Abstract
Nedd8 is a ubiquitin-like modifier that is attached to the cullin components of E3 ubiquitin ligases. More recently, p53 has also been shown to be Nedd8-modified. Nedd8 attachment occurs in a manner similar to that observed for other ubiquitin-like modifiers. In the present study, we report on the characterization of Nep1, a deneddylating enzyme in fission yeast (Schizosaccharomyces pombe). Unlike loss of ned8, deletion of the nep1 gene is not lethal, although nep1.d cells are heterogeneous in length, suggesting a defect in cell-cycle progression. Viability of nep1.d cells is dependent on a functional spindle checkpoint but not on the DNA integrity checkpoint. Deletion of a related gene (nep2), either alone or in combination with nep1.d, also has little effect on cell viability. We show that Nep1 can deneddylate the Pcu1, Pcu3 and Pcu4 cullins in vitro and that its activity is sensitive to N-ethylmaleimide, consistent with the idea that it is a member of the cysteine protease family. nep1.d cells accumulate Nedd8-modified proteins, although these do not correspond to modified forms of the cullins, suggesting that, although Nep1 can deneddylate cullins in vitro, this is not its main function in vivo. Nep1 can be co-precipitated with the signalosome subunit Csn5. Nep1 itself is present in a high-molecular-mass complex, but the presence of this complex is not dependent on the production of intact signalosomes. Our results suggest that, in vivo, Nep1 may be responsible for deneddylating proteins other than cullins.
Keywords: cullin, deneddylation, Nedd8, Nepl, signalosome, spindle checkpoint
Abbreviations: CSN, COP9 signalsome; HA, haemagglutin; JAMM, Jub1/MPM/Mov34 metalloenzyme; Nedd, neuronal precursor of cell-expressed developmentally down-regulated gene; NEM, N-ethylmaleimide; ORF, open reading frame; SUMO, small ubiquitin-like modifier; TAP tag, tandem affinity purification tag; TRITC, tetramethylrhodamine β-isothiocyanate; Ubls, ubiquitin-like proteins
INTRODUCTION
Ubiquitin and Ubls (ubiquitin-like proteins) are small proteins that can be covalently attached to target proteins to modify their function or stability. Ubiquitin is a highly conserved 76-amino-acid protein, the main function of which is to target proteins for proteasome-mediated destruction. Ubiquitin-dependent protein degradation of target proteins requires their modification by attachment of ubiquitin chains linked through the side chain of Lys48. However, ubiquitin also has additional roles that do not involve proteolysis. For example, mono-ubiquitylation of PCNA (proliferating-cell nuclear antigen) is involved in the recruitment of DNA polymerase, and poly-ubiquitylation of PCNA through the side chain of Lys63 promotes alternative DNA repair mechanisms in a manner that is not yet understood [1,2]. Several Ubls have been identified and characterized. These include SUMO (small ubiquitin-like modifier), Nedd8 (neuronal precursor of cell-expressed developmentally down-regulated gene 8)/Den1, ISG15 (interferon-stimulated gene 15) and Hub1 [3–6]. The functions of these Ubls are quite distinct from those of ubiquitin.
The attachment of most Ubls to target proteins is mechanistically similar to that of ubiquitylation (see e.g. [4]). Similar to ubiquitin, most Ubls are expressed as precursors that are processed by C-terminal proteolysis to the mature forms that have a C-terminal glycine residue that forms the covalent linkage to the side chain of a target lysine residue. The processing requires the action of specific Ubl proteases. The mature form of the Ubl is subsequently activated through the formation of a thioester bond with a cysteine residue on a Ubl-specific activator protein. The Ubl is next passed on to a conjugating protein, where it again forms a thioester link. Finally, the Ubl is covalently attached to the target proteins through an ε-amino link. One or more lysine residues on the target protein can be modified and at least some Ubls are believed to form chains in a manner similar to that described for ubiquitin. In some cases, Ubls require the activity of specific ligases for attachment to target proteins.
Perhaps the most extensively studied Ubl is SUMO, which shows approx. 17% similarity to ubiquitin and also has an extended N-terminal region that appears unstructured when studied by NMR [7]. In some cases, both sumoylation and ubiquitylation can occur on the same lysine residue of a target protein, but SUMO modification does not target proteins for proteasome-mediated proteolysis. Although the molecular basis for the effects of sumoylation of target proteins remains unknown, sumoylation has been shown to affect protein–protein interactions, protein localization, enzyme activity and to be antagonistic to ubiquitination. Other Ubls studied seem to have more specific biological roles. For example, ISG15 is encoded by ISG15 and modifies key regulators of signal transduction such as the JAK (Janus kinase)/STAT (signal transduction and activators of transcription) pathway [8] and is mainly confined to the immunological response in higher eukaryotes. Another Ubl, Hub1, plays a role in cell morphogenesis in Saccharomyces cerevisiae and has been reported to modify cell polarity factors [6]. Hub1 lacks the C-terminal glycine residue, and its ability to modify targets covalently has recently been disputed [9].
The Nedd8 Ubl was first identified in mice. Homologues of Nedd8 are found in all eukaryotes, the homologue in S. cerevisiae being known as Rub1 (as a modifier related to ubiquitin). Nedd8 is 57% identical with ubiquitin and has been shown to modify a family of proteins known as cullins that are present in all organisms [10]. Cullins are the scaffold subunit for several related families of E3 ubiquitin ligases. The subunit composition of the canonical SCF (Skp1–Cullin–F box) family of E3 ligases is well understood: the N-terminus of cullin1 associates with a RING protein HRT/RBX1/ROC1, whereas the C-terminus associates with Skp1, which adapts various F-box proteins [11]. A similar modular association of HRT/RBX1/ROC1 and alternatives to Skp1 appears to underlie additional families of E3 ubiquitin ligases associated with Cullins 2, 3 and possibly 4 [12,13].
In fission yeast (Schizosaccharomyces pombe), Nedd8 is essential for cell viability [14]. Germinated ned8.d cells are capable of undergoing two or three rounds of cell division and arrest after forming elongated cells. Similar to ned8, the Schizo. pombe Nedd8 activator and conjugator genes (uba3 and ubc12 respectively) are also highly conserved and are essential for viability [14]. Nedd8 has been shown to be attached covalently to the three Schizo. pombe cullins Pcu1, Pcu3 and Pcu4 [14,15], and modification of Pcu1 by Nedd8 has been shown to be required for the essential function of Pcu1. Thus it is probable that the processes of neddylation and deneddylation are highly regulated. A Nedd8-specific protease activity is associated with the signalosome complex [CSN (COP9 signalsome)]. Specifically, immunopurified CSN can cleave Nedd8 from cullins [16] in a manner that is dependent on the integrity of a conserved JAMM (Jab1/MPM/Mov34 metalloenzyme) domain within subunit 5 (Csn5) that is proposed to define a novel isopeptidase [17]. The signalosome is a multisubunit complex, originally identified as being required for photomorphogenesis in Arabidopsis [18]. Since then, it has been purified from a range of eukaryotic cells, including humans [19] and Schizo. pombe [16]. In Schizo. pombe, null mutants of csn1, csn2, csn3, csn4 and csn5 accumulate neddylated forms of the cullins Pcu1 and Pcu3 [15,16]. The JAMM motif within Csn5 protein has been shown to be required for cleavage of Nedd8 from Cul1 in vivo [17], but this remains to be confirmed using recombinant proteins.
