Abstract
We have identified and characterized a novel NADP+-dependent D-arabitol dehydrogenase and the corresponding gene from the rust fungus Uromyces fabae, a biotrophic plant pathogen on broad bean (Vicia faba). The new enzyme was termed ARD1p (D-arabitol dehydrogenase 1). It recognizes D-arabitol and mannitol as substrates in the forward reaction, and D-xylulose, D-ribulose and D-fructose as substrates in the reverse reaction. Co-factor specificity was restricted to NADP(H). Kinetic data for the major substrates and co-factors are presented. A detailed analysis of the organization and expression pattern of the ARD1 gene are also given. Immunocytological data indicate a localization of the gene product predominantly in haustoria, the feeding structures of these fungi. Analyses of metabolite levels during pathogenesis indicate that the D-arabitol concentration rises dramatically as infection progresses, and D-arabitol was shown in an in vitro system to be capable of quenching reactive oxygen species involved in host plant defence reactions. ARD1p may therefore play an important role in carbohydrate metabolism and in establishing and/or maintaining the biotrophic interaction in U. fabae.
Keywords: D-arabitol dehydrogenase, biotrophy, carbohydrate storage, polyol, reactive oxygen species, Uromyces fabae
Abbreviations: ARD1, NADP-dependent D-arabitol dehydrogenase 1 from U. fabae; ARD1fp, ARD1 fusion protein; ARD1p, ARD1 protein; EST, expressed sequence tag; KMB, α-oxo-γ-methiol-butyric acid; MAD1fp, NADP-dependent mannitol dehydrogenase 1 from U. fabae His-tagged fusion protein; ROS, reactive oxygen species; TBS, Tris-buffered saline
INTRODUCTION
Polyols are secondary metabolites mainly associated with the fungal kingdom [1]. They are frequently found in spores, which was taken as evidence for having a role as storage compounds [2,3]. Other evidence points at a role of polyols in fungal osmoprotection [4,5]. Koide et al. [6] also suggested that mannitol, at least, functions in the translocation of carbohydrates from plant roots to their fungal symbionts in mycorrhizal interactions. Furthermore, it has been suggested that some fungi are able to switch between anabolic and catabolic pathways and storage of reducing power by interconversion of NADH and NADPH in the so-called mannitol cycle [7]. It was also shown that some of these polyols are powerful radical scavengers in vitro [8] and in vivo [9,10], a property which makes them potential protectants against ROS (reactive oxygen species) released by the host organism as part of their defence response.
Besides the hexitols mannitol and D-sorbitol, the pentitol D-arabitol is one of the polyols found most frequently in fungi. Serum D-arabitol levels, for example, are used as a diagnostic marker for invasive candidiasis [11,12]. Although directly linked to Candida infection, it is not known what role D-arabitol plays in the infection process of Candida spp. Basidiospores of Schizophyllum commune were reported to contain significant amounts of both mannitol and D-arabitol [13]. The latter disappeared rapidly during germination, which can be taken as evidence for a role of D-arabitol in carbohydrate storage. Maclean and Scott [2] reported the presence of mannitol and D-arabitol in both ungerminated and germinated uredospores of Puccinia graminis f. sp. tritici, a finding which may be interpreted as both polyols having a role in stress tolerance.
Until now, only a small number of enzymes utilizing arabitol as a main substrate have been described. There are the NAD+-dependent D-arabitol:NAD+ 4-oxidoreductases (EC 1.1.1.11) functioning mainly in the reduction of D-xylulose [14]. L-Arabitol:NAD+ 4-oxidoreductases (EC 1.1.1.12) perform the above reaction with L-xylulose as the substrate [15]. There are the ribulose-forming arabitol:NAD+ 2-oxidoreductases (EC 1.1.1.13 and EC 1.1.1.250) acting on the L- and D-isomers respectively [16,17]. Broad-substrate-spectrum alditol:NAD(P)+ 1-oxidoreductases (EC 1.1.1.21) [18], and D- and L-xylulose reductases (EC 1.1.1.9 and EC 1.1.1.10 respectively) were found to exhibit side activity with arabitol [19,20]. We also found one description of a NADP+-dependent D-arabitol 4-oxidoreductase [21]; however, no follow-up studies or classifications of this reaction were found.
The rust fungi (Uredinales) are major pathogens of many plants, including economically important crops [22,23]. They are obligate biotrophic pathogens which do not kill their hosts, but claim nutrients from living plant cells [24]. The interface through which most of this metabolite transfer takes place is called the haustorium [25–27]. Until recently, however, there was no conclusive proof of such a re-location. Voegele et al. [28] were able to show that haustoria are indeed the sites of hexose uptake in the rust fungus Uromyces fabae.
Part of our research extending from these findings now focuses on the fate of carbohydrates once they are taken up by the fungus. In the present paper, we show that, in U. fabae, D-arabitol is produced from D-xylulose and D-ribulose by a novel NADP+-dependent D-arabitol dehydrogenase, ARD1p, which is localized in the lumen of haustoria. Possible functions of D-arabitol in the U. fabae–Vicia faba pathosystem will be discussed.
EXPERIMENTAL
Cultivation of plants and micro-organisms
Cultivation of V. faba cv. con Amore, inoculation with U. fabae uredospores, germination of spores, and the generation of in vitro grown infection structures on artificial surfaces were performed as described in [29,30]. Saccharomyces cerevisiae strains were grown in SC (synthetic complete) medium with 2% glucose as the carbon source and drop-out mix lacking uracil for selection of transformants [31].
