terminal transferase activities are differentially coordinated by proliferating cell nuclear antigen and replication protein A. J. Biol. Chem. 2005;280:1971–1981. doi: 10.1074/jbc.M411650200. [DOI] [PubMed] [Google Scholar]
- 31.Prasad R., Widen S. G., Singhal R. K., Watkins J., Prakash L., Wilson S. H. Yeast open reading frame YCR14C encodes a DNA beta-polymerase-like enzyme. Nucleic Acids Res. 1993;21:5301–5307. doi: 10.1093/nar/21.23.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiser T., Gassmann M., Thommes P., Ferrari E., Hafkemeyer P., Hübscher U. Biochemical and functional comparison of DNA polymerases alpha, delta, and epsilon from calf thymus. J. Biol. Chem. 1991;266:10420–10428. [PubMed] [Google Scholar]
- 33.Mizushina Y., Kamisuki S., Kasai N., Ishidoh T., Shimazaki N., Takemura M., Asahara H., Linn S., Yoshida S., Koiwai O., et al. Petasiphenol: a DNA polymerase lambda inhibitor. Biochemistry. 2002;41:14463–14471. doi: 10.1021/bi020476q. [DOI] [PubMed] [Google Scholar]
- 34.Shevelev I., Blanca G., Villani G., Ramadan K., Spadari S., Hubscher U., Maga G. Mutagenesis of human DNA polymerase lambda: essential roles of Tyr505 and Phe506 for both DNA polymerase and terminal transferase activities. Nucleic Acids Res. 2003;31:6916–6925. doi: 10.1093/nar/gkg896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fiala K. A., Abdel-Gawad W., Suo Z. Pre-steady-state kinetic studies of the fidelity and mechanism of polymerization catalyzed by truncated human DNA polymerase lambda. Biochemistry. 2004;43:6751–6762. doi: 10.1021/bi049975c. [DOI] [PubMed] [Google Scholar]
- 36.Blanca G., Shevelev I., Ramadan K., Villani G., Spadari S., Hubscher U., Maga G. Human DNA polymerase lambda diverged in evolution from DNA polymerase beta toward specific Mn(++) dependence: a kinetic and thermodynamic study. Biochemistry. 2003;42:7467–7476. doi: 10.1021/bi034198m. [DOI] [PubMed] [Google Scholar]
- 37.Mizushina Y., Hirota M., Murakami C., Ishidoh T., Kamisuki S., Shimazaki N., Takemura M., Perpelescu M., Suzuki M., Yoshida H., et al. Some anti-chronic inflammatory compounds are DNA polymerase lambda-specific inhibitors. Biochem. Pharmacol. 2003;66:1935–1944. doi: 10.1016/s0006-2952(03)00551-3. [DOI] [PubMed] [Google Scholar]