Recently, another deneddylating enzyme has been identified in human cells (NEDP1/Den1) [20,21]. This protein has been shown to have both Nedd8 processing activity and the ability to remove Nedd8 from cullins.
We report here on two Schizo. pombe proteins related to NEDP1/Den1, which, similar to the SUMO proteases, fall under the C48 class of cysteine proteases as defined by Rawlings and Barrett [22]. We show that one of the proteins, Nep1, has the ability to remove Nedd8 from cullins in vitro. Deletion analysis indicates that neither of the genes is essential for viability. The nep1 null strain accumulates high-molecular-mass neddylated species. However, the neddylation status of the cullins in the nep1.d strain resembles that observed in wild-type cells. The Nep1 protein is present in a high-molecular-mass complex that is independent of the CSN complex. Immunoprecipitation experiments indicate that Nep1 can be co-precipitated with Csn5 but not with Csn1, suggesting that Nep1 may be interacting with that fraction of the Csn5 pool that is not associated with the signalosome. In contrast with the signalosome, which is located in the nucleus, Nep1 is located predominantly in the cytoplasm. We propose that, although Nep1 can deneddylate cullins in vitro, its in vivo activity is probably directed against other neddylated targets.
EXPERIMENTAL
Strains and plasmids
The wild-type strain used was sp.011 (ade6.704, leu1.32, ura4.Δ18, h−). The ulp1.d and ulp2.d strains were described previously ([23] and J. C. Y. Ho, L. Zhou, D. L. Taylor and F. Z. Watts, unpublished work). nep1.d and nep2.d were created by the method of Bahler et al. [24] using the following primers: Nep1-PF, 5′-AGCTGTTTTGTTTGCGGACCTATTGTTTGTTTAAATTCCTTTTTTTCCCCATTTCGTACTTACTTTTAAACAATTTTTAACGGATCCCCGGGTTAATTAA-3′; Nep1-PR, 5′-GATAAAACTCAGATTGTTTGAGTTGAAACAAGCCTTCCCGCTAGTTTGTATATTCGAAACTAAACAACGCAAGTGTAGCAGAATTCGAGCTCGTTTAAAC-3′; Nep2-PF, 5′-AATTCGATGATGAAGCAAAAAATAAATCAGACATCCAAACTTCACTTCAAAGCAAGTTGCATTTTGGGAGACAGAGGAAACGGATCCCCGGGTTAATTAA-3′; and Nep2-PR, 5′-CCTGGTAAAAAACGCGTTTCTAAAAACTTGTATTCGTCTTTTTCTCTTTCTCCCACTGTCCTATATTCCCACTTTTAAAAAGAATTCGAGCTCGTTTAAAC-3′. The rad3.d, csn1.d and csn5.d strains were obtained from A. Carr (University of Sussex) [25–27] and mad2.d was provided by S. Sazer (Baylor College of Medicine, Houston, TX, U.S.A.) [28]. Integrated Myc-tagged nep1 and nep2 strains were created using the following primers: Nep1-CTF, 5′-AGTTACCGAAATTTCATAATAGCACTGATAATCCTTTTCTCACTCCTCCGGAAGAACTTGTATCTGGTGATTTCCCTTTTCGGATCCCCGGGTTAATTAA; Nep1-PR, described above; Nep2-CTF, 5′-TACTTCAGAGACCACCCTCAATCGTACCGAGACCGGAAACCGCAGCTATACAACATACACAACAATCCATTGAGATTCATCGGATCCCCGGGTTAATTAA; and Nep2-CTR, 5′-AATTCGATGATGAAGCAAAAAATAAATCAGACATCCAAACTTCACTTCAAAGCAAGTTGCATTTTGGGAGACAGAGGAAAGAATTCGAGCTCGTTTAAAC. Integrated Myc-tagged pcu1 was obtained from T. Toda (Cancer Research UK, London, U.K.) [14], Myc-tagged pcu3 and TAP (tandem affinity purification)-tagged pcu4 strains were from D. Wolf (Harvard School of Public Health, Boston, MA, U.S.A.) [29] and the TAP-tagged Csn1 and Csn5 strains were obtained from C. Liu (University of Sussex) [13]. The nep1 ORF (open reading frame) was amplified from cDNA using the following primers: Nep1-ORFF, 5′-CTCGAGATGAGCAGTTCTCCAA-3′; and Nep1-ORFR, 5′-GGATCCAACAACGCAAGTGTAG-3′. The nep2 ORF was amplified from genomic DNA using the primers: Nep2-ORFF, 5′-CATATGCGCTCCAATTCCATTTTC-3′; and Nep2-ORFR, 5′-CTCGAGTTTGGGAGACAGAGGA-3′. PCR fragments were cloned into the T vector and pUC19 respectively, and sequenced before subcloning. pREP41HA was used for expression in Schizo. pombe and pET15b was used for expression in Escherichia coli. pREP41HA-Nedd8 was obtained from T. Toda [14].
Protein purification methods
Nep1 and Nep2 were purified as follows: 250 ml of BL21 λDE3 pLysS cells, transformed with plasmid pET15b-ORF, were harvested and lysed by sonication. The lysate was centrifuged at 50000 g for 1 h at 4 °C. The supernatant was loaded on to a TALON column (2 ml matrix), which was equilibrated with 50 mM Tris/HCl (pH 8.0) and 5 mM 2-mercaptoethanol. Protein was eluted using 50 mM Tris/HCl (pH 8.0), 150 mM NaCl, 200 mM imidazole and 10% (v/v) glycerol and fractions were collected. All fractions were checked by SDS/PAGE and Western blotting. Protein concentrations were estimated after Coomassie Blue staining of the gels.