Nucleic acid manipulations
Isolation of RNA and total DNA extraction were performed according to Hahn and Mendgen [29]. Haustorium-specific cDNA and genomic DNA libraries are described in [29]. Screening of libraries and other nucleic acid manipulations were performed using standard molecular-biology techniques [32]. Yeast transformation was performed according to Elble [33]. Sequencing was done on an ABI 377 HT automated sequencer (GATC, Konstanz, Germany) using the Big-Dye Terminator Cycle Sequencing Ready Reaction Mix (PE Applied Biosystems, Foster City, CA, U.S.A.). Sequencing data were analysed using Chromas1.45 (Technelysium Pty., Helensvale, Queensland, Australia) and DNASTAR (Madison, WI, U.S.A.) software. Non-radioactive hybridization experiments were performed according to Engler-Blum et al. [34]. Genomic Southern and Northern hybridizations were carried out using homologous probes at hybridization temperatures between 65 °C and 68 °C. Signal detection was performed using alkaline-phosphatase-conjugated anti-digoxigenin Fab fragments, CSPD [disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]-decan}-4-yl)phenyl phosphate] (Roche Diagnostics GmbH, Mannheim, Germany) as substrate, and autoradiography.
Plasmid constructions
For expression in S. cerevisiae strain 23344c [35], a recombinant λgt10 cDNA clone carrying a full-length ARD1 cDNA was digested with NotI, and the cDNA insert was ligated into vector pDR195 [36] yielding plasmid pDR195::ARD1. Ampicillin-resistant transformants were screened for the correct orientation of the insert using colony-PCR with vector- and gene-specific primers.
For the generation of anti-ARD1p antibodies, the complete open reading frame of ARD1 without the native start codon was cloned into expression vector pET28a(+) (Novagen, Madison, WI, U.S.A.) by introducing unique BamHI and SacI restriction sites via PCR using primers SDH1-5′, 5′-GTCGCGGATCCG17CCACTCAAACTCTACCCG-3′ and SDH1-3′, 5′-GTCGACGGAGCTCC2239ATGTGATTCCTTGAAAG-3′ (the introduced restriction sites are underlined and superscript numbers give the position of the preceding nucleotide in GenBank® sequence AJ809335). The resulting plasmid pET28a(+)::ARD1 encoded a His-tagged fusion protein consisting of 34 newly introduced amino acids at the N-terminus (including a His6-cluster) and amino acids 2–349 of ARD1p. All plasmid constructs were verified by sequencing.
Expression of His-tagged ARD1p fusion protein and antibody generation
Overexpression of the His-tagged fusion protein encoded by plasmid pET28a(+)::ARD1 (ARD1fp) was carried out using Escherichia coli strain BL21(DE3) and induction with IPTG (isopropyl β-D-thiogalactoside) [37]. Purification of the fusion proteins was performed using immobilized metal-ion-affinity chromatography under denaturing conditions according to the manufacturer's protocol (The QIAexpressionist, Qiagen, Hilden, Germany). Antibodies were obtained by repeated fortnightly injection of a female New Zealand White rabbit with 500 μg of purified fusion protein together with Freund's adjuvant. Serum S757 was purified in a two-step procedure as described in [28]. The fusion protein used for negative adsorption was encoded by pET28a(+)::MAD1 (where MAD1 is NADP-dependent mannitol dehydrogenase 1 from U. fabae) [38]. The MAD1fp fusion protein is identical with ARD1fp with respect to the vector portion of the fusion protein including the His6-tag. Adsorbing S757 against MAD1fp therefore effectively removes antibodies that were generated against the vector portion of ARD1fp.
Enzyme extraction
Protein was prepared from infected and non-infected plants, in vitro infection structures and uredospores by grinding the tissue in extraction buffer together with 1/10 of its wet weight of acid-washed sea sand. Extraction from transgenic yeast used extensive vortex-mixing of cells in extraction buffer, together with an equal volume of 1-mm-diameter glass beads after washing the cells once with double-distilled water. This was followed by a high-speed centrifugation (16000 g for 5 min at 4 °C) and an ultracentrifugation (35000 rev./min for 15 min at 4 °C using a RP45A rotor in a Sorvall RC M120 instrument) step. Extraction buffer for maximum ARD1p activity and stability was 50 mM Tris/HCl, pH 7.5, and 5% glycerol.
Enzyme assays
ARD1p activity was assayed following the absorbance change of NAD(P)+/NAD(P)H at 340 nm. A molar absorption coefficient for NAD(P)H of 6.22×106 cm2·mol−1 was used for calculations. Reactions were monitored with a Hitachi U-2000 spectro-photometer, or a TECAN Spectra Classic ELISA reader, operated with TECAN Magellan2 software (TECAN, Grödig, Austria). Standard reaction buffer for the forward reaction was 100 mM glycine/NaOH, pH 9, 50 mM NaCl, and 5% glycerol. For the reverse reaction, 100 mM sodium phosphate buffer, pH 6, was used. For kinetic experiments, substrate concentrations were varied between 1 mM and 1 M, and co-factor concentrations were between 12.5 μM and 700 μM.
Protein concentrations were determined by the Bradford method [39] using the Bio-Rad protein assay dye (Bio-Rad, Hercules, CA, U.S.A.) following the micro assay procedure with dilutions of BSA as standard.
Extraction of metabolites and metabolite analyses
Metabolites were extracted from infected and non-infected plants, in vitro grown infection structures and uredospores, and the subsequent sugar and sugar alcohol analyses were performed as described in [38].
ROS quenching assay
The ability of D-arabitol to quench ROS downstream of H2O2 was assayed by coupling the Fenton reaction to the cleavage of KMB (α-oxo-γ-methiol-butyric acid) as described in [38].
