Abstract
For neuronal migration to occur, the cell must undergo morphological changes that require modifications of the cytoskeleton. Several different MAPs (microtubule-associated proteins) or actin-binding proteins are proposed to be involved in the migration of neurons. Therefore we have specifically analysed how two members of the MAP family, MAP1B and LIS1 (lissencephaly-related protein 1), interact with one another and participate in neuronal migration. Our results indicate that, in hippocampal neurons, MAP1B and LIS1 co-localize, associate and interact with each another. The interaction between these two MAPs is regulated by the phosphorylation of MAP1B. Furthermore, this interaction interferes with the association between LIS1 and the microtubule-dependent molecular motor, dynein. Clearly, the differential binding of these cytoskeletal proteins could regulate the functions attributed to the LIS1–dynein complex, including those related to extension of the neural processes necessary for neuronal migration.
Keywords: dynein, lissencephaly-related protein 1 (LIS1), microtubule-associated protein 1B (MAP1B), microtubule, migration, transgenic mouse
Abbreviations: DCX, doublecortin; E18, embryonic day 18; GSK, glycogen synthase kinase; GST, glutathione S-transferase; IC, intermediate chain; KO, knockout; LC, light chain; LIC, light IC; LIS1, lissencephaly-related protein 1; MAP, microtubule-associated protein; OA, okadaic acid; SBH, subcortical band heterotopia; WT, wild-type
INTRODUCTION
MAP1B (microtubule-associated 1B protein) is the first MAP to be expressed during the development of the nervous system [1]. Since the expression of this protein is down-regulated during brain development, MAP1B may play an important role during the morphological differentiation of neurons [2,3]. The results of studies performed both in the fruitfly [4] and in different loss-of-function mouse models [5–8] suggest that MAP1B is involved in axonal growth. Indeed, in one such mouse model, the mice die at a perinatal stage [5] and the structural abnormalities observed in all the laminated brain structures in these mice suggest that MAP1B could play a role in neuronal migration and axonal guidance [5,9]. Furthermore, the axons of the central nervous system and peripheral nervous system neurons derived from a mutant MAP1B mouse were shorter than WT (wild-type) neurons in culture [10,11].
The biochemical properties and subcellular distribution of MAP1B are modified by protein phosphorylation [12–14]. However, little is known about the functional consequences of MAP1B phosphorylation in neural function.
MAP1B is not the only MAP believed to be involved in neuronal migration. Previously, two other MAPs, DCX (doublecortin) [15] and LIS1 (lissencephaly-related protein 1) [16,17], were implicated in neuronal migration. Similarly, deficits in these proteins provoke abnormalities in the migration of neurons into the embryonic cortex, which, in extreme cases, lead to the loss of the normal convolutions of the cortex in humans, a disorder known as lissencephaly (‘smooth brain’).
Lissencephaly is a severe abnormality of neuronal migration characterized by the absence of (agyria) or a decrease in (pachygyria) the convolutions, resulting in a smooth cerebral cortex [18]. Furthermore, SBH (subcortical band heterotopia) is a related disorder in which there are bilateral bands of grey matter interposed in the white matter, between the cortex and the lateral ventricles [19]. SBH is very common among females with mutations in DCX [15,20]. Indeed, lissencephaly and SBH have been observed in different regions of the same brain, indicating a wide spectrum of ‘agyria–pachygyria band’.
LIS1 co-immunoprecipitates with the protein dynein, with which it co-localizes in tissue culture and in the brain [21–23]. In fact, LIS1 interacts with three distinct subunits of the dynein complex [24]. Overexpression of LIS1 [21–23], as well as impairing its expression [23,25], interferes with dynein function. In addition, LIS1 appears to be a regulatory subunit of PAF-AH (plateletactivating factor acetylhydrolase) 1b [15,26,26a,27]. Cytoplasmic dynein is composed of two heavy chains, two or more ICs (intermediate chains, with a molecular mass of ∼74 kDa), some LICs (light ICs, with a molecular mass of ∼55 kDa) [28,29] and several LCs (light chains). These LCs include two copies of the Tctex1/rp3 LC and a dimer of the highly conserved LC8 protein [24,30,31]. Furthermore, dynein is capable of forming a complex with dynactin subunits with which LIS1 can also interact [32].
Given that both LIS1 and MAP1B are MAPs that play an important role in neuronal migration, it is possible that they interact with one another. We show that this is indeed the case, and in addition to examining this novel interaction between MAP1B and LIS1, we have documented some of the functional consequences with respect to intracellular localization of the Golgi apparatus [33].
MATERIALS AND METHODS
Animals
Generation, using the gene-trapping approach, of the map1b mutant used here has been reported previously [34].
Cell cultures
The mouse neuroblastoma cell line N2A was grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum, 2 mM glutamine and antibiotics, and incubated in a humidified atmosphere with 7% CO2.
Hippocampal neurons from E18 (embryonic day 18) embryos were dissected in Hanks balanced salt solution without calcium and magnesium (Invitrogen); after treatment with trypsin for 20 min, the cells were gently dissociated, counted and then seeded in dishes coated with 1 mg/ml poly(L-lysine) and 20 μg/ml laminin. After culturing in the Neurobasal medium (Invitrogen) containing 10% (v/v) horse serum for 3 h, the medium was removed and Neurobasal medium containing N2 (Invitrogen) was added. After 18 h, the cultured cells were fixed in 4% (w/v) paraformaldehyde and 4% (w/v) sucrose for further processing.
To analyse which kinases might be involved in mode I MAP1B phosphorylation, the GSK3 (glycogen synthase kinase 3) inhibitor LiCl (10 mM) or a cdk5 inhibitor roscovitin (200 nM) was added to the cultures. In other experiments, the phosphatase inhibitor OA (okadaic acid; 1 μM; mainly inhibiting protein phosphatase 2A) was added to block MAP1B dephosphorylation.
Cell transfection
N2A neuroblastoma cells were transfected with a GSK3 cDNA [35,36] containing a c-Myc peptide tag. Cells were transfected with the Lipofectamine™ reagent (Invitrogen).