Gel filtration was undertaken using 100 ml cultures grown overnight at 30 °C and harvested at A595 0.5. The cell pellet was resuspended in 1 ml of buffer A (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.5% Triton, 1 mM PMSF, 5 mM 2-mercaptoethanol, 10 μg/ml apoprotinin and 10 μg/ml leupeptin). An equal volume of glass beads was added to the cells, which were then lysed using a Ribolyser. The lysate was clarified by centrifugation for 2 min at 420 g and the resulting supernatant was spun for 2 h at 30000 rev./min in a Sorvall AH650 rotor at 4 °C. The extract was then passed through an equilibrated Superdex 200HR 10/30 column (Amersham Biosciences) and 0.5 ml fractions were collected. The column was calibrated using the low- and high-molecular-mass gel-filtration calibration kit (Amersham Biosciences).
Immunoprecipitation was performed on the soluble, whole cell extracts prepared in buffer A by lysing cells with a Ribolyser as described above. The extract (450 μl, 5 mg of protein/ml) was precleared by incubation with prewashed Protein G beads for 30 min at 4 °C. The supernatant was transferred to a fresh test tube, and 8 μl of primary mouse anti-Myc antibody was added for 1 h at 4 °C. The extract was centrifuged at 18000 g for 5 min in a Microfuge and transferred to a fresh test tube containing 20 μl of prewashed Protein G beads. The immune complexes were left to be absorbed by the Protein G beads by incubating at 4 °C for 30 min, and they were subsequently washed twice with buffer A. They were resuspended in 60 μl of 50 mM Tris/HCl (pH 7.5) and 5 mM 2-mercaptoethanol.
TAP-tagged proteins were purified from soluble whole cell extracts in buffer A prepared as described above. The extract (450 μl, 5 mg of protein/ml) was centrifuged for 5 min at 18000 g in a Microfuge. The supernatant was transferred to a fresh test tube and incubated with 20 μl of prewashed Protein G beads for 30 min at 4 °C. After incubation, the complexes were washed twice with ice-cold buffer A. The Protein G beads–Pcu4–TAP pellets were resuspended in 50 μl of 50 mM Tris/HCl (pH 7.5) and 2 mM dithiothreitol, and 5 μl of the suspension was taken for each deneddylation assay.
The production of anti-Pmt3 antisera has been described in a previous study [30]. Monoclonal anti-Myc antibodies were purified from the cell supernatant (CRL1729 cell line; from the A.T.C.C.) using Protein G–Sepharose. Anti-His antisera were obtained from Amersham Biosciences. Anti-HA antibodies (where HA stands for haemagglutinin) were obtained from Babco (Covance Research Products, Denver, PA, U.S.A.) and anti-tubulin antibodies were from Sigma.
Deneddylation assay
Deneddylation assays were undertaken as follows: 20 μl of the reaction mixture contained 0.8 mM MgCl2, 1 nM ATP, 5 mM Tris/HCl (pH 7.5), 2 mM dithiothreitol and 4 μl of the immunoprecipitated protein beads from the different mutants containing Myc-tagged versions of Pcu1 or Pcu3, or TAP-tagged Pcu4. Nep1 (1.2 μg) or Nep2 (0.8 μg) was added to the reaction. After a 1 h incubation at 37 °C, 5 μl of 5×SDS sample buffer (60 mM Tris/HCl, pH 6.8, 25% glycerol, 2% SDS, 5% 2-mercaptoethanol and 0.1% Bromophenol Blue) was added to the mixture, boiled at 100 °C for 3 min, and separated by SDS/PAGE (7.5% polyacrylamide). Western-blot analysis was then performed using anti-Myc antisera and detection was by chemiluminescence.
Immunofluorescence microscopy
Cells were fixed in 3.7% (w/v) paraformaldehyde for 10 min and stained with primary antibody (anti-Myc; 1/2000) followed by TRITC (tetramethylrhodamine β-isothiocyanate)-conjugated goat anti-mouse IgG. Cells were observed using a Deltavision Spectris microscope.
RESULTS
Identification of the sequences related to deneddylators
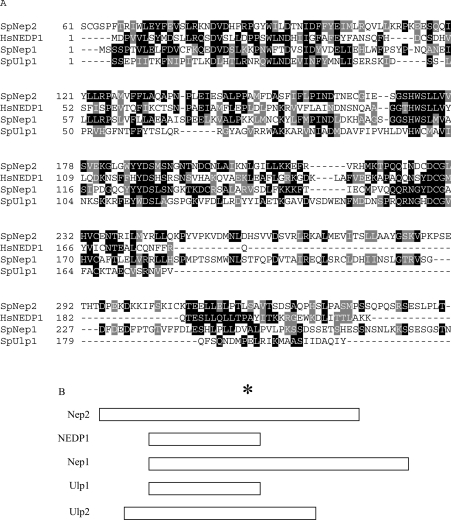
During our search of the Schizo. pombe protein databases for SUMO proteases, we identified two related sequences SPBC17D11.01 (Nep1) and SPBC32H8.02c (Nep2), which encode proteins of 420 and 415 amino acids having predicted molecular masses of 47.2 and 46.4 kDa respectively. While both these ORFs have approx. 16% sequence identity with Schizo. pombe Ulp1 [23] and Ulp2 (J. C. Y. Ho, L. Zhou, D. L. Taylor and F. Z. Watts, unpublished work), they are also related to the recently characterized deneddylating enzymes NEDP1/Den1p [20,21] (Figure 1A). The SUMO and Den1p/NEDP1 proteases fall into a class of cysteine proteases containing conserved histidine, aspartate and cysteine residues at their catalytic sites [22]. Overall, Nep1 and Nep2 are 23% identical with each other, and this increases to 35% between amino acids 1 and 176 of Nep1 and between amino acids 64 and 237 of Nep2.
Figure 1. Nep1 and Nep2 have identity with deneddylating enzymes.
(A) Sequence alignment of Schizo. pombe Ulp1, Nep1 (amino acids 1–285) and Nep2 (amino acids 61–351) proteins with the human deneddylating enzyme NEDP1 created using CLUSTAL W. Black shading indicates identical residues and grey shading indicates conserved amino acids. (B) Comparison of the masses of the proteins relative to the position of the conserved cysteine residue (the asterisk indicates Cys163 in NEDP1). Ulp1, 568 amino acids; Nep1, 420 amino acids; Nep2, 415 amino acids; and NedP1, 212 amino acids.