Immunocytochemistry
Rust-infected leaf pieces (8 days post-infection) were infiltrated with ethanol/ethanoic (acetic) acid (1:3, v/v). Specimens were treated with ethanol (three times for 1 day) and embedded in a resin consisting of 75% butyl methacrylate, 25% methyl methacrylate, 1% benzoyl peroxide and 0.15% dithiothreitol, mixed with 20 μl/ml N,N-dimethylaniline. The resin was added to mixtures with ethanol (25, 50, 75 and 100% resin) for 1 day, each at 4 °C. Sections, 0.5 μm thick, were floated on water and dried on microscope slides. Slides with sections were treated with acetone for 30 s, and three times for 15 min with blocking buffer [0.1% BSA in TBS (Tris-buffered saline: 10 mM Tris/HCl, pH 7.4, and 150 mM NaCl)] and incubated with S757p antibody (1:100) in TBS for 2 h. To detect non-specific binding, the pre-immune serum was used as a control. After three washes for 15 min in TBS, sections were incubated with secondary antibody (cyanin-3-conjugated goat anti-rabbit) (Rockland, Gilbertsville, PA, U.S.A.), diluted 1:400 with TBS, for 1 h at 20 °C. After washing for 15 min in TBS, staining with 0.0001% bisbenzimide (Hoechst 33342; Sigma, Taufkirchen, Germany) and three more washes in TBS, samples were examined with a Zeiss Axioplan 2 microscope equipped for epifluorescence (filters BP 490, FT 510 and LP 565) with phase contrast or fluorescence. Images were acquired with a Zeiss Axiocam camera and superimposed with Photoshop software (Adobe Systems, Mountain View, CA, U.S.A.).
RESULTS
Characterization of U. fabae ARD1
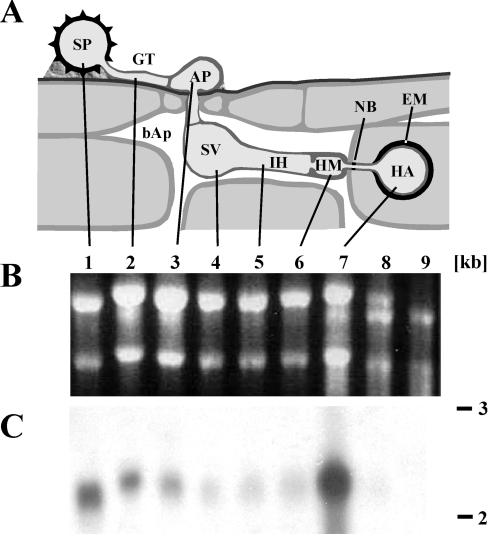
Sequence information from a small EST (expressed sequence tag) sequencing project (M. Hahn, unpublished work) was used to screen a haustorium-specific U. fabae cDNA library for genes linked to polyol production/utilization. Using a partial cDNA sequence exhibiting homology with a putative zinc-binding dehydrogenase as a probe, a full-length clone containing a single open reading frame comprising 1050 bp was isolated. The predicted polypeptide consists of 349 amino acids, with an expected molecular mass of 38.4 kDa. The corresponding gene, ARD1, was amplified from U. fabae genomic DNA and sequenced. Southern Blot analysis of U. fabae genomic DNA digested with five different restriction enzymes indicated clearly that ARD1, like most other genes from U. fabae described so far, is a single-copy gene (results not shown). RNA blot analysis indicated that ARD1 is expressed at all stages of early rust development (spores, germlings and infection structures developed on an artificial surface), with highest transcript levels present in haustoria (Figure 1). No hybridization signal was observed in non-infected plants, which served as negative control.
Figure 1. Expression pattern of ARD1.
(A) Schematic representation of rust infection structures. (B) Ethidium-bromide-stained denaturing agarose gel (loading control). (C) Northern blot of the gel depicted in (B). 1, uredospore (SP); 2, germ tube (GT) after 4 h germination; 3–6, in vitro infection structures harvested at the following stages: 3, appressorium (AP) stage (6 h); 4, substomatal vesicle (SV) stage (12 h); 5, infection hyphae (IH) stage (18 h); 6, haustorial mother cell (HM) stage (21 h); 7, isolated haustoria (HA); 8, infected leaves; 9, non-infected leaves (structures for samples 2–6 were obtained in vitro). bAp, bulk apoplast; NB, neckband; EM, extrahaustorial matrix. Numbers on the right give the size estimate in kb.
Characterization of U. fabae ARD1p
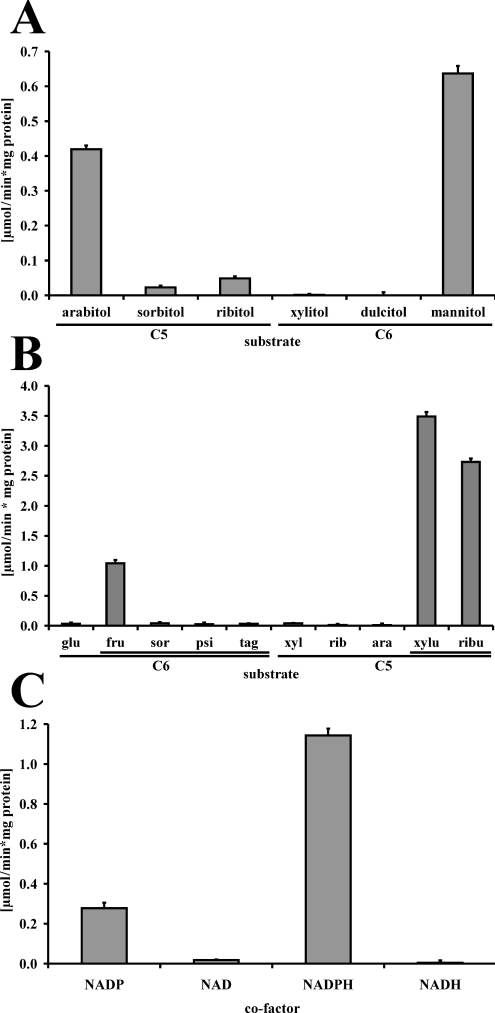
Rust fungi cannot be grown in culture, and there is currently no functional stable transformation system available for them. Biochemical analysis of the ARD1 gene product (ARD1p) was therefore carried out by heterologous expression of ARD1 in S. cerevisiae. Cell-free extracts of strain 23344c transformed with the expression vector pDR195 alone did not show activity with any of the substrates under all conditions tested. However, extracts of transformants bearing plasmid pDR195::ARD1 exhibited considerable alcohol dehydrogenase activity with the polyols D-arabitol and mannitol (Figure 2A). None of the other D- or L-polyols tested showed noteworthy activity with either NAD+ or NADP+ as a co-factor (Figure 2A, and results not shown). For the reverse reaction, we found activity with D-fructose, D-ribulose and D-xylulose, but not with any other sugar tested (Figure 2B, and results not shown). ARD1p therefore clearly acts on the C5-ketoses D-xylulose and D-ribulose, and the C6-ketose D-fructose, but not on aldoses or L-isoforms of the tested sugars. Figure 2(C) shows that the enzyme exhibits specificity for NADP(H) as a co-factor. No ARD1p activity could be detected using NAD(H) as a co-factor (Figure 2C, and results not shown). To our knowledge, this is the first description of an alcohol dehydrogenase exhibiting such substrate and co-factor specificity.
