Abstract
Length variants within a CA-repeat-rich region of intron 4 of the human SP-B (pulmonary surfactant protein-B) gene are associated with several lung diseases. The hypothesis that SP-B intron 4 affects mRNA splicing was studied. SP-B minigenes containing exons 1–6 with a normal-sized intron 4 (pBi4normal) or intron 4 containing deletions (pBi4del) of 193, 211, 264 or 340 bp were expressed in CHO (Chinese hamster ovary) cells by transient transfection. Two forms of SP-B transcripts, normal and incompletely spliced, were detected. With pBi4normal, normal-sized SP-B mRNA was the predominant form and a very low amount of incompletely spliced mRNA was present, whereas with the pBi4del variants the amount of normal SP-B mRNAs was lower and the amount of incompletely spliced mRNA was relatively high. Reverse transcription–PCR results and sequencing data indicated that the incompletely spliced SP-B RNA contained intron 4 sequence, and this incompletely spliced RNA was also observed in normal lung. Lung cancer tissues with intron 4 deletions exhibited a larger amount of abnormally spliced RNAs compared with normal lung tissue or cancerous tissue with normal-sized intron 4. The results indicate that intron 4 length variants affect SP-B mRNA splicing, and that this may contribute to lung disease.
Keywords: deletion variant, exonic splicing enhancer (ESE), lung disease, pulmonary surfactant protein B (SP-B), RNA splicing
Abbreviations: ARDS, acute respiratory distress syndrome; BGHpA, bovine growth hormone polyadenylation; BPD, bronchopulmonary dysplasia; CHO, Chinese hamster ovary; COPD, chronic obstructive pulmonary disease; ESE, exonic splicing enhancer; ESS, exonic splicing silencer; FBS, fetal bovine serum; GMEM, Glascow's modified Eagle's medium; HNF, hepatocyte nuclear factor; HSD11B2, 11β-hydroxysteroid dehydrogenase type 2; M-MLV, Moloney murine leukaemia virus; pBi4normal, normal-sized SP-B intron 4; pBi4del, SP-B intron 4 deletion construct; RDS, respiratory distress syndrome; RT-PCR, reverse transcription–PCR; SC35, splicing component 35; SF2/ASF, splicing factor 2/alternative splicing factor; SP-B, pulmonary surfactant protein-B; SRP40 (etc.), serine/arginine-rich protein 40 (etc.)
INTRODUCTION
Approximately 3% of the sequences in the human genome are microsatellite repeats. CA/TG dinucleotide repeats are distributed in the mammalian genome, on average, every 20–30 kb. Dinucleotide repeats are widely used for genetic mapping, but their function is not entirely known. Microsatellite repeats play very important roles in genetic instability and gene function [1]. Genetic instability of microsatellite repeats has been found in a variety of human carcinomas [2,3], including primary small cell lung cancer [4] and non-small-cell lung carcinomas [5]. Gene dysfunction and human diseases caused by microsatellite repeats have been well documented [6,7]. A study in yeast indicated that 12 repeats of CTG, GAA or GTG exhibit differences in chromatin structure and have an impact on gene expression [8]. Moreover, the length of the GAA repeat in the region upstream of the transcription start site has been shown to regulate gene expression of the pMGA gene in Mycoplasma gallisepticum [9,10]. Inhibition of gene expression by dinucleotide repeats has been observed where the dinucleotide repeats were located in the promoter region of the nucleolin gene [11], as well as in the introns of the tilapia prolactin 1 [12], HSD11B2 (11β-hydroxysteroid dehydrogenase type 2) [13,14] and epidermal growth factor receptor [15] genes. For the latter, inhibition of gene expression was as high as 80% [15].
A length variation polymorphism has been identified in intron 4 of the gene encoding SP-B (pulmonary surfactant protein-B). This polymorphism occurs in a CA-repeat-rich region of 382 nucleotides in which more than 65% of the nucleotides are CAs; these are distributed in 13 stretches of CA repeats [16]. At least eight deletions and nine insertions have been identified in this region. SP-B intron 4 length variants have been shown to be associated with RDS (respiratory distress syndrome) [16–19], ARDS (acute RDS) [20], BPD (bronchopulmonary dysplasia) [21], COPD (chronic obstructive pulmonary disease) [22] and lung cancer [23]. However, the impact of SP-B intron 4 length variants on SP-B mRNA or gene function has not been studied.