Immunostaining
Cells grown on coverslips were incubated with PBS/0.1% Triton X-100 for 5 min and then with PBS/5% (w/v) BSA for 1 h. Subsequently, primary antibodies raised against the following proteins were diluted in PBS/1% BSA and used for labelling the cells: for LIS1, H-300 antibody diluted 1/100 (Santa Cruz Biotechnology); for dynein, DIC antibody 1/20 (Sigma); for MAP1B, N19 antibody 1/20 (Santa Cruz Biotechnology), SMI31 antibody 1/100 (Sternberger Monoclonals; [36,37]) and 125 antibody 1/20 [37] and, as a Golgi marker, LL-FITC antibody 1/2 (Sigma). The secondary antibodies used were: anti-mouse–Oregon Green 1/200 (Molecular Probes), anti-goat–Texas Red 1/400 (Molecular Probes) and anti-rabbit–Texas Red 1/400 (Molecular Probes).
All images were captured by either light or confocal laser-scanning microscopy (Zeiss, Cologne, Germany).
Western-blot analysis
Protein samples were separated on 10% (v/v) acrylamide gel for dynein, LIS1 and actin and on 6% acrylamide gel for MAP1B. After electrophoresis, the fractionated proteins were transferred on to nitrocellulose membranes as indicated previously [5]. The filters were saturated in a solution containing 5% (w/v) powdered milk in PBS and incubated with the following antibodies: SMI31 antibody 1/1000, 125 antibody 1/20, N19 antibody 1/1000, antiactin antibody 1/1000 (Sigma), anti-LIS1 antibody 1/1000 and anti-dynein 1/2000. Secondary antibodies (Gibco BRL, Gaithersburg, MD, U.S.A.) were used at a concentration of 1/2000 and the labelling was visualized using ECL® (PerkinElmer, Norwalk, CT, U.S.A.).
Polymerization of microtubules
Microtubules were isolated by the method of Shelanski et al. [38] from the brains of WT or mutant E18 mouse embryos. The isolated microtubule proteins were characterized by gel electrophoresis.
Immunoprecipitation
A mouse brain (E18) was homogenized in 0.7 ml of cold immunoprecipitation buffer (1% Triton X-100, 150 mM NaCl, 10 mM Tris, pH 7.4, 1 mM EDTA, 1 mM EGTA, pH 8.0, 0.2 mM sodium orthovanadate, 0.2 mM PMSF and 0.5% Nonidet P40). The homogenate was centrifuged at 16000 g for 15 min at 4 °C and the supernatant was considered as the total cell lysate.
To 400 μg of the supernatant, 5 μg of a specific antibody was added in a final volume of 1 ml. The solution was vortex-mixed and incubated for another 1 h at 4 °C. Subsequently, 10 μg of rabbit anti-mouse IgG was added and the mixture was incubated for a further 30 min at 4 °C. Then, 20 μl of 50% Protein A–agarose bead solution was added, mixed and incubated with agitation for 30 min at 4 °C. The beads were pelleted by centrifugation at 16000 g for 15 min at 4 °C, and the supernatant was removed. The pellet was washed twice with the immunoprecipitation buffer and resuspended in 30 μl of 2× concentrated electrophoresis sample buffer [250 mM Tris, pH 6.8, 4% (w/v) SDS, 10% glycerol, 0.006% Bromophenol Blue and 2% (w/v) 2-mercaptoethanol]. The proteins were separated by gel electrophoresis, and the fractionated proteins were then characterized by Western-blot analysis.
Pull-down assay
DNAs encoding GST (glutathione S-transferase) and GST–LIS1 recombinant proteins were inserted into the pGEX-4T plasmid (Amersham Biosciences) and the proteins were expressed in Escherichia coli BL21 cells. Transformed cells were cultured in SOC medium (20 g/l Bacto Tryptone, 5 g/l Bacto yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4 and 20 mM glucose) and recombinant protein expression was induced by the addition of 0.1 mM isopropyl β-D-thiogalactoside for 2 h at 30 °C. The cell lysates were centrifuged and the supernatant was then incubated with glutathione–Sepharose 4B beads (Amersham Biosciences) for 2 h at room temperature (22 °C). This mixture was then incubated with brain extracts prepared in a buffer containing 20 mM Tris/HCl (pH 6.8), 100 mM NaCl, 1 mM EDTA, 0.1% Triton X-100 and protease inhibitors. Afterwards, the proteins bound to glutathione–Sepharose beads were washed four times and then resuspended in 2× electrophoresis sample buffer, boiled and separated by gel electrophoresis.
Cross-linking assays
Protein (200 μg) from soluble E18 brain extracts was incubated with increasing concentrations of formaldehyde (0, 0.01% and 0.1%) for 30 min at room temperature. The reaction was stopped by adding 1% glycine and the electrophoresis sample buffer. The mixture was boiled for 5 min at 90 °C, and the proteins were characterized by electrophoresis and Western blotting.
RESULTS
LIS1 interacts with MAP1B
Given the important role of LIS1 in the development of the brain, we were interested in testing the hypothesis that the interaction of LIS1 with other MAPs can affect neuronal migration. Therefore we initially focused our attention on the possible interaction between LIS1 and MAP1B. To address this, we analysed the relationship between these proteins using a MAP1B-deficient transgenic mouse, performing our experiments during the late-prenatal stage of the development of the nervous system (E17–E18).