Figure 2. Substrate and co-factor specificity of ARD1p.
(A) The forward reaction was assayed at a co-factor concentration of 0.4 mM NADP+ and a substrate concentration of 500 mM, except for dulcitol which was added to saturation. All substrates were in the D-conformation. (B) The reverse reaction was assayed at a co-factor concentration of 0.2 mM NADPH and substrate concentrations of 500 mM for glucose (glu), xylose (xyl), ribose (rib) and arabinose (ara), and 100 mM for fructose (fru), sorbose (sor), psicose (psi), tagatose (tag), xylulose (xylu) and ribulose (ribu). Sugars are ordered in groups of C6 and C5, with single lines indicating aldoses and double lines showing ketoses. All substrates were in the D-conformation. (C) Co-factor specificity of ARD1p was assayed with 500 mM D-mannitol and 0.5 mM NAD+ or NADP+ for the forward reaction, and 500 mM D-fructose and 0.2 mM NADH or NADPH for the reverse reaction. Results are the means±S.E.M. (n=5).
Enzyme extraction procedures were adapted further to yield high and stable ARD1p activity in order to determine the kinetic parameters for the main reactions. ARD1p activity for the forward reaction was highest at pH values slightly above 9, with a fairly broad optimum curve (results not shown). The pH-optimum curve for the reverse reaction was also broad, with a maximum around pH 6.0 (results not shown). Neither the addition of different bivalent cations nor that of EDTA had any significant effect on ARD1p activity (results not shown). Under optimal conditions for the respective reactions, we determined the kinetic parameters shown in Table 1.
Table 1. Kinetic parameters for ARD1p.
| Substrate or co-factor* | Vmax [μmol/(min·mg of protein)] | Km (mM) | Vmax/Km [1/(min·mg of protein)] |
|---|---|---|---|
| D-Arabitol | 1.92 | 834 | 2.30×10−6 |
| NADPara | 1.20 | 0.08 | 1.50×10−2 |
| D-Xylulose | 9.91 | 7.80 | 1.27×10−3 |
| NADPHxyl | 15.31 | 0.26 | 5.89×10−2 |
| D-Ribulose | 4.41 | 53.9 | 8.18×10−5 |
| NADPHrib | 5.15 | 0.20 | 2.58×10−2 |
| Mannitol | 0.86 | 425 | 2.02×10−6 |
| NADPman | 0.68 | 0.07 | 9.7×10−3 |
| D-Fructose | 1.65 | 160 | 1.03×10−5 |
| NADPHfru | 2.83 | 0.16 | 1.76×10−2 |
* Indices for the co-factors indicate the substrate with which they were determined.
In order to determine the equilibrium between D-ribulose, D-xylulose and D-arabitol, we also established the kinetic parameters for the forward reaction (oxidation of D-arabitol) at pH 6. These are: Vmax(D-arabitol)=117 nmol/(min·mg of protein), Vmax(NADP+)=97 nmol/(min·mg of protein), Km(D-arabitol)=706 mM and Km(NADP+)=150 μM. We used the Haldane equation to calculate the equilibrium constants (Keq) for the conversion of D-arabitol into D-xylulose or D-ribulose. The constants calculated are: 1.43×10−12 M for the formation of D-xylulose and 5.36×10−11 M for the formation of D-ribulose. This means that in the equilibrium, concentrations of D-arabitol and NADP+ are much higher than those of NADPH and D-xylulose/D-ribulose.
Localization of U. fabae ARD1p
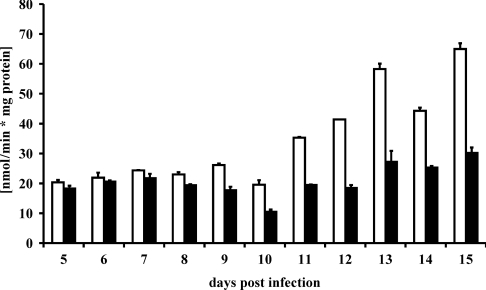
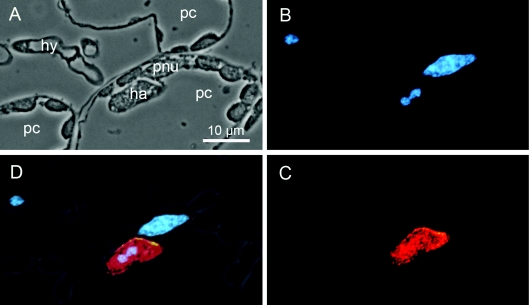
No significant NADP+-dependent D-arabitol dehydrogenase activity could be found in spores or in vitro infection structures (results not shown). However, a small, but distinct, activity was found in infected leaves as infection progressed (Figure 3). Control samples from uninfected leaves showed only a constant low background activity. We therefore conclude that the NADP+-dependent D-arabitol dehydrogenase activity found in infected leaves is of fungal origin and most likely at least in part a result of the action of ARD1p. To determine the exact localization of ARD1p within the parasitic rust mycelium, we used immunofluorescence microscopy. Purified anti-ARD1p antibody (S757p) revealed a localization of ARD1p exclusively in the lumen of haustoria (Figure 4). No other fungal or plant structures were labelled. Pre-immune serum used as a control did not reveal any signal in any fungal or plant structure.