Although Nep1 and Nep2 are similar in molecular mass, they differ in the position of the conserved cysteine relative to the rest of the protein (Figure 1B). Nep1 has a long C-terminal portion, which has a histidine-rich region between amino acids 284 and 318 that comprises 18/34 (53%) histidine residues. In contrast, Nep2 has an extended N-terminus. The functions of these C- and N-terminal regions is unknown. However, this heterogeneity in molecular mass is typical of this family of proteases (for a review, see [31]), with additional sequences being proposed to be necessary for determining the intracellular localization of the proteins.
nep1 and nep2 are not required for cell viability
To determine whether the nep1 and nep2 genes are essential for cell viability, we deleted them by gene-targeting as described in the Experimental section. Viable colonies were obtained after transformation and selection of a haploid strain, indicating that neither nep1 nor nep2 is essential. To determine whether the two genes have redundant functions, we created the nep1.d nep2.d double mutant. This was also viable, demonstrating that the two genes are not essential for viability. Analysis of the phenotypes of the two single null mutants and of the double mutant indicated that there is no temperature-sensitive growth defect and no sensitivity to DNA-damaging agents or other inhibitors was evident. Included in this analysis were UV (50–200 J/m2), ionizing radiation (250–1500 Gy; 1 Gy=100 rads), methyl methanesulphonate (0.005%), hydroxyurea (10 mM) and camptothecin (2 μM). We also tested sensitivity to growth conditions that impose osmotic stress (1 M sorbitol and 0.9% NaCl) and to the microtubule inhibitor thiabendazole (15 μg/ml; results not shown).
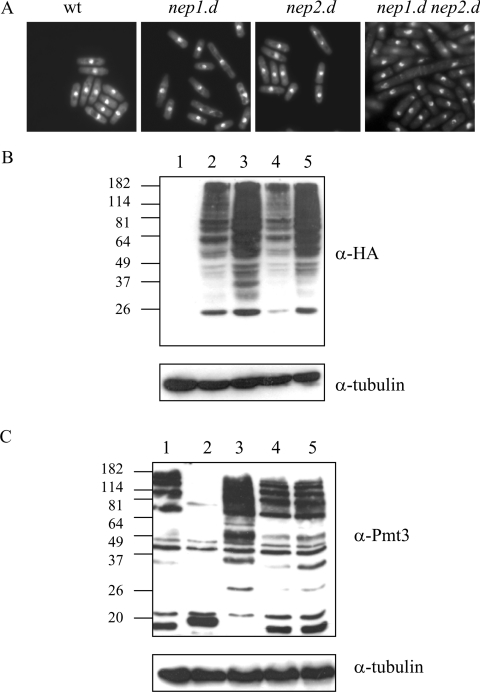
Microscopic analysis of the cells indicated that nep2.d cells resemble the wild-type. In contrast, nep1.d cells were heterogeneous in length (Figure 2A). In the nep1.d nep2.d double mutant, some (∼5%) of the cells were highly elongated and exhibited misplaced septa. This implies that the nep1 and nep2 genes have non-overlapping functions and that nep1 has a role in promoting normal cell-cycle timing. To determine whether the defect in cell-cycle timing resulted from defects in DNA metabolism or defects in microtubule dynamics during mitosis, we crossed nep1.d and the nep1.d nep2.d double mutant with rad3.d and mad2.d that are deleted for genes that are required for the DNA structure and spindle checkpoints respectively [28,32]. The nep.1d nep2.d rad3.d triple mutant was viable, with a cell morphology reminiscent of the nep1.d nep2.d phenotype. However, whereas the nep2.d mad2.d double mutant was viable, the nep1.d mad2.d and nep1.d nep2.d mad2.d mutants were not, indicating that nep1.d is synthetically lethal with the mad2 null mutant. This indicates that the elongated cell phenotype is not dependent on the DNA integrity checkpoint, but is dependent on the spindle checkpoint.
Figure 2. Phenotype of nep1 and nep2 null mutants.
(A) nep1.d and nep1.d nep2.d cells are heterogeneous in length. Cells in an exponentially growing culture were stained with DAPI (4,6-diamidino-2-phenylindole; 1 μg/ml). (B) nep1 null mutants accumulate high molecular mass Nedd8-containing species. Extracts of cells transformed with pREP41HA (lane 1) or pREP41HA-Nedd8 (lanes 2–5) were analysed by Western blotting using anti-HA antisera. Lanes 1 and 2, wild-type (wt); lane 3, nep1.d; lane 4, nep2.d; lane 5, nep1.d nep2.d. (C) nep1 and nep2 null mutants do not affect the levels of SUMO-containing species. Western-blot analysis of strains was performed using anti-Pmt3 antisera. Lane 1, wild-type (wt); lane 2, ulp1.d; lane 3, ulp2.d; lane 4, nep1.d; lane 5, nep2.d. Blots were also probed with anti-tubulin antibodies to show equal loading.
We next investigated whether nep1.d, nep2.d and the nep1.d nep2.d strains were defective in deneddylation in vivo. Total cell extracts were prepared from each strain after transformation with pREP41HA or pREP41HA-Nedd8 (to enable us to detect Nedd8-modified protein species). Extracts were analysed by Western blotting with anti-HA antisera (Figure 2B). We first established that the anti-HA antisera did not cross-react with cell extract proteins in nep+ cells transformed with empty vector (lane 1) and that they recognize high molecular mass species in cells transformed with pREP41HA-Nedd8 (lane 2). We then analysed the nep1 and nep2 single and double mutants. The results indicate that nep1.d (lane 3) accumulates significantly increased levels of high molecular mass anti-HA-containing material when compared with the equivalent nep+ control (lane 2). In contrast, the profiles observed in extracts derived from nep2.d cells (lane 4) resemble that of the nep+ control extract. The nep1.d nep2.d double mutant extract (lane 5) also exhibited a significantly increased level of high molecular mass anti-HA-containing material, with a profile similar to that observed for the nep1.d single mutant extract.
Since Nep1 and Nep2 show significant similarity to the SUMO proteases Ulp1 and Ulp2, we were interested in determining whether the nep1.d and nep2.d strains accumulate high molecular mass SUMO (Pmt3)-containing species. We have previously reported that ulp1.d is defective in both processing SUMO to its mature form and in deconjugating SUMO from target proteins [23]. In total cell extracts derived from ulp1.d cells, a lower level of high molecular mass SUMO-containing species is observed when compared with ulp1+ cell extracts (Figure 2C, compare lane 2 with lane 1) because unprocessed SUMO cannot be conjugated with target proteins (note the increased mass of unconjugated SUMO in lane 2) [23]. In contrast, extracts derived from a strain deleted for ulp2 (lane 3) contain an increased level of high molecular mass SUMO-containing species relative to extracts derived from ulp+ cells. This indicates that a major defect of ulp2.d cells is SUMO deconjugation. When extracts were prepared from strains deleted for either nep1 or nep2 or from a strain deleted for both the genes (lanes 4–6), the profile of anti-pmt3 reactive species resembles the nep+ ulp+ (wild-type) extract. These results indicate that nep1 and nep2 are not required for SUMO processing or deconjugation.