SP-B is essential for maintaining normal surface tension at the air–liquid interface in the alveolus of the lung. The absence of SP-B protein is incompatible with life, and dysfunction of SP-B compromises lung function [24–27]. Evidence indicates that SP-B exhibits an anti-inflammatory function in the lung [28,29]. The SP-B gene consists of 11 exons [30], and maps on chromosome 2 [31]. It encodes a 42 kDa precursor protein [32]. The SP-B precursor undergoes several post-translational processing steps to produce a mature protein of 8 kDa, with the last two protein processing steps being Type II-cell-specific [33–35]. The mature SP-B protein is encoded by exons 6 and 7 of the gene. Based on associations of SP-B intron 4 deletion/insertion variations with lung diseases, we hypothesized that these deletions/insertions affect the levels of correctly spliced/functional SP-B mRNA by affecting mRNA splicing. This hypothesis was tested in the present work by studying the impact of various SP-B intron 4 constructs on SP-B mRNA content and RNA splicing in CHO (Chinese hamster ovary) and H441 cells, as well as by investigating the pressence of incompletely spliced SP-B RNA containing intron 4 sequence in normal human lung tissues and in tissues with lung disease.
EXPERIMENTAL
Lung tissue
Normal lung tissue (n=5) was obtained from the Gift of Life Donor Program (Philadelphia, PA, U.S.A.). RNA from cancerous lung tissue was available in our laboratory. The protocol for the use of the human lung tissue in the present study was approved by The Human Subjects Protection Office of The Pennsylvania State University College of Medicine.
DNA isolation
Genomic DNA from human blood samples and plasmid DNA from transformed Escherichia coli cells were isolated using a QIAamp DNA Kit and a QIAprep Spin Mini Kit (Qiagen, Valencia, CA, U.S.A.) respectively. The isolation was performed according to the manufacturer's instructions.
RNA isolation
The tissues were ground to powder in liquid nitrogen. CHO or H441 cells were collected following various periods of incubation as indicated in the Figure legends. From the powdered tissue and cell extracts, total RNA was isolated using an RNeasy kit (QIAgen) according to the manufacturer's instructions. The isolated RNA was treated with DNase I (RNase-free) to eliminate possible contamination with DNA.
Northern analysis
Samples of total RNA (5 μg from transfected cells, or 10 μg from human lung tissue) were separated on a 1% (w/v) agarose gel containing formaldehyde (0.22 M) and transferred on to a GeneSceen Plus membrane (Perkin Elmer Life Science, Boston, MA, U.S.A.) using a NorthernMax™ kit (Ambion, Austin, TX, U.S.A.). The blot was fixed using a UV Stratalinker 2400 (Stratagene, La Jolla, CA, U.S.A.) [36]. RNA markers (Promega, Madison, WI, U.S.A.) were used to estimate the sizes of SP-B transcript bands on the Northern blot. The markers were mixed with ethidium bromide and run with RNA samples side by side, and the gel slice was cut off and photographed under the UV light.
Each oligo was labelled to high specific radioactivity by the addition of a poly[32P]ATP tail using terminal transferase (Stratagene). For detection of the expression of the SP-B minigene in the transfected cells, 32P-tailed oligos 603 and 190 (see Table 1) were used as indicated in the Figure legends [36].
Table 1. Primers used in the study.
Lower-case letters indicate non-SP-B gene sequence. The hSP-B (human SP-B) DNA sequence was taken from GenBank accession no. M24461. S, sense; AS, antisense; UTR, untranslated region.