Figure 3. NADP+-dependent D-arabitol dehydrogenase activity in infected plants.
The reverse reaction was assayed with D-xylulose as a substrate under standard conditions using extracts of infected (□) and non-infected leaves (■). Results are means±S.E.M. (n=3).
Figure 4. Localization of ARD1p.
(A) Phase-contrast image depicting a fungal hypha (hy) contacting a plant cell (pc). In the contacted host cell, a fungal haustorium (ha) is visible. The plant cell nucleus (pnu) is located in close proximity to the haustorium. Plastids are visible in all three plant cells (pc) depicted in the cross section. (B) Nucleic acid stain using bisbenzimide (Hoechst 33342) highlighting the prominent plant and three smaller fungal nuclei. (C) Immunofluorescence localization of ARD1p in the lumen of haustoria using S757p primary antibody and cyanin-3-labelled goat anti-rabbit secondary antibody. (D) Superimposed images of (A)–(C).
D-Arabitol in the U. fabae–V. faba pathosystem
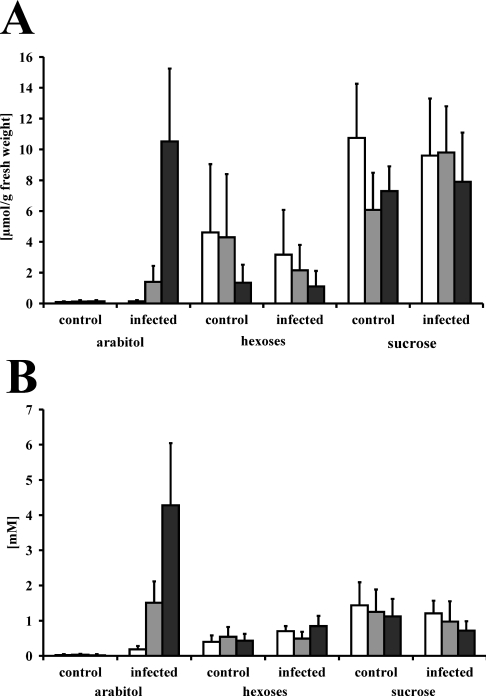
Having found an enzyme which most likely produces D-arabitol in haustoria of the biotrophic rust fungus U. fabae, we set out to analyse sugar and sugar alcohol levels throughout the course of infection. Of all the compounds analysed, only two showed considerable changes following infection in a time-dependent manner. One compound was identified as mannitol. The changes in mannitol content and the source for this sugar alcohol were described separately [38]. The other compound was unequivocally identified as D-arabitol by a combination of HPLC, NMR and polarimetry (results not shown). Figure 5 shows that levels of free hexoses (D-glucose and D-fructose) and sucrose varied over time, but not in response to pathogen attack. D-Arabitol, however, rose from virtually non-detectable levels in non-infected leaves to concentrations of up to 10.5 μmol/g of fresh weight in cell extracts (Figure 5A) and up to 4.3 mM in apoplastic fluids (Figure 5B) of rust-infected leaves. In contrast, in control plants, the level of D-arabitol remained below the detection limit. This indicates strongly that D-arabitol is synthesized by the biotrophic rust mycelium and is released into the apoplast.
Figure 5. Sugar and sugar alcohol concentrations in infected and non-infected (control) V. faba leaf extracts (A) and apoplastic fluids (B).
Metabolites were analysed by HPLC 3 (white bars), 5 (grey bars) and 12 days (black bars) post infection. Concentrations are given as μmol/g of fresh weight for leaf extracts and mM for apoplastic fluids. Results are means±S.E.M. (n=6 for leaf extracts; n=10–15 for apoplastic fluids). ‘hexoses’ represents the sum of glucose and fructose.
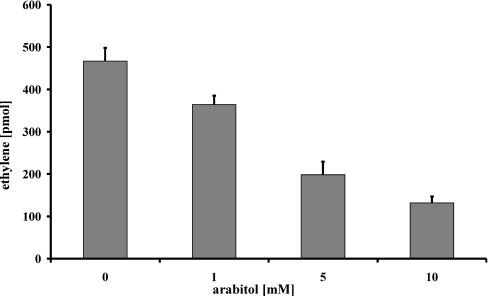
It has been shown that some polyols are able to react with and thereby detoxify ROS [8]. The presence of D-arabitol in the apoplast suggests that this polyol could carry out a protective function for the rust fungus against host-derived ROS. We therefore tested if the D-arabitol produced during the pathogenic interaction would be sufficient to quench ROS derived from H2O2. This was carried out in an in vitro system coupling the Fenton reaction to the conversion of KMB into ethylene and monitoring product formation via gas chromatography. Figure 6 shows that the concentrations of D-arabitol found in the apoplast of U. fabae-infected V. faba are sufficient to quench ROS effectively.
Figure 6. ROS quenching capacity of D-arabitol.
Different concentrations of D-arabitol were added to the in vitro reaction containing KMB, and the reduction in ethylene production was followed on a gas chromatograph. Results are means±S.E.M. (n=4).
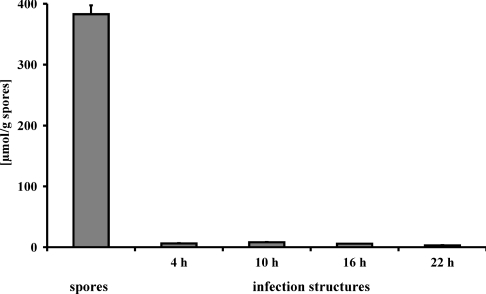
Uredospores and in vitro grown infection structures (germlings, appressoria, infection hyphae and haustorial mother cells [30]) of U. fabae were also tested for the presence of D-arabitol and sugars. Sucrose, fructose and glucose were below the detection limit of 10 μM in spores and all infection structures analysed (results not shown). D-Arabitol, however, was found in substantial quantities in spores, but disappeared rapidly upon infection- structure formation (Figure 7). This strongly suggests a role of D-arabitol as storage compound.