Expression and purification of the proteins
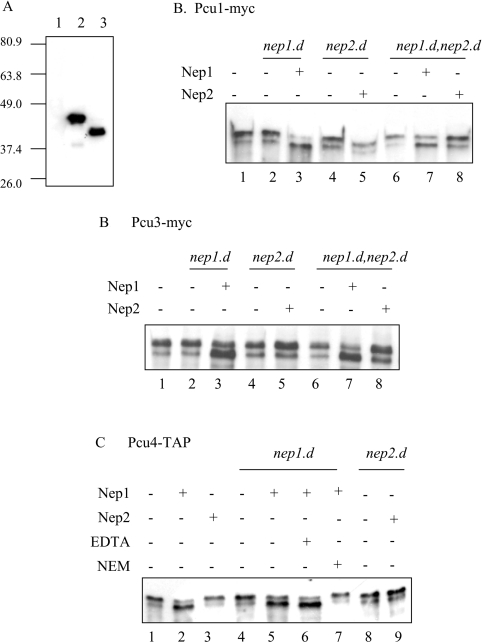
To assay the activity of recombinant Nep1 and Nep2 proteins, the two respective ORFs were amplified, sequenced and cloned into pET15b as described in the Experimental section. Comparison of the genomic and cDNA sequences indicates that the nep1 gene contains two introns, of 80 and 74 nt, consistent with the predicted splicing pattern reported in the Sanger database. After expression in E. coli (see the Experimental section), the proteins were purified by Ni2+ affinity and subjected to Western-blot analysis using anti-His antisera. Figure 3(A) shows that the Nep1 and Nep2 proteins migrate close to the predicted molecular masses of 47 and 46 kDa respectively.
Figure 3. Nep1 is capable of deneddylating cullins in vitro.
(A) Expression of Nep1 and Nep2. Lane 1, empty pET15b; lane 2, pET15b-nep1; and lane 3, pET15b-nep2 expressed in E. coli from pET15b. Proteins were separated by SDS/PAGE and Western blotted with anti-His antisera. (B–D) Deneddylation of cullins. Tagged cullins were immunoprecipitated from wild-type, nep1.d, nep2.d or nep1.d nep2.d cells containing integrated tagged pcu1-myc (B), pcu3-myc (C) or pcu4-TAP (D). Immuneprecipitates were incubated in 20 μl with 1.2 μg of purified Nep1 or 0.8 μg of Nep2 protein as indicated. Where added, the concentrations of EDTA and NEM were 100 mM.
Nep1 and Nep2 can deneddylate cullins in vitro
We next determined whether Nep1 and Nep2 are capable of removing Nedd8 from neddylated cullins. To achieve this, we first introduced Myc-tagged or TAP-tagged versions of the cullin genes pcu1, pcu3 and pcu4 into nep1.d, nep2.d and the nep1.d nep2.d double mutant backgrounds. The tagged cullins were subsequently immunoprecipitated from the different strains and the precipitated material was used to assay the deneddylating activity of recombinant Nep1 and Nep2. The cullins are present predominantly in a slower migrating form in all the strains used in the assay (lanes 1, 2, 4 and 6 of Figures 3B and 3C and lanes 1, 4 and 8 of Figure 3D). Addition of Nep1 (60 ng/μl) to immuneprecipitates of Myc-tagged Pcu1 derived from nep1.d cells (Figure 3B, lane 3) or double mutant nep1.d nep2.d cells (lane 7) results in the conversion of most of the slower migrating species into the faster migrating species, consistent with deneddylation of Pcu1. In contrast, the addition of Nep2 (40 ng/μl) to immuneprecipitates derived from nep2.d cells (lane 5) or nep1.d nep2.d double mutant cells (lane 8) has no effect on the higher molecular mass species. Similar results were obtained by adding Nep1 or Nep2 to immuneprecipitates containing Myc-tagged Pcu3 (Figure 3C) and TAP-tagged Pcu4 (Figure 3D).
Since Nep1 has identity with previously characterized cysteine proteases that are inhibited by the thiol reagent NEM (N-ethylmaleimide), we investigated whether the same agent would inhibit the activity observed. Figure 3(D) indicates that the deneddylating activity of Nep1 (observed in lane 5) is inhibited by 100 mM NEM (lane 7), but not by 100 mM EDTA (lane 6).
Neddylated cullins do not accumulate in nep1.d or nep2.d mutants
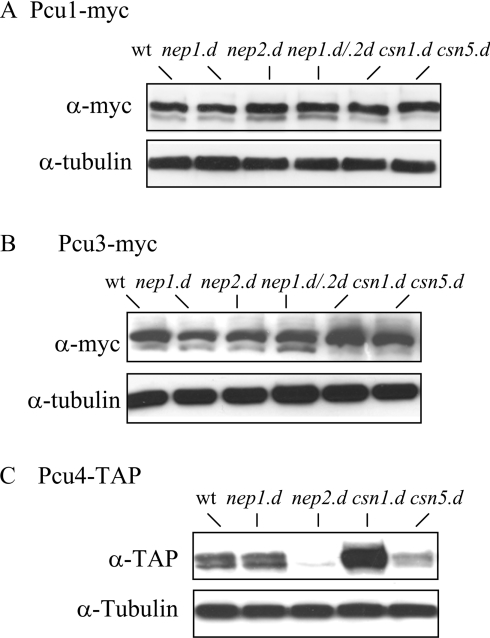
Since Nep1 is capable of deneddylating cullins in vitro, we next investigated whether nep1.d cells accumulate neddylated forms of the cullins in vivo. Total cell extracts were prepared from nep+, nep1.d, nep2.d and double-mutant nep1.d nep2.d strains that also harboured either a Myc- or TAP-tagged cullin. Extracts were prepared by the trichloroacetic acid extraction method to prevent potential deconjugation during extract preparation and these were subjected to Western-blot analysis using either anti-Myc or rabbit anti-mouse IgG antisera as appropriate. Figure 4(A) indicates that the levels of modified Pcu1 are very similar in nep+, nep1.d, nep2.d and the nep1.d nep2.d strains. As demonstrated previously [16], somewhat decreased levels of unmodified forms of Pcu1 were observed in extracts prepared from csn1.d and csn5.d cells. Similar results were obtained for Pcu3 in extracts derived from the nep1.d, nep2.d and nep1.d nep2.d strains (Figure 4B). Again, this contrasts with the absence of the unmodified form of Pcu3 seen in csn1.d and csn5.d [27]. We also analysed the modification status of Pcu4. Previously, it has been reported that Pcu4 is mostly Nedd8-modified in csn1.d and csn2.d mutant backgrounds, but that deletion of csn3, csn4 and csn5 does not affect the profile of neddylation when compared with csn+ cells [13]. Thus it was possible that the Csn5-dependent protease is not responsible for deneddylation of Pcu4 and that Nep1 and/or Nep2 may be suitable candidates. However, when we analysed Pcu4 modification levels in the relevant extracts, the results indicated that nep1.d cells contain the same ratio of modified to unmodified Pcu4 as that seen in nep+ and csn5.d cells. Intriguingly, Pcu4–TAP is very unstable in the nep2.d strain, precluding analysis of its Nedd8 modification status. As observed previously [13], Pcu4 appeared to be more abundant and was present mainly in the Nedd8-modified form in the csn1.d cell extract.