| Primer | Gene | S/AS | Nucleotide positions | Gene location | Sequence (5′ to 3′) |
|---|---|---|---|---|---|
| 50 | hSP-B | AS | 3346–3327 | Intron 6 | ggggaattcACAATAGCCACTGCAGGTCT |
| 70A | hSP-B | S | 445–464 | Exon 2 | CAAAGCCTGGAGCAAGCATT |
| 71A | hSP-B | AS | 2824–2843 | Exon 6 | ggggaattCAATTGCTGCTCGGAGAGATC |
| 94 | hSP-B | S | 7–26 | Exon 1 | GAGGTGCCATGGCTGAGTCA |
| 110 | hSP-B | AS | 8648–8668 | 3′ UTR | gaattcACCAGTGGAAGTGACTCCCGAGG |
| 171 | hSP-B | AS | 1541–1522 | Exon 4 | GTCGTCAAGCACTTGGTTGCA |
| 172 | hSP-B | S | 1552–1571 | Exon 4 | CTGGTCATCGACTACTTCCA |
| 190 | hSP-B | AS | 1082–1064 | Exon 3 | GCCTCCTTGGCCATCTTGT |
| 535 | hSP-B | AS | 9786–9760 | 3′ Flanking | gcgactagtCTATGACGTCTGCTTCTCTGCCAAGGGAG |
| 536 | hSP-B | S | −938–966 | 5′ Flanking | gcggtcgacTCATCATGGTACTAATTCCTGCCCGTCCA |
| 603 | hSP-B | AS | 2458–2439 | Exon 5 | TGGCTGCCGGGATTTGCACA |
| 689 | hSP-B | AS | 2885–2866 | Exon 6 | ATCAGAGCCCTGCAGAGCCA |
| 998 | hSP-B | AS | 1129–1161 | Intron 3 | TCAGCTCTTCCCTGCTCTGTCtCTCATCTCTTGG |
| 1009 | hSP-B | AS | 2135–2158 | Intron 4 | TGTGTGTGAGAGTGAGGGTGTAAG |
| 1188 | hSP-B | AS | 1596–1615 | Intron 4 | gacatggactagTGGAGGCAGGCAGGAGGTGAG |
| 1193 | hSP-B | S | 1417–1436 | Intron 4 | gacatggactagtCACGCAGGCCCCTGTGCCCA |
| 1194 | hSP-B | AS | 2028–2047 | Intron 4 | gacatggactagtGCCCTTTGTGGACAGGGTGT |
| 1219 | hSP-B | AS | 445–464 | Exon 2 | AATGCTTGCTCCAGGCTTTG |
| 1221 | pcDNA3.1 | S | 893–1013 | tggctagcgtttaaacttaag | |
| 1285 | hSP-B | S | 71–81/392–400 | Exons 1–2 | tacgactcactatagggcgaCCCAGGCACTGCTGCCTGGA |
| 1286 | hSP-B | AS | 4839–4848/6083–6093 | Exons 8–9 | taggtgacactatagaatacAATTGCTTGCACTTTTCCCTG |
| 1044 | T7 promoter | S | gccagtgaattgtaatacgactcactatagggcga | ||
| 1045 | SP6 promoter | AS | ttacgccaagctatttaggtgacactatagaatac |
For the detection of SP-B mRNA in lung tissues, the blot was hybridized with a [32P]UTP-labelled SP-B antisense RNA (exons 1–9) probe. The RNA probes were prepared by in vitro transcription with a Strip-EZ RNA kit (Ambion). The templates used for in vitro transcription were PCR fragments that were amplified by two steps of RT-PCR (reverse transcription–PCR). For the first step of PCR, the primers used were 1285 (exons 1–2) and 1286 (exons 8–9) (all primers used in the present study are given in Table 1 and Figure 1). T7 or SP6 promoter sequences were attached to primers 1285 and 1286 respectively. These RT-PCR products were subsequently amplified with primer pair 1044/1045 and used for in vitro transcription [36].
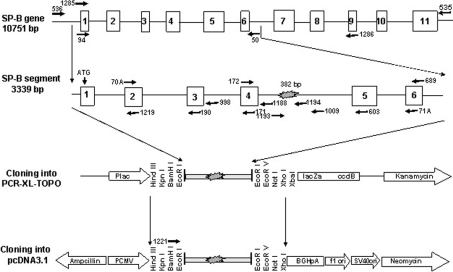
Figure 1. Schematic illustration of cloning of the SP-B minigene.
The entire SP-B gene (10751 bp) was amplified from genomic DNA by long template PCR amplification with primers 535 and 536 (see Table 1). The long PCR products were used as a template to amplify a 3339 bp SP-B fragment with primers 94 and 50. The 3339 bp PCR products were purified and cloned into the PCR-XL-Topo vector, and then subcloned into the pcDNA3.1 vector under the control of pCMV, and were followed by the BGHpA polyadenylation signal, as shown.
Northern hybridization was performed in Ultrahyb solution (Ambion). Hybridization (for RNA probes at 68 °C and for oligo probes at 42 °C) and washing procedures were performed according to the manufacturer's instructions (Ambion). The blot was exposed to Kodak film (XAR 5) at −80 °C for 12 h (for CHO cells) or 2 min (for human lung). The density of the bands on the Northern blot was quantified by densitometry using a Molecular Dynamics model 100A Scanner (Amersham Pharmacia Biotech) and Quantity One software (Discovery Services, Bio-Rad, Hercules, CA, U.S.A.).