Figure 7. D-Arabitol as a storage compound.
The amount of D-arabitol present in spores and infection structures was analysed by HPLC of chloroform/methanol extracts. Results are means±S.E.M. (n=6 for spores; n=2 for infection structures).
DISCUSSION
ARD1p, a novel D-arabitol dehydrogenase
We have identified a NADP+-dependent D-arabitol dehydrogenase, ARD1p, from the biotrophic plant pathogen U. fabae. Its ability to utilize D-arabitol, D-xylulose and D-ribulose in an NADP(H)-dependent reaction clearly sets it apart from other enzymes in this class described to date. A number of different enzymes acting on arabitol have been described in the literature. D-Arabitol 4-dehydrogenases (EC 1.1.1.11) with similar substrate specificity to ARD1p have been found, but they solely utilize NAD(H) as a co-factor [40,41]. D-Arabitol 2-dehydrogenases (EC 1.1.1.250) also exclusively use NAD(H) as a co-factor [17,42]. Other arabitol dehydrogenases exhibit stereoselectivity for the L-isomer only (EC 1.1.1.12 and EC 1.1.1.13), but essentially exhibit similar substrate specificity to ARD1p [15,16]. Xylulose reductases (EC 1.1.1.9 and EC 1.1.1.10) have also been implicated in the oxidation of arabitol [19,20]. However, none of these enzymes matches the specificities determined for ARD1p. The closest functional homologue for ARD1p is a NADP+-dependent D-arabitol dehydrogenase from Saccharomyces mellis described by Weimberg [21]. However, no follow-up studies and classifications of the described activity were carried out.
Sequence analysis of the ARD1 gene also gave no indication of a relatedness of ARD1p to other known arabitol dehydrogenases. Initial NCBI BLASTP searches [43] indicated the presence of putative conserved domains. Highest scores were obtained with ‘zinc-binding dehydrogenases’ (gnl|CDD|25397) and ‘threonine dehydrogenase and related zinc-dependent dehydrogenases’ (gnl|CDD|10788). However, no dependence of ARD1p activity on Zn2+ or other cations could be found in the present study, nor did EDTA inhibit the reaction. The scores found could be due to structural similarities of dehydrogenases in general. However, recent screens of the databases revealed close relatives of ARD1p, all being hypothetical proteins predicted from completely sequenced fungal genomes. Accession numbers EAL21954 from the human pathogen Cryptococcus neoformans, EAA49456 from the rice pathogen Magnaporthe grisea and EAA66542 from the saprophytic mould Aspergillus nidulans show a high degree of similarity to ARD1p. Future studies will have to show if the sequence similarity extends to functional similarity. However, the sequence similarities already indicate that ARD1p belongs to a new group of enzymes which appear to be restricted to the fungal kingdom.
Physiological roles for ARD1p
Whereas ARD1 transcripts were detected at low levels in virtually all structures of the rust fungus (Figure 1), ARD1p could only be detected in the lumen of haustoria (Figure 4). This apparent discrepancy might be due to the different limits of detection. We therefore cannot exclude that low amounts of ARD1p are present in intercellular hyphae. However, both experiments indicated the presence of highest amounts of ARD1p in haustoria. Haustoria have been implicated in the supply of rust fungi with nutrients ever since their discovery [44]. However, only recently was it shown that haustoria are indeed the site of hexose uptake in the biotrophic mycelium of U. fabae [28]. From an EST sequencing project, there is good evidence that genes encoding enzymes of glycolysis and the pentose phosphate pathway are expressed in haustoria (M. Hahn, unpublished work). ARD1p could therefore act on D-ribulose and/or D-xylulose arising from the pentose phosphate pathway as described for D-arabitol:NAD+ 4-oxidoreductases and D-arabitol:NAD+ 2-oxidoreductases [17,41]. This hypothesis would be consistent with our finding that D-arabitol is a major constituent of the biotrophic mycelium (Figure 5) and with the equilibrium constants calculated, which suggest that ARD1p is arabitol-forming rather than arabitol-consuming. We therefore suggest that D-arabitol is formed in haustoria by the action of ARD1p. Utilizing NADPH, rather than NADH, as a co-factor would circumvent co-factor interconversion if one of the main functions of the pentose phosphate cycle is the production of D-arabitol. Mobilization of D-arabitol at least in spores does not seem to proceed via ARD1p, since the substantial amounts of D-arabitol deposited in spores rapidly diminish without detectable ARD1p.
Apart from D-arabitol, D-xylulose and D-ribulose, ARD1p can also use D-fructose and mannitol as a substrate. Mannitol was shown to be another polyol that is synthesized and excreted from U. fabae during the biotrophic interaction [38]. Given the localization and substrate specificity of ARD1p, it cannot be ruled out that this enzyme contributes to the mannitol pool produced by the fungus.
Physiological roles for D-arabitol
The rise of D-arabitol in the course of infection of V. faba by the biotrophic fungus U. fabae might most easily be explained with a role of this polyol in carbohydrate storage, as suggested by Lewis and Smith [1]. However, our findings do not rule out roles in osmoprotection [45] or in translocation, as suggested for mannitol in some mycorrhizal interactions [6]. Actually, given the localization of ARD1p in haustoria, accumulation of D-arabitol in spores has to be preceded by a translocation of the polyol to these structures. A carbohydrate-storage function is also supported by the large quantities of D-arabitol found in spores, which diminished rapidly upon germination. Niederpruem and Hunt [13] reported a similar scenario during the germination of basidiospores of Schizophyllum commune. In P. graminis f. sp. tritici, on the other hand, arabitol was found in both ungerminated and germinated uredospores, a finding interpreted as arabitol having a primary role in stress tolerance [2]. However, the fact that D-arabitol was found in large quantities in apoplastic fluids also hints at a potential role as radical scavenger. D-Arabitol could act in a similar way as described for the C6-polyol mannitol [9,10,38].