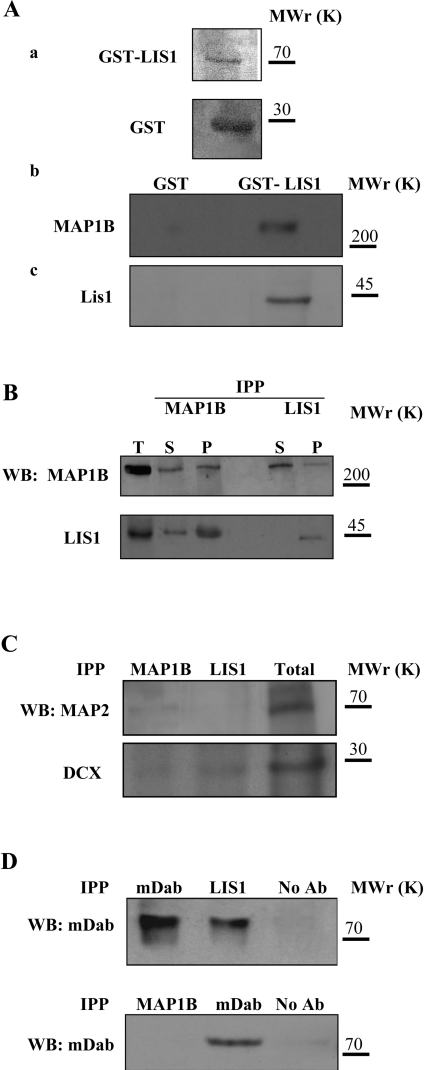
The possibility that LIS1 and MAP1B might interact was examined using several biochemical assays. Thus we found that GST–LIS1 was capable of pulling down MAP1B from an extract isolated from E18 mouse brains, but no proteins associated with GST alone (Figure 1A). These results were further confirmed in co-immunoprecipitation assays, where antibodies raised against MAP1B but not the preimmune serum were capable of coimmunoprecipitating LIS1 as well as MAP1B itself (Figure 1B, upper panel). Likewise, antibodies against LIS1 specifically co-precipitated MAP1B (Figure 1B, lower panel). The specificity of this interaction was demonstrated by the failure to detect an interaction between MAP1B and other neuronal MAPs, such as MAP2 and DCX (Figure 1C). DCX and phosphorylated mDab1 did co-precipitate with LIS1, as described elsewhere (Figures 1C and 1D; [39,40]); however, MAP1B did not appear to interact with mDab1 (Figure 1D). These results indicate that MAP1B interacts specifically with LIS1.
Figure 1. MAP1B interacts with LIS1.
(A) Proteins from brain extracts were pulled down using GST–LIS1. (a) GST and GST–LIS1 were expressed and characterized (see the Materials and methods section). (b) MAP1B is specifically pulled-down with GST–LIS1; (c) the control of LIS1 is shown. The electrophoretic mobilities of the 200, 70, 45 and 30 kDa markers are indicated. (B) MAP1B and LIS1 proteins co-immunoprecipitate. Upper panel: immunoprecipitation experiments performed with antibodies raised against MAP1B (IPP MAP1B) or LIS1 (IPP LIS1), and proteins were visualized by Western blot (WB) with anti-MAP1B antibody. Lower panel: a similar experiment was conducted but the proteins were visualized with an anti-LIS1 antibody. The total protein (T) before immunoprecipitation and the immunoprecipitated (P) and non-immunoprecipitated (S) proteins are indicated. (C) To analyse the specificity of the interaction described in (A, B), a pellet fraction obtained after immunoprecipitation with MAP1B and LIS1 antibodies was revealed using antibodies against MAP2 and DCX. (D) The interaction between LIS1 and MAP1B is also specific when compared with mDab, a protein involved in the reelin signalling pathway. Molecular mass (kDa) is indicated on the right.
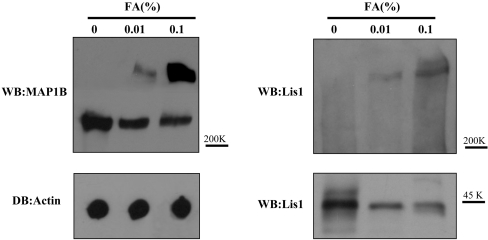
To determine whether this interaction between LIS1 and MAP1B was direct or whether it required another molecule, we performed cross-linking experiments (Figure 2). In the presence of formaldehyde, a protein complex with electrophoretic mobility lower than that of MAP1B was observed. This protein complex was recognized by antibodies raised against MAP1B and LIS1, indicating that both proteins are present in the complex and suggesting that the complex is formed by the direct binding between the two proteins.
Figure 2. Cross-linking of MAP1B with LIS1.
Protein from a mouse brain extract was incubated for 30 min at room temperature in the presence or absence of formaldehyde (FA). After this incubation, the protein was separated by gel electrophoresis and characterized by Western blotting (WB) using an antibody raised against MAP1B or LIS1. Additionally, the amount of actin in each sample was determined by dot blot (DB). The electrophoretic mobilities for MAP1B and LIS1 as well as the 200 and 40 kDa markers are indicated.
LIS1–MAP1B interaction is regulated by MAP1B phosphorylation
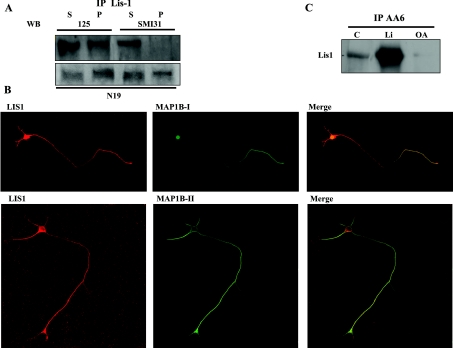
MAP1B is a phosphoprotein [14] and this transient post-translational modification plays an important role in its regulation. Therefore we tested whether MAP1B phosphorylation influences its interaction with LIS1. MAP1B phosphorylation can be classified into two modes. Mode I phosphorylation involves proline-directed protein kinases, whereas mode II phosphorylation is dependent on the kinase CK2 [12,14,37]. These different modes of phosphorylation can be distinguished by the use of two different phospho-specific antibodies (SMI31 and 125 antibodies). Mode I phosphorylation occurs mainly due to modification of MAP1B by GSK3 [13,14] and it can be detected by the reaction of phospho-MAP1B with the antibody SMI31. Mode II phosphorylation can be detected using the antibody 125 [14,41].
When proteins were immunoprecipitated with an antibody raised against LIS1, only mode II-phosphorylated MAP1B was seen to associate with LIS1 (Figure 3A). These results were in sharp contrast with those observed when the N19 antibody, which does not recognize phosphorylated MAP1B, was used. These results were further substantiated by immunocytochemistry. A high degree of co-localization was observed between mode II-phosphorylated MAP1B and LIS1 (Figure 3B), whereas mode I-phosphorylated MAP1B and LIS1 only weakly co-localized (Figure 3B). Whereas mode I phospho-MAP1B is mainly located in the distal axonal region, mode II phospho-MAP1B (that interacts with LIS1) is found in the proximal axonal and somatodendritic regions (Figure 3B). The results of these experiments strongly suggest that MAP1B and LIS1 interact and that this interaction is disturbed by mode I MAP1B phosphorylation.