Figure 4. Deletion of nep1 and nep2 does not affect the neddylation of cullins in vivo.
Total cell extracts were prepared from strains containing pcu1-myc (A), pcu3-myc (B) and pcu4-TAP (C) and analysed by Western blotting with anti-Myc antibodies (A, B), anti-mouse IgG (C) or anti-tubulin antibodies (as control). wt, wild type.
Nep1 is present in a high molecular mass complex
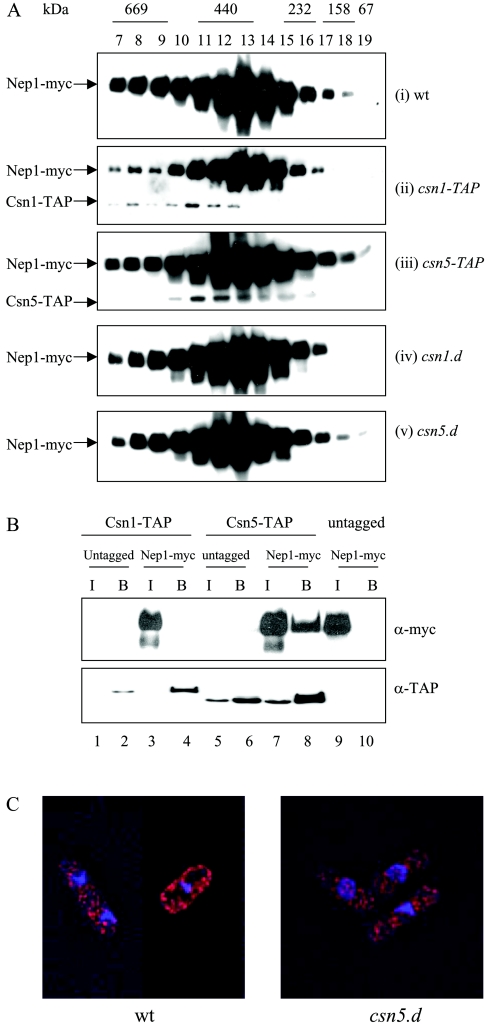
As discussed above, the purified signalosome complex has been reported to have deneddylating activity [16], which is dependent on the JAMM motif of the Csn5 subunit [17]. We were therefore interested in determining whether Nep1 is associated with the signalosome. Gel filtration using a strain containing Myc-tagged nep1 as the sole copy of the nep1 gene indicates that Nep1 is present in a high molecular mass complex, of mass in the range 200–400 kDa (Figure 5A), with maximal levels in fraction 13. Although the migration of Nep1 is similar to that of the signalosome complex ([15,27] and Figure 5A, ii), it is not completely coincident with it. In our experiments, peak levels of Csn1–TAP are observed at a mass corresponding to approx. 500 kDa (in fractions 10–13), similar to that observed by Zhou et al. [15] and Mundt et al. [27]. As observed by Mundt et al. [27], we also see that a subpopulation of Csn5 is present in lower molecular mass fractions (fractions 14–16), corresponding to a subcomplex that is independent of Csn1 and Csn2 [27].
Figure 5. Nep1 is present in a high molecular mass complex.
(A) Gel filtration of extracts of strains [(i)–(v)] using a Superdex 200HR 10/30 column; 200 μl of each fraction was analysed by SDS/PAGE and Western blotted using anti-Myc antisera. (B) Nep1 coimmunoprecipitates with Csn5 but not with Csn1. TAP-tagged proteins were purified with IgG-coupled magnetic beads and analysed by Western blotting with anti-Myc antisera. I, input; and B, bound. (C) Nep1 localizes to the cytoplasm in wild-type (wt) and csn5.d cells. Cells containing Myc-tagged Nep1 were incubated with anti-Myc monoclonal antibodies followed by TRITC-conjugated goat anti-mouse antisera. Red, Myc (Nep1); blue, DAPI (4,6-diamidino-2-phenylindole) (DNA).
To confirm whether the Nep1 in fractions 10–12, which correspond to the mass of the signalosome, is dependent on the presence of the intact signalosome, we next investigated whether Nep1 is still present in these high molecular mass complexes in strains deleted for individual CSN subunits. Figure 5(A) shows that the mass distribution of the Myc–Nep1-containing complex is identical in csn1.d (iv), csn5.d (v) and wild-type cells (i). Whereas Csn5 is not required for the integrity of the signalosome complex, deletion of csn1 has been shown to disrupt the complex [27,33]. This indicates that the formation of the Nep1-containing complex(es) is independent of the signalosome complex.
Csn5 and Nep1 co-immunoprecipitate
To analyse further the relationship between Csn5 and Nep1, we also used immunoprecipitation to determine whether Csn5 and Nep1 interact. Precipitation of Csn5–TAP with IgG-coupled magnetic beads pulls down a fraction of the Nep1 present in whole cell extract (Figure 5B, lane 8, upper panel). This precipitation was dependent on the presence of TAP-tagged Csn5 (lane 10), demonstrating specificity. Interestingly, co-precipitation of Nep1 with Csn1–TAP was not observed (lane 4, upper panel). Since Csn5 is believed to be present in both a small ‘mini-signalosome’ complex and in the larger ‘holo-signalosome’ complex, and these are distinguished by the absence and presence of Csn1 respectively, these results suggest that a small fraction of the Nep1 is capable of interacting with Csn5 when it is present in the mini-signalosome, but not when it is present in the holo-signalosome.
The holo-signalosome has been shown to be mainly nuclear in both Schizo. pombe [27] and higher eukaryotic cells [34]. There are reports that the Csn5-containing mini-signalosome is largely present in the cytoplasm. We therefore examined the localization of Nep1. Figure 5(C) shows that Myc-tagged Nep1 is located predominantly in the cytoplasm. However, the localization of Nep1 is unchanged in a csn5.d strain.