In summary, we have identified a novel NADP+-dependent D-arabitol dehydrogenase, which is localized in the feeding structures of the rust fungus U. fabae, the haustoria. The enzyme might be linked to the production, but apparently not the utilization, of D-arabitol in this biotrophic parasite. The present study and related work [38] indicate functions for the polyols D-arabitol and mannitol in carbohydrate storage and translocation, and in the suppression of host defences.
Acknowledgments
We are grateful to Christine Giele, Annette Schmid and Heinz Vahlenkamp for expert technical assistance. This work was supported by a grant provided by the Deutsche Forschungsgemeinschaft to K.M. and R.T.V. (Me 523/24-1).
References
- 1.Lewis D. H., Smith D. C. Sugar alcohols (polyols) in fungi and green plants. I. Distribution, physiology and metabolism. New Phytol. 1967;66:143–184. [Google Scholar]
- 2.Maclean D. J., Scott K. J. Identification of glucitol (sorbitol) and ribitol in a rust fungus, Puccinia graminis f. sp. tritici. J. Gen. Microbiol. 1976;97:83–89. doi: 10.1099/00221287-97-1-83. [DOI] [PubMed] [Google Scholar]
- 3.Reisener H. J., Goldschmid H. R., Ledingham G. A., Perlin A. S. Formation of trehalose and polyols by wheat stem rust (Puccinia graminis tritici) uredospores. Can. J. Biochem. Physiol. 1962;40:1248–1251. [PubMed] [Google Scholar]
- 4.Shen B., Hohmann S., Jensen R. G., Bohnert H. Roles of sugar alcohols in osmotic stress adaptation: replacement of glycerol by mannitol and sorbitol in yeast. Plant Physiol. 1999;121:45–52. doi: 10.1104/pp.121.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark A. J., Blissett K. J., Oliver R. P. Investigating the role of polyols in Cladosporium fulvum during growth under hyper-osmotic stress and in planta. Planta. 2003;216:614–619. doi: 10.1007/s00425-002-0833-2. [DOI] [PubMed] [Google Scholar]
- 6.Koide R. T., Shumway D. L., Stevens C. M. Soluble carbohydrates of red pine (Pinus resinosa) mycorrhizas and mycorrhizal fungi. Mycol. Res. 2000;104:834–840. [Google Scholar]
- 7.Hult K., Veide A., Gatenbeck S. The distribution of the NADPH regenerating mannitol cycle among fungal species. Arch. Microbiol. 1980;128:253–255. doi: 10.1007/BF00406168. [DOI] [PubMed] [Google Scholar]
- 8.Smirnoff N., Cumbes Q. J. Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry. 1989;28:1057–1060. [Google Scholar]
- 9.Shen B., Jensen R. G., Bohnert H. J. Increased resistance to oxidative stress in transgenic plants by targeting mannitol biosynthesis to chloroplasts. Plant Physiol. 1997;113:1177–1183. doi: 10.1104/pp.113.4.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen B., Jensen R. G., Bohnert H. J. Mannitol protects against oxidation by hydroxyl radicals. Plant Physiol. 1997;115:527–532. doi: 10.1104/pp.115.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gold J. W., Wong B., Bernard E. M., Kiehn T. E., Armstrong D. Serum arabinitol concentrations and arabinitol/creatinine ratios in invasive candidiasis. J. Infect. Dis. 1983;147:504–513. doi: 10.1093/infdis/147.3.504. [DOI] [PubMed] [Google Scholar]
- 12.Wong B., Bernard E. M., Gold J. W., Fong D., Silber A., Armstrong D. Increased arabinitol levels in experimental candidiasis in rats: arabinitol appearance rates, arabinitol/creatinine ratios, and severity of infection. J. Infect. Dis. 1982;146:346–352. doi: 10.1093/infdis/146.3.346. [DOI] [PubMed] [Google Scholar]
- 13.Niederpruem D. J., Hunt S. Polyols in Schizophyllum commune. Am. J. Bot. 1967;54:241–245. [Google Scholar]
- 14.Blakley E. R., Spencer J. F. Studies on the formation of D-arabitol by osmophilic yeasts. Can. J. Biochem. Physiol. 1962;40:1737–1748. [PubMed] [Google Scholar]
- 15.Chiang C., Knight S. G. A new pathway of pentose metabolism. Biochem. Biophys. Res. Commun. 1960;3:554–559. doi: 10.1016/0006-291x(60)90174-1. [DOI] [PubMed] [Google Scholar]
- 16.Chiang C., Knight S. G. L-Arabinose metabolism by cell-free extracts of Penicillium chrysogenum. Biochim. Biophys. Acta. 1961;46:271–278. doi: 10.1016/0006-3002(61)90750-8. [DOI] [PubMed] [Google Scholar]
- 17.Wong B., Murray J. S., Castellanos M., Croen K. D. D-Arabitol metabolism in Candida albicans: studies of the biosynthetic pathway and the gene that encodes NAD-dependent D-arabitol dehydrogenase. J. Bacteriol. 1993;175:6314–6320. doi: 10.1128/jb.175.19.6314-6320.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scher B. M., Horecker B. L. Pentose metabolism in Candida. 3. The triphosphopyridine nucleotide-specific polyol dehydrogenase of Candida utilis. Arch. Biochem. Biophys. 1966;116:117–128. doi: 10.1016/0003-9861(66)90020-8. [DOI] [PubMed] [Google Scholar]
- 19.Richard P., Putkonen M., Vaananen R., Londesborough J., Penttila M. The missing link in the fungal L-arabinose catabolic pathway, identification of the L-xylulose reductase gene. Biochemistry. 2002;41:6432–6437. doi: 10.1021/bi025529i. [DOI] [PubMed] [Google Scholar]