Figure 3. The interaction between MAP1B and LIS1 is regulated by MAP1B phosphorylation.
(A) Immunoprecipitation of brain extracts with LIS1 was performed using specific antibodies and the phosphorylated MAP1B was analysed. The MAP1B recovered in the pellet fraction was rich in mode II-phosphorylated MAP1B, whereas mode I-phosphorylated MAP1B was recovered mostly in the supernatant fraction. (B) Double immunofluorescence of hippocampal neurons in culture, showing the distribution of LIS1 and mode I-phosphorylated MAP1B or mode II-phosphorylated MAP1B. (C) Immunoprecipitation with A6 (an antibody that recognizes MAP1B irrespective of its level of phosphorylation). The precipitated proteins were fractionated by gel electrophoresis and visualized with an antibody raised against LIS1. The immunoprecipitated proteins from cultured neurons incubated in the presence of lithium (a specific inhibitor of mode I MAP1B phophorylation) or in the presence of OA (which prevents the dephosphorylation of MAP1B phosphorylated by mode I) are shown, together with those from untreated cells (control, C).
In support of this conclusion, we found that the presence of lithium, an inhibitor of GSK3 and mode I MAP1B phosphorylations, augmented the association between MAP1B and LIS1 observed by immunoprecipitation (Figure 3C). However, no differences were detected after treatment with roscovitin, an inhibitor of another proline-directed kinase, cdk5 (results not shown). Furthermore, the interaction between MAP1B and LIS1 was impaired in the presence of OA that blocks MAP1B mode I dephosphorylation (Figure 3C).
To investigate further how mode I phosphorylation of MAP1B may affect its binding to LIS1, we augmented the phosphorylation of MAP1B by expressing GSK3 in the neuroblastoma N2A cells containing MAP1B and LIS1, and transfecting these cells with GSK3β cDNA. A high proportion of N2A cells were transfected, as seen by the expression of the Myc label co-expressed with GSK3β; upon GSK3 expression, mode I phosphorylation of MAP1B increased (Figures 4A and 4B). As a consequence of MAP1B hyperphosphorylation, less phospho-MAP1B co-immunoprecipitated with LIS1 (Figure 4C).
Figure 4. GSK3 regulates the interaction of MAP1B with LIS1.
(A) N2A neuroblastoma cells were transfected with DNA encoding GSK3β bearing a c-Myc peptide tag. The amounts of LIS1, MAP1B, c-Myc, total GSK3, mode I-phosphorylated MAP1B (MAP1B-P) and actin were measured in untransfected (C) and GSK3-transfected cells. (B) The proportion of transfected cells was measured using the reaction with an antibody raised against the c-Myc peptide. (C) A protein extract from untransfected cells (C) or transfected (GSK3) cells was immunoprecipitated (IPP) with an antibody raised against LIS1, and the presence of MAP1B or LIS1 in the immunoprecipitated (P) or soluble (S) fraction was determined by Western blotting.
Dynein does not bind to MAP1B
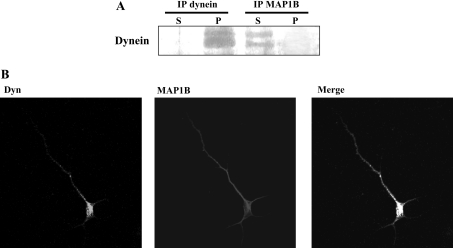
LIS1 has been shown to interact with the cytoplasmic motor protein dynein [21,23,25]. It is known that the dynein–LIS1 connection is conserved in yeast, fungi, flies and mammals, and hence it is possible that dynein interacts with MAP1B. Dynein was visualized with a serum that recognizes its LICs, but these two dynein proteins did not co-immunoprecipitate with MAP1B antibodies (Figure 5A). In addition, immunocytochemistry demonstrated that the two proteins did not co-localize. Staining for dynein revealed a punctate pattern proximal to the soma with no significant overlap with MAP1B staining (Figure 5B). These results suggest that MAP1B does not bind to dynein.
Figure 5. Dynein does not bind to MAP1B.
(A) Protein from mouse brain extracts was immunoprecipitated (IP) with antibodies raised against LIC dynein or MAP1B, and then visualized with an anti-LIC dynein antibody. (B) Double immunofluorescence analysed by confocal microscopy, showing the distribution of dynein, of mode II-phosphorylated MAP1B and the merged image.
Absence of MAP1B can modify the LIS1–dynein interaction
Since LIS1 interacts with both MAP1B and dynein [32], we have tested whether decreasing the amount of MAP1B available will affect the interaction between LIS1 and dynein. The overall levels of LIS1 or dynein in the presence or absence of MAP1B were initially established by comparing the amounts of protein in brain extracts obtained from WT and from MAP1B-deficient mice [KO (knockout) mice]. No differences were observed in the amount of LIS1 or dynein on comparing WT and KO mice (Figure 6A).
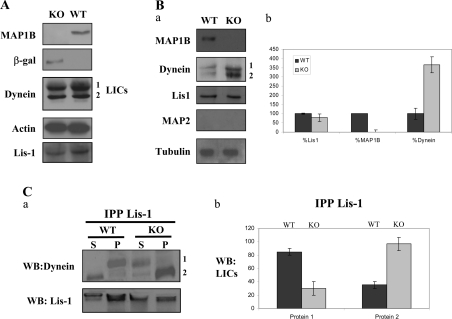
Figure 6. The absence of MAP1B does not affect the overall levels of dynein or LIS1 proteins, but results in an increase in LIS1–dynein interaction.