DISCUSSION
Neddylation is one of the many post-translational modifications involving members of the Ubl family. Until recently, the only proteins that had been observed to be modified by Nedd8 were the cullins. Recently, p53 has also been shown to be neddylated [35]. In the present study, using Western-blot analysis of total cell extracts overexpressing HA-tagged Nedd8, we show that, in addition to the cullins, there are probably numerous proteins modified by neddylation in Schizo. pombe.
In the present study, we characterize a Schizo. pombe Nedd8 protease, Nep1. This protein has sequence identity with cysteine proteases, in particular to the human deneddylator Den1/NEDP1. Curiously, Schizo. pombe contains not just Nep1 but also a related sequence, Nep2. In contrast, mammals appear to have only one Nep1-related protein [20,36]. No obvious homologues of the NEDP1 subfamily of cysteine proteases are evident in S. cerevisiae.
We have demonstrated that Nep1 is capable of removing Nedd8 from cullins in vitro. By analogy with the human deneddylator, we would expect Nep1 also to have Nedd8 processing activity, at least in vitro. To date, we have not been able to detect significant processing activity using either a His-tagged or a GST-tagged version of Nep1. The reason for this is unknown. It may be that Nep1, while retaining deneddylating activity during the purification process, has lost the ability to process Nedd8. Alternatively, it may be that the related protein, Nep2, is involved in the processing of Nedd8. We could not produce sufficient recombinant Nep2 from E. coli to allow characterization of its activity and therefore we have not established either deneddylating or processing activity for this protein. However, since high molecular mass Nedd8-containing species are observed in nep1.d, nep2.d and nep1.d nep2.d mutants, it seems probable that processing of Nedd8 is occurring in these strains, since unprocessed Nedd8 cannot be conjugated with target proteins. This suggests that protein(s) other than Nep1 and Nep2 can process Nedd8 in vivo. However, such functions may be redundant with Nep1 and Nep2, and we cannot exclude these proteins as the main Nedd8 processors.
nep1 and nep2 are not essential for viability. The absence of a strong phenotype in the null mutants is consistent with there being other Nedd8 proteases in Schizo. pombe. Csn5 has been shown to be associated with deneddylating activity [16], although recombinant Csn5 itself has not been demonstrated to have deneddylating activity. The identity of these additional Nedd8 proteases remains to be determined.
The role of Nep2 remains elusive. The levels of the cullins, Pcu1, Pcu3 and Pcu4, are similar in wild-type, nep1.d, csn1.d and csn5.d strains. However, the level of Pcu4–TAP is low in nep2.d cells compared with that observed in wild-type and nep1.d cells. In contrast, Pcu4–TAP levels are increased in csn1.d cells. In addition, we were unable to create a TAP-tagged nep1.d nep2.d pcu4 strain. The reason for the altered levels of Pcu4–TAP in nep2.d and csn5.d genetic backgrounds is unknown. Although we have not excluded the possibility that the TAP tag may be interfering with the stability of Pcu4, it may be the case that Nep2 and Csn1 have roles in controlling the levels of Pcu4.
The Nep1 protein is present in a high molecular mass complex in Schizo. pombe. Formation of this complex is not dependent on an intact signalosome since the complex is still present in mutants defective in Csn subunits. In Schizo. pombe, Csn5 exists in two complexes of different masses [15]. The fact that Nep1 immunoprecipitates with Csn5, but not with Csn1 (most of which is present in the signalosome complex), suggests that Nep1 may be interacting with the fraction of Csn5 that is not associated with the holo-signalosome. This holo-signalosome-independent Csn5 has been observed in the cytoplasm. The localization of Nep1 to the cytoplasm would be consistent with its interaction with the non-nuclear fraction of Csn5.
Although Nep1 is capable of deneddylating cullins in vitro, this does not appear to be the primary role of the protein, since the modification status of the cullins is the same in wild-type cells and in cells deleted for nep1. This suggests that Nep1 may remove Nedd8 from proteins other than the cullins. The lethality of nep1.d when in combination with mad2.d suggests that Nep1 may have a role in deneddylating a microtubule- or kinetochore-associated protein(s). Since nep1.d cells are not sensitive to the microtubule inhibitor thiabendazole, the implication is that nep1.d cells are not defective in microtubule dynamics but may, for example, have decreased ability to establish sister chromatid cohesion. Alternatively, nep1.d may affect the function of spindle pole bodies. In this respect, it is interesting to note that neddylated Cul1 (in association with an SCF ubiquitin ligase) has been observed to localize to the centrosome [37]. Since there are no homologues of Nep1 and Nep2 in S. cerevisiae, it is tempting to speculate that the Nep1- and Nep2-dependent targets are proteins that are not present in S. cerevisiae or are regulated differently. Comparison of the predicted proteins in Schizo. pombe and S. cerevisiae indicates that approx. 16% of the predicted proteins in Schizo. pombe do not have homologues in S. cerevisiae [38]. Use of the nep1.d strain might facilitate identification of such protein(s) because of the accumulation of the modified forms in this strain.
Acknowledgments
This work was supported by a BBSRC project grant (85/G15586, to F.Z.W.). We thank T. Carr and C. Liu (University of Sussex) for helpful discussions and access to reagents.