- 20.Verho R., Putkonen M., Londesborough J., Penttila M., Richard P. A novel NADH-linked L-xylulose reductase in the L-arabinose catabolic pathway of yeast. J. Biol. Chem. 2004;279:14746–14751. doi: 10.1074/jbc.M312533200. [DOI] [PubMed] [Google Scholar]
- 21.Weimberg R. Mode of formation of D-arabitol by Saccharomyces mellis. Biochem. Biophys. Res. Commun. 1962;8:442–445. doi: 10.1016/0006-291x(62)90293-0. [DOI] [PubMed] [Google Scholar]
- 22.Long D. L. 2003 Final Cereal Rust Bulletin, Cereal Disease Laboratory. U.S. Department of Agriculture; 2003. http://www.cdl.umn.edu/crb/2003crb/03crbfin.html. [Google Scholar]
- 23.Kawuki R. S., Adipala E., Tukamuhabwa P. Yield loss associated with soya bean rust (Phakopsora pachyrhizi Syd.) in Uganda. J. Phytopathol. 2003;151:7–12. [Google Scholar]
- 24.Voegele R. T., Mendgen K. Rust haustoria: nutrient uptake and beyond. New Phytol. 2003;159:93–100. doi: 10.1046/j.1469-8137.2003.00761.x. [DOI] [PubMed] [Google Scholar]
- 25.Heath M. C., Skalamera D. Cellular interactions between plants and biotrophic fungal parasites. Adv. Bot. Res. 1997;24:195–225. [Google Scholar]
- 26.Bushnell W. R. Physiology of fungal haustoria. Annu. Rev. Phytopathol. 1972;10:151–176. [Google Scholar]
- 27.Mendgen K., Struck C., Voegele R. T., Hahn M. Biotrophy and rust haustoria. Physiol. Mol. Plant Pathol. 2000;56:141–145. [Google Scholar]
- 28.Voegele R. T., Struck C., Hahn M., Mendgen K. The role of haustoria in sugar supply during infection of broad bean by the rust fungus Uromyces fabae. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8133–8138. doi: 10.1073/pnas.131186798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hahn M., Mendgen K. Characterization of in planta-induced rust genes isolated from a haustorium-specific cDNA library. Mol. Plant Microbe Interact. 1997;10:427–437. doi: 10.1094/MPMI.1997.10.4.427. [DOI] [PubMed] [Google Scholar]
- 30.Deising H., Jungblut P. R., Mendgen K. Differentiation-related proteins of the broad bean rust fungus Uromyces viciae-fabae, as revealed by high resolution two-dimensional polyacrylamide gel electrophoresis. Arch. Microbiol. 1991;155:191–198. [Google Scholar]
- 31.Sherman F. Getting started with yeast. In: Guthrie C., Fink G. R., editors. Methods in Enzymology: Guide to Yeast Genetics and Molecular Biology, vol. 194. San Diego: Academic Press; 1991. pp. 3–21. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J., Russel D. W. Molecular Cloning: a Laboratory Manual. 3rd edn. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 33.Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- 34.Engler-Blum G., Meier M., Frank J., Müller G. A. Reduction of background problems in nonradioactive Northern and Southern blot analyses enables higher sensitivity than 32P-based hybridizations. Anal. Biochem. 1993;210:235–244. doi: 10.1006/abio.1993.1189. [DOI] [PubMed] [Google Scholar]
- 35.Marini A. M., Springael J. Y., Frommer W. B., Andre B. Cross-talk between ammonium transporters in yeast and interference by the soybean SAT1 protein. Mol. Microbiol. 2000;35:378–385. doi: 10.1046/j.1365-2958.2000.01704.x. [DOI] [PubMed] [Google Scholar]
- 36.Rentsch D., Laloi M., Rouhara I., Schmelzer E., Delrot S., Frommer W. B. NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett. 1995;370:264–268. doi: 10.1016/0014-5793(95)00853-2. [DOI] [PubMed] [Google Scholar]
- 37.Studier F. W., Moffat B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 38.Voegele R. T., Hahn M., Lohaus G., Link T., Heiser I., Mendgen K. Possible roles for mannitol and mannitol dehydrogenase in the biotrophic plant pathogen Uromyces fabae. Plant Physiol. 2005;137:190–198. doi: 10.1104/pp.104.051839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 40.Lin E. C. An inducible D-arabitol dehydrogenase from Aerobacter aerogenes. J. Biol. Chem. 1961;236:31–36. [PubMed] [Google Scholar]
- 41.Wood W. A., McDonough M. J., Jacobs L. B. Ribitol and D-arabitol utilization by Aerobacter aerogenes. J. Biol. Chem. 1961;236:2190–2195. [PubMed] [Google Scholar]
- 42.Quong M. W., Miyada C. G., Switchenko A. C., Goodman T. C. Identification, purification, and characterization of a D-arabinitol-specific dehydrogenase from Candida tropicalis. Biochem. Biophys. Res. Commun. 1993;196:1323–1329. doi: 10.1006/bbrc.1993.2397. [DOI] [PubMed] [Google Scholar]
- 43.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Bary A. Recherches sur le developpement de quelques champignons parasites. Ann. Sci. Nat. Bot. 1863;20:5–148. [Google Scholar]
- 45.Chaturvedi V., Bartiss A., Wong B. Expression of bacterial mtlD in Saccharomyces cerevisiae results in mannitol synthesis and protects a glycerol-defective mutant from high-salt and oxidative stress. J. Bacteriol. 1997;179:157–162. doi: 10.1128/jb.179.1.157-162.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]