(A) Western-blot analyses of brain extracts derived from control (WT) and MAP1B-deficient (KO) mice are shown. Actin was tested as a loading control. (B, a) These experiments are similar to the pull-down experiments indicated in Figure 1(A) using GST–LIS1 recombinant protein mixed with brain extracts from control (WT) and from MAP1B-deficient (KO) animals. The first panel shows that MAP1B was recovered from the WT brain extracts after pull down with GST–LIS1, whereas no MAP1B was recovered from KO mice brain extracts. An increase in the amount of dynein subunits recovered after GST–LIS1 pull down was observed when the proteins were mixed with KO mice brain extracts. No MAP2 was recovered after pull down. Tubulin was used as an internal control to compare the protein concentration in the two brain extracts. (B, b) Quantification of the proteins recovered after pull-down experiments. (C, a) Co-immunoprecipitation experiments show that dynein LICs bind to LIS1 in the presence (WT) or absence (KO) of MAP1B. In the absence of MAP1B, a decrease of protein 1 was detected as well as an increase of protein 2, corresponding to the dynein LICs. (C, b) Quantification of the results by densitometry analysis.
To test whether the absence of MAP1B affects the association between LIS1 and dynein, GST–LIS1 was used to recover dynein from the brain cell extracts derived from mice lacking MAP1B [5]. The amounts of dynein recovered in this way were compared with that bound by GST–LIS1 in brain extracts from WT littermates. In the absence of MAP1B, more dynein was associated with LIS1 (Figure 6Ba), and when this increase was quantified, in the absence of MAP1B, the interaction between dynein and LIS1 was augmented by 3–4-fold (Figure 6Bb).
These results suggest that MAP1B and dynein proteins may compete for LIS1 and, to test this possibility, brain extracts from MAP1B-deficient or WT mice were immunoprecipitated with an antibody raised against LIS1. The proportion of dynein that co-immunoprecipitated with LIS1 was compared in control and MAP1B-deficient brains. As shown previously, in the absence of MAP1B, the association of LIC dynein with LIS1 increased. When we estimated the amount of individual LICs of dynein (1 and 2) that had interacted with LIS1, significant changes were observed in both proteins between control and MAP1B-deficient extracts (Figure 6C). Our results strongly suggest that MAP1B competes with dynein for LIS1 binding and that the presence of MAP1B could interfere with the subunit composition of the dynein complex.
Association of LIS1 with microtubules in the presence or absence of MAP1B
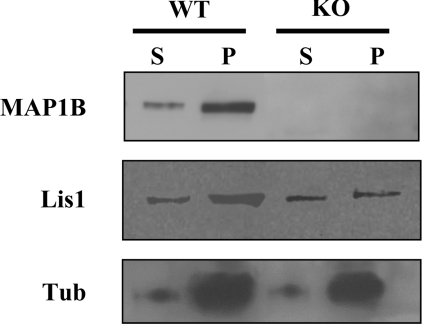
On the basis of these results, we examined the co-polymerization of LIS1 with microtubules in the presence or absence of MAP1B. We found that, in the presence of MAP1B, more LIS1 was bound to microtubules (Figure 7). This is highly suggestive that LIS1 can bind to microtubules not only through a direct association with tubulin, but also through its interaction with MAP1B.
Figure 7. Association of LIS1 with neuronal microtubules in the presence or absence of MAP1B.
Microtubule protein was polymerized in vitro and the association of LIS1 with the polymers was examined. The first panel shows that MAP1B was present in the microtubule fraction derived from control animals (WT) but was absent from those from mutant animals (KO). Furthermore, LIS1 was enriched in the pellet fraction after microtubule polymerization. The level of assembled tubulin is also shown. A decrease (30%) in the amount of LIS1 bound to microtubules was found in the KO-derived polymer compared with that found in microtubules from WT mice.
The lack of MAP1B may affect the function of the LIS1–dynein complex
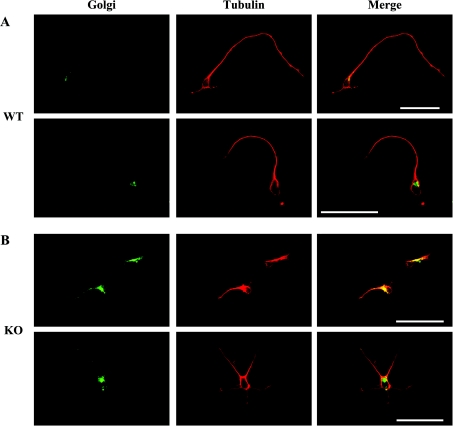
The localization and organization of the Golgi complex is dependent on microtubule assembly. In the absence of polymerized microtubules, the Golgi apparatus becomes dispersed [42] and it has been suggested [33] that the dynein–LIS1 complex could play an important role in maintaining the Golgi apparatus close to the cell nucleus [29]. Since the presence of MAP1B appears to affect the dynein–LIS1 complex, the localization of the Golgi apparatus was analysed in hippocampal neurons containing or lacking MAP1B. In WT neurons (Figure 8A), the Golgi apparatus is located close to the nucleus and the microtubule-organizing centre, whereas in neurons lacking MAP1B, the Golgi apparatus is much more dispersed (Figure 8B).
Figure 8. Golgi localization in neurons with or without MAP1B.
Hippocampal neurons at culture stage 2 (see [5]) from WT (A) or mutant MAP1B (B) mice were incubated with the antibody LL-FITC to localize the Golgi complex (see the Material and methods section). In neurons from WT mice, it is localized to a specific site close to the nucleus, whereas it appears to be dispersed in neurons lacking MAP1B. Scale bars, 50 μm.
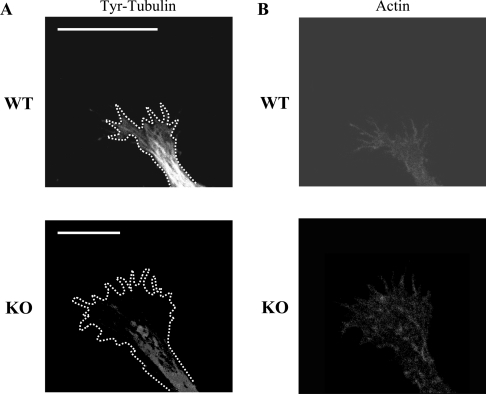
We also examined whether there were differences in the microtubule network in neurons from MAP1B–/– and WT mice. We focused our attention on the axonal growth cones where microtubules could be detected. Differences were indeed detected in the organization of the microtubule network between WT and MAP1B–/– neurons (Figure 9). The loss of MAP1B caused a decrease in the proportion of microtubules present in the peripheral growth cone region when compared with that observed in WT cells. This observation is also in accordance with previous results [10,11].