Reverse transcription
Total RNA (1 μg) was used as template for reverse transcription in vitro by M-MLV (Moloney murine leukaemia virus) reverse transcriptase (Invitrogen, Carlsbad, CA, U.S.A.) with a poly(T) primer. A portion of 1 μl of reverse transcription product was used as template for PCR amplification [37].
PCR amplification
PCR was carried out in a volume of 30 μl containing 0.1 mM of each dNTP, 60 ng of each primer, 1×mix of Buffers 1 and 2, and 1 unit of Taq polymerase (reagents for PCR were from Roche Boehringer Mannheim Diagnostics, Indianapolis, IN, U.S.A.). The long PCR profile for amplification of a 10751 bp fragment of SP-B with primers 535 and 536 consisted of denaturation at 95 °C for 2 min, 10 cycles of 95 °C for 30 s, 58 °C for 1 min and 68 °C for 10 min, then 20 cycles of 95 °C for 30 s, 62 °C for 1 min and 68 °C for 12 min, followed by a final extension step at 72 °C for 20 min. For other PCR amplifications, the profile consisted of denaturation at 95 °C for 2 min, 35 cycles of 95 °C for 30 s, 55 °C for 1 min and 72 °C for 1 min per kb of PCR fragment to be amplified, and a final extension step at 72 °C for 5 min [38].
Cloning of the SP-B minigene
As illustrated in Figure 1, the entire SP-B gene region (10751 bp) was amplified from genomic DNA by long template PCR with PCR primers 535 and 536 [38]. The 10751 bp SP-B fragment was used as a template to amplify, with primers 94 and 50, a 3339 bp segment that included 8 bp upstream of the start ATG codon in exon 1 and ended 433 bp downstream of exon 6. The 3339 bp SP-B segment, or SP-B minigene, was separated on a 0.8% agarose gel, purified and cloned into a PCR-XL-Topo vector, according to the manufacturer's instructions for the Topo-XL-PCR cloning kit (Invitrogen). The positive clones were sequenced. Sequence alignment was done with Lasergene software (DNAStar, Inc., Madison, WI, U.S.A.). The SP-B minigene was removed from the Topo vector as a HindIII/XhoI fragment and cloned into the pcDNA3.1 vector (Invitrogen) under the control of the cytomegalovirus enhancer–promoter (pCMV), and followed by the BGHpA (bovine growth hormone polyadenylation) signal and the transcription termination sequence. Exon 4 and intron 4 sequencing, and partial sequencing of the rest of the minigene regions, was performed for all of the constructs. Five SP-B minigenes with different lengths of intron 4 were characterized (see Figure 2) and used for further study.
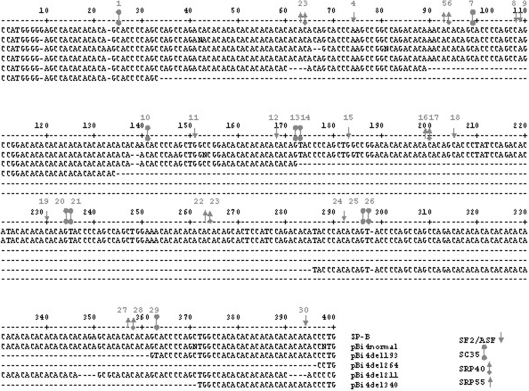
Figure 2. Sequence alignment of intron 4 length variants of the SP-B gene.
The published SP-B sequence shown (nt 2677–3074) is from GenBank (accession no. M24461) [30]. pBi4normal is the SP-B normal allele and served as a control; pBi4del193, pBi4del264, pBin4del211 and pBi4del340 are SP-B deletion variants with deletion of 193, 264, 211 and 340 bp respectively in intron 4. The locations of various ESE motifs (SF2/ASF, SRP40, SC35 and SRP55) are indicated by the indicated symbols on the first nucleotide of the respective consensus sequences. The individual consensus sequences are shown in the text. The grey numbers (1–30) indicate the locations of ESEs, and the black numbers indicate nucleotide numbering.
DNA sequencing
Sequencing of plasmid DNAs and PCR products was carried out using an Applied Biosystems ABI 377 DNA sequencer in the Molecular Genetic Core Facility, The Pennsylvania State University College of Medicine. The PCR products for sequencing were separated on a 1.2% (w/v) agarose gel and then purified using a Qiaquick kit (Qiagen).
Cell culture
The mammalian CHO-K1 cell line and the human lung adenocarcinoma cell line NCI-H441 were used for transfection. Both cell lines were