References
- 1.Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature (London) 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 2.Stelter P., Ulrich H. D. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature (London) 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 3.Johnson E. S. Protein modification by sumo. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 4.Yeh E. T., Gong L., Kamitani T. Ubiquitin-like proteins: new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- 5.Malakhov M. P., Kim K. I., Malakhova O. A., Jacobs B. S., Borden E. C., Zhang D. E. High-throughput immunoblotting. Ubiquitin-like protein ISG15 modifies key regulators of signal transduction. J. Biol. Chem. 2003;278:16608–16613. doi: 10.1074/jbc.M208435200. [DOI] [PubMed] [Google Scholar]
- 6.Dittmar G. A., Wilkinson C. R., Jedrzejewski P. T., Finley D. Role of a ubiquitin-like modification in polarized morphogenesis. Science. 2002;295:2442–2446. doi: 10.1126/science.1069989. [DOI] [PubMed] [Google Scholar]
- 7.Bayer P., Arndt A., Metzger S., Mahajan R., Melchior F., Jaenicke R., Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 1998;280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 8.Malakhova O. A., Yan M., Malakhov M. P., Yuan Y., Ritchie K. J., Kim K. I., Peterson L. F., Shuai K., Zhang D. E. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 2003;17:455–460. doi: 10.1101/gad.1056303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luders J., Pyrowolakis G., Jentsch S. The ubiquitin-like protein HUB1 forms SDS-resistant complexes with cellular proteins in the absence of ATP. EMBO Rep. 2003;4:1169–1174. doi: 10.1038/sj.embor.7400025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osaka F., Kawasaki H., Aida N., Saeki M., Chiba T., Kawashima S., Tanaka K., Kato S. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deshaies R. J. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- 12.Geyer R., Wee S., Anderson S., Yates J., Wolf D. A. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol. Cell. 2003;12:783–790. doi: 10.1016/s1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
- 13.Liu C., Powell K. A., Mundt K., Wu L., Carr A. M., Caspari T. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint-dependent and -independent mechanisms. Genes Dev. 2003;17:1130–1140. doi: 10.1101/gad.1090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osaka F., Saeki M., Katayama S., Aida N., Toh E. A., Kominami K., Toda T., Suzuki T., Chiba T., Tanaka K., et al. Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 2000;19:3475–3484. doi: 10.1093/emboj/19.13.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou C., Seibert V., Geyer R., Rhee E., Lyapina S., Cope G., Deshaies R. J., Wolf D. A. The fission yeast COP9/signalosome is involved in cullin modification by ubiquitin-related Ned8p. BMC Biochem. 2001;2:7. doi: 10.1186/1471-2091-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyapina S., Cope G., Shevchenko A., Serino G., Tsuge T., Zhou C., Wolf D. A., Wei N., Deshaies R. J. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- 17.Cope G. A., Suh G. S., Aravind L., Schwarz S. E., Zipursky S. L., Koonin E. V., Deshaies R. J. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 18.Wei N., Deng X. W. The role of the COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 1996;112:871–878. doi: 10.1104/pp.112.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seeger M., Kraft R., Ferrell K., Bech-Otschir D., Dumdey R., Schade R., Gordon C., Naumann M., Dubiel W. A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J. 1998;12:469–478. [PubMed] [Google Scholar]
- 20.Mendoza H. M., Shen L. N., Botting C., Lewis A., Chen J., Ink B., Hay R. T. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J. Biol. Chem. 2003;278:25637–25643. doi: 10.1074/jbc.M212948200. [DOI] [PubMed] [Google Scholar]
- 21.Wu K., Yamoah K., Dolios G., Gan-Erdene T., Tan P., Chen A., Lee C. G., Wei N., Wilkinson K. D., Wang R., et al. DEN1 is a dual function protease capable of processing the C-terminus of Nedd8 deconjugating hyper-neddylated CUL1. J. Biol. Chem. 2003;278:28882–28891. doi: 10.1074/jbc.M302888200. [DOI] [PubMed] [Google Scholar]
- 22.Rawlings N. D., Barrett A. J. MEROPS: the peptidase database. Nucleic Acids Res. 2000;28:323–325. doi: 10.1093/nar/28.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor D. L., Ho J. C., Oliver A., Watts F. Z. Cell-cycle-dependent localisation of Ulp1, a Schizosaccharomyces pombe Pmt3 (SUMO)-specific protease. J. Cell Sci. 2002;115:1113–1122. doi: 10.1242/jcs.115.6.1113. [DOI] [PubMed] [Google Scholar]
- 24.Bahler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., III, Steever A. B., Wach A., Philippsen P., Pringle J. R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 25.Bentley N. J., Holtzman D. A., Flaggs G., Keegan K. S., DeMaggio A., Ford J. C., Hoekstra M., Carr A. M. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- 26.Mundt K. E., Porte J., Murray J. M., Brikos C., Christensen P. U., Caspari T., Hagan I. M., Millar J. B., Simanis V., Hofmann K., et al. The COP9/signalosome complex is conserved in fission yeast and has a role in S phase. Curr. Biol. 1999;9:1427–1430. doi: 10.1016/s0960-9822(00)80091-3. [DOI] [PubMed] [Google Scholar]
- 27.Mundt K. E., Liu C., Carr A. M. Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol. Biol. Cell. 2002;13:493–502. doi: 10.1091/mbc.01-10-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He X., Patterson T. E., Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou C., Wee S., Rhee E., Naumann M., Dubiel W., Wolf D. A. Fission yeast COP9/signalosome suppresses cullin activity through recruitment of the deubiquitylating enzyme Ubp12p. Mol. Cell. 2003;11:927–938. doi: 10.1016/s1097-2765(03)00136-9. [DOI] [PubMed] [Google Scholar]
- 30.Ho J. C., Warr N. J., Shimizu H., Watts F. Z. SUMO modification of Rad22, the Schizosaccharomyces pombe homologue of the recombination protein Rad52. Nucleic Acids Res. 2001;29:4179–4186. doi: 10.1093/nar/29.20.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watts F. Z. SUMO Proteases. In: Wilson V. G., editor. Sumoylation: Molecular Biology and Biochemistry. Wymondham, U.K.: Horizon Bioscience; 2004. pp. 113–130. [Google Scholar]
- 32.Caspari T., Carr A. M. DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochimie. 1999;81:173–181. doi: 10.1016/s0300-9084(99)80050-9. [DOI] [PubMed] [Google Scholar]
- 33.Wang X., Kang D., Feng S., Serino G., Schwechheimer C., Wei N. CSN1 N-terminal-dependent activity is required for Arabidopsis development but not for Rub1/Nedd8 deconjugation of cullins: a structure-function study of CSN1 subunit of COP9 signalosome. Mol. Biol. Cell. 2002;13:646–655. doi: 10.1091/mbc.01-08-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaefer L., Beermann M. L., Miller J. B. Coding sequence, genomic organization, chromosomal localization, and expression pattern of the signalosome component Cops2: the mouse homologue of Drosophila alien. Genomics. 1999;56:310–316. doi: 10.1006/geno.1998.5728. [DOI] [PubMed] [Google Scholar]
- 35.Xirodimas D. P., Saville M. K., Bourdon J. C., Hay R. T., Lane D. P. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell (Cambridge, Mass.) 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 36.Gan-Erdene T., Kolli N., Yin L., Wu K., Pan Z. Q., Wilkinson K. D. Identification and characterization of DEN1, a deneddylase of the ULP family. J. Biol. Chem. 2003;19:19. doi: 10.1074/jbc.M302890200. [DOI] [PubMed] [Google Scholar]
- 37.Freed E., Lacey K. R., Huie P., Lyapina S. A., Deshaies R. J., Stearns T., Jackson P. K. Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes Dev. 1999;13:2242–2257. doi: 10.1101/gad.13.17.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood V., Gwilliam R., Rajandream M. A., Lyne M., Lyne R., Stewart A., Sgouros J., Peat N., Hayles J., Baker S., et al. The genome sequence of Schizosaccharomyces pombe. Nature (London) 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]