Figure 9. Microtubule network in growth cones in the presence or absence of MAP1B.
(A) Hippocampal neurons from WT or KO mice were cultured, and the presence of microtubules in their growth cones was studied by immunofluorescence analysis using an antibody raised against Tyr-tubulin. A difference in the amounts of peripheral microtubules was found. Scale bars, 5 μm. The outline of the growth cones is indicated in each case by white dots. (B) Same as (A), but the growth cones were analysed by immunofluorescence using phalloidin to identify their shape.
DISCUSSION
The interaction between MAP1B and LIS1 is dependent on phosphorylation
It has been shown previously that LIS1 interacts with the MAP CLIP170 [43], a MAP that binds mainly to the growing tips of microtubules [44]. In the present study, we have shown that LIS1 can also interact with another MAP, MAP1B. Interestingly, this interaction is negatively regulated by phosphorylation of MAP1B (mode I) and, in fact, this is the third interaction involving LIS1 that is sensitive to phosphorylation. Whereas the interaction between LIS1 and CLIP170 is sensitive to the status of LIS1 phosphorylation [43], the interaction between LIS1 and mDab1 is regulated by mDab1 phosphorylation [40]. We show here that the interaction between LIS1 and MAP1B is regulated by MAP1B phosphorylation and this could have important functional consequences, especially if we bear in mind that the protein reelin regulates mDab phosphorylation. Reelin plays a key role in cerebral cortex lamination, and signalling through this protein is responsible for the tyrosine phosphorylation of the adaptor protein mDab1 [40]. Thus, based on the results presented here, we suggest that reelin could influence the LIS1–dynein interaction through mDab1 phosphorylation and through MAP1B mode I phosphorylation that will intensify this interaction.
MAP1B–LIS1 versus dynein–LIS1
MAP1B could compete with dynein for binding to LIS1 since it is known that LIS1 can interact with dynein ICs in non-mammalian and mammalian cells [21,23]. We have now found that, upon binding to LIS1, MAP1B may modify the proportions of specific LIC dynein subunits bound to LIS1. We also found that these dynein proteins bind differently to LIS1 depending on whether MAP1B is present or not. On the other hand, LIS1 binding to microtubules also seems to be affected by the presence of MAP1B. In MAP1B-deficient cells, there is a shift in the proportion of LIS1 found in the soluble and pelleted fractions after microtubule polymerization. Therefore a consequence of the loss of MAP1B function will be to promote the LIS1–dynein interaction.
Cytoplasmic dynein is important for the organization of the Golgi apparatus [33] and for cell migration. In this context, modification of the neuronal cytoskeleton could have a key role in generating the correct movement of neurons [45].
MAP1B and dynein compete for LIS1
MAP1B is involved in neuronal outgrowth [10,30] and recently it has been reported that MAP1B could play a role in neuronal migration, depending on the presence of Netrin-1 [46] or that of reelin [14]. Therefore it is interesting that MAP1B binds to LIS1, another MAP involved in neuronal migration, as discussed above.
On the other hand, the novel LIS1–MAP1B interaction described in the present study, and specifically the competition between MAP1B and dynein to associate with LIS1, could be another of several control points at which LIS1 and/or dynein influences neuronal migration. This concept is reinforced by the fact that, in the absence of MAP1B, the LIS1–dynein interaction is altered. One explanation for such a behaviour could be related to the fact that the microtubule-binding domain of dynein is composed of at least three different sequences in the central domain of the protein. Two of these sequences are highly similar to the microtubule-binding domain of MAP1B [47]. Additionally, the peptides that displace MAP1B binding from microtubules are also capable of interfering with the dynein–microtubule interaction [47]. Taking into account all these results, we hypothesize that these proteins, MAP1B and dynein, compete for the same microtubule-binding site [47].
It will be of interest to determine the effects of depleting MAP1B on the function of the LIS1–dynein complex, since it could affect axonal transport. Future experiments will address the possible role of MAP1B in the impairment of axonal transport regulated by dynein.
Acknowledgments
This work was supported by grants from the Comisión Interministerial de Ciencia y Tecnología, MCyT, MSyC, F. Lilly and an institutional grant from the Fundación Ramon-Areces. O.R. is the Incumbent of the Berstein-Mason Professorial Chair of Neurochemistry and is supported by the March of Dimes (grant no. 6-FY01-5), Foundation Jerome Lejeune, Y. Leon Benozyio Institute for Molecular Medicine, the Kekst Center, and by the Leo and Julia Forchheimer Center for Molecular Genetics. The technical assistance of S. Soto-Largo and R. Cuadros and the advice of M. Pérez and C. Sánchez are acknowledged. We also thank B. Alarcon for providing the antibodies used for localizing the Golgi apparatus and A. Goffinet (Developmental Neurobiology Unit, University of Louvain Medical School, Belgium) for providing the antibodies raised against mDab1.
References
- 1.Tucker R. P., Garner C. C., Matus A. In situ localization of microtubule-associated protein mRNA in the developing and adult rat brain. Neuron. 1989;2:1245–1256. doi: 10.1016/0896-6273(89)90309-7. [DOI] [PubMed] [Google Scholar]
- 2.Diaz-Nido J., Avila J. Characterization of proteins immunologically related to brain microtubule-associated protein MAP-1B in non-neural cells. J. Cell Sci. 1989;92:607–620. doi: 10.1242/jcs.92.4.607. [DOI] [PubMed] [Google Scholar]
- 3.Schoenfeld T. A., McKerracher L., Obar R., Vallee R. B. MAP 1A and MAP 1B are structurally related microtubule associated proteins with distinct developmental patterns in the CNS. J. Neurosci. 1989;9:1712–1730. doi: 10.1523/JNEUROSCI.09-05-01712.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roos J., Hummel T., Ng N., Klambt C., Davis G. W. Drosophila Futsch regulates synaptic microtubule organization and is necessary for synaptic growth. Neuron. 2000;26:371–382. doi: 10.1016/s0896-6273(00)81170-8. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Billault C., Demandt E., Wandosell F., Torres M., Bonaldo P., Stoykova A., Chowdhury K., Gruss P., Avila J., Sanchez M. P. Perinatal lethality of microtubule-associated protein 1B-deficient mice expressing alternative isoforms of the protein at low levels. Mol. Cell. Neurosci. 2000;16:408–421. doi: 10.1006/mcne.2000.0880. [DOI] [PubMed] [Google Scholar]
- 6.Edelmann W., Zervas M., Costello P., Roback L., Fischer I., Hammarback J. A., Cowan N., Davies P., Wainer B., Kucherlapati R. Neuronal abnormalities in microtubule-associated protein 1B mutant mice. Proc. Natl. Acad. Sci. U.S.A. 1996;93:1270–1275. doi: 10.1073/pnas.93.3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meixner A., Wiche G., Propst F. Analysis of the mouse MAP1B gene identifies a highly conserved 4.3 kb 3′ untranslated region and provides evidence against the proposed structure of DBI-1 cDNA. Biochim. Biophys. Acta. 1999;1445:345–350. doi: 10.1016/s0167-4781(99)00062-7. [DOI] [PubMed] [Google Scholar]
- 8.Takei Y., Kondo S., Harada A., Inomata S., Noda T., Hirokawa N. Delayed development of nervous system in mice homozygous for disrupted microtubule-associated protein 1B (MAP1B) gene. J. Cell Biol. 1997;137:1615–1626. doi: 10.1083/jcb.137.7.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meixner A., Haverkamp S., Wassle H., Fuhrer S., Thalhammer J., Kropf N., Bittner R. E., Lassmann H., Wiche G., Propst F. MAP1B is required for axon guidance and is involved in the development of the central and peripheral nervous system. J. Cell Biol. 2000;151:1169–1178. doi: 10.1083/jcb.151.6.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Billault C., Avila J., Caceres A. Evidence for the role of MAP1B in axon formation. Mol. Biol. Cell. 2001;12:2087–2098. doi: 10.1091/mbc.12.7.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Billault C., Owen R., Gordon-Weeks P. R., Avila J. Microtubule-associated protein 1B is involved in the initial stages of axonogenesis in peripheral nervous system cultured neurons. Brain Res. 2002;943:56–67. doi: 10.1016/s0006-8993(02)02534-9. [DOI] [PubMed] [Google Scholar]
- 12.Ulloa L., Diaz-Nido J., Avila J. Depletion of casein kinase II by antisense oligonucleotide prevents neuritogenesis in neuroblastoma cells. EMBO J. 1993;12:1633–1640. doi: 10.1002/j.1460-2075.1993.tb05808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Perez J., Avila J., Diaz-Nido J. Implication of cyclin-dependent kinases and glycogen synthase kinase 3 in the phosphorylation of microtubule-associated protein 1B in developing neuronal cells. J. Neurosci. Res. 1998;52:445–452. doi: 10.1002/(SICI)1097-4547(19980515)52:4<445::AID-JNR8>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez-Billault C., Jimenez-Mateos E. M., Caceres A., Diaz-Nido J., Wandosell F., Avila J. Microtubule-associated protein 1B function during normal development, regeneration, and pathological conditions in the nervous system. J. Neurobiol. 2004;58:48–59. doi: 10.1002/neu.10283. [DOI] [PubMed] [Google Scholar]
- 15.Gleeson J. G., Lin P. T., Flanagan L. A., Walsh C. A. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- 16.Sapir T., Elbaum M., Reiner O. Reduction of microtubule catastrophe events by LIS1, platelet-activating factor acetylhydrolase subunit. EMBO J. 1997;16:6977–6984. doi: 10.1093/emboj/16.23.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiner O., Carrozzo R., Shen Y., Wehnert M., Faustinella F., Dobyns W. B., Caskey C. T., Ledbetter D. H. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature (London) 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- 18.Dobyns W. B., Reiner O., Carrozzo R., Ledbetter D. H. Lissencephaly, A human brain malformation associated with deletion of the LIS1 gene located at chromosome 17p13. JAMA, J. Am. Med. Assoc. 1993;270:2838–2842. doi: 10.1001/jama.270.23.2838. [DOI] [PubMed] [Google Scholar]
- 19.Barkovich A. J., Guerrini R., Battaglia G., Kalifa G., N'Guyen T., Parmeggiani A., Santucci M., Giovanardi-Rossi P., Granata T., D'Incerti L. Band heterotopia: correlation of outcome with magnetic resonance imaging parameters. Ann. Neurol. 1994;36:609–617. doi: 10.1002/ana.410360409. [DOI] [PubMed] [Google Scholar]
- 20.des Portes V., Pinard J. M., Billuart P., Vinet M. C., Koulakoff A., Carrie A., Gelot A., Dupuis E., Motte J., Berwald-Netter Y., et al. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell (Cambridge, Mass.) 1998;92:51–61. doi: 10.1016/s0092-8674(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 21.Faulkner N. E., Dujardin D. L., Tai C. Y., Vaughan K. T., O'Connell C. B., Wang Y., Vallee R. B. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki S., Shionoya A., Ishida M., Gambello M. J., Yingling J., Wynshaw-Boris A., Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- 23.Smith D. S., Niethammer M., Ayala R., Zhou Y., Gambello M. J., Wynshaw-Boris A., Tsai L. H. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat. Cell Biol. 2000;2:767–775. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- 24.Tai A. W., Chuang J. Z., Sung C. H. Cytoplasmic dynein regulation by subunit heterogeneity and its role in apical transport. J. Cell Biol. 2001;153:1499–1509. doi: 10.1083/jcb.153.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z., Steward R., Luo L. Drosophila Lis1 is required for neuroblast proliferation, dendritic elaboration and axonal transport. Nat. Cell Biol. 2000;2:776–783. doi: 10.1038/35041011. [DOI] [PubMed] [Google Scholar]
- 26.Hattori M., Adachi H., Tsujimoto M., Arai H., Inoue K. Miller-Dieker lissencephaly gene encodes a subunit of brain platelet-activating factor acetylhydrolase. Nature (London) 1994;370:216–218. doi: 10.1038/370216a0. [DOI] [PubMed] [Google Scholar]
- 26a.Erratum. Nature (London) 1994. p. 391.
- 27.Tanaka T., Serneo F. F., Higgins C., Gambello M. J., Wynshaw-Boris A., Gleeson J. G. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J. Cell Biol. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perrone C. A., Tritschler D., Taulman P., Bower R., Yoder B. K., Porter M. E. A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in chlamydomonas and Mammalian cells. Mol. Biol. Cell. 2003;14:2041–2056. doi: 10.1091/mbc.E02-10-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- 30.Terada S., Hirokawa N. Moving on to the cargo problem of microtubule-dependent motors in neurons. Curr. Opin. Neurobiol. 2000;10:566–573. doi: 10.1016/s0959-4388(00)00129-x. [DOI] [PubMed] [Google Scholar]
- 31.Chuang J. Z., Milner T. A., Sung C. H. Subunit heterogeneity of cytoplasmic dynein: differential expression of 14 kDa dynein light chains in rat hippocampus. J. Neurosci. 2001;21:5501–5512. doi: 10.1523/JNEUROSCI.21-15-05501.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tai C. Y., Dujardin D. L., Faulkner N. E., Vallee R. B. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J. Cell Biol. 2002;156:959–968. doi: 10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allan V. J., Thompson H. M., McNiven M. A. Motoring around the Golgi. Nat. Cell Biol. 2002;4:E236–E242. doi: 10.1038/ncb1002-e236. [DOI] [PubMed] [Google Scholar]
- 34.Chowdhury K., Bonaldo P., Torres M., Stoykova A., Gruss P. Evidence for the stochastic integration of gene trap vectors into the mouse germline. Nucleic Acids Res. 1997;25:1531–1536. doi: 10.1093/nar/25.8.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dominguez I., Itoh K., Sokol S. Y. Role of glycogen synthase kinase 3 beta as a negative regulator of dorsoventral axis formation in Xenopus embryos. Proc. Natl. Acad. Sci. U.S.A. 1995;92:8498–8502. doi: 10.1073/pnas.92.18.8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez C., Perez M., Avila J. GSK3beta-mediated phosphorylation of the microtubule-associated protein 2C (MAP2C) prevents microtubule bundling. Eur. J. Cell Biol. 2000;79:252–260. doi: 10.1078/s0171-9335(04)70028-x. [DOI] [PubMed] [Google Scholar]
- 37.Ulloa L., Dombradi V., Diaz-Nido J., Szucs K., Gergely P., Friedrich P., Avila J. Dephosphorylation of distinct sites on microtubule-associated protein MAP1B by protein phosphatases 1, 2A and 2B. FEBS Lett. 1993;330:85–89. doi: 10.1016/0014-5793(93)80925-k. [DOI] [PubMed] [Google Scholar]
- 38.Shelanski M. L., Gaskin F., Cantor C. R. Microtubule assembly in the absence of added nucleotides. Proc. Natl. Acad. Sci. U.S.A. 1973;70:765–768. doi: 10.1073/pnas.70.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caspi M., Atlas R., Kantor A., Sapir T., Reiner O. Interaction between LIS1 and doublecortin, two lissencephaly gene products. Hum. Mol. Genet. 2000;9:2205–2213. doi: 10.1093/oxfordjournals.hmg.a018911. [DOI] [PubMed] [Google Scholar]
- 40.Assadi A. H., Zhang G., Beffert U., McNeil R. S., Renfro A. L., Niu S., Quattrocchi C. C., Antalffy B. A., Sheldon M., Armstrong D. D., et al. Interaction of reelin signaling and Lis1 in brain development. Nat. Genet. 2003;35:270–276. doi: 10.1038/ng1257. [DOI] [PubMed] [Google Scholar]
- 41.Ulloa L., Diez-Guerra F. J., Avila J., Diaz-Nido J. Localization of differentially phosphorylated isoforms of microtubule-associated protein 1B in cultured rat hippocampal neurons. Neuroscience. 1994;61:211–223. doi: 10.1016/0306-4522(94)90225-9. [DOI] [PubMed] [Google Scholar]
- 42.Thyberg J., Moskalewski S. Role of microtubules in the organization of the Golgi complex. Exp. Cell Res. 1999;246:263–279. doi: 10.1006/excr.1998.4326. [DOI] [PubMed] [Google Scholar]
- 43.Coquelle F. M., Caspi M., Cordelieres F. P., Dompierre J. P., Dujardin D. L., Koifman C., Martin P., Hoogenraad C. C., Akhmanova A., Galjart N., et al. LIS1, CLIP-170's key to the dynein/dynactin pathway. Mol. Cell. Biol. 2002;22:3089–3102. doi: 10.1128/MCB.22.9.3089-3102.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ligon L. A., Shelly S. S., Tokito M., Holzbaur E. L. The microtubule plus-end proteins EB1 and dynactin have differential effects on microtubule polymerization. Mol. Biol. Cell. 2003;14:1405–1417. doi: 10.1091/mbc.E02-03-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rivas R. J., Hatten M. E. Motility and cytoskeletal organization of migrating cerebellar granule neurons. J. Neurosci. 1995;15:981–989. doi: 10.1523/JNEUROSCI.15-02-00981.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Del Rio J. A., Gonzalez-Billault C., Urena J. M., Jimenez E. M., Barallobre M. J., Pascual M., Pujadas L., Simo S., La Torre A., Wandosell F., et al. MAP1B is required for Netrin 1 signaling in neuronal migration and axonal guidance. Curr. Biol. 2004;14:840–850. doi: 10.1016/j.cub.2004.04.046. [DOI] [PubMed] [Google Scholar]
- 47.Koonce M. P., Tikhonenko I. Functional elements within the dynein microtubule-binding domain. Mol. Biol. Cell. 2000;11:523–529. doi: 10.1091/mbc.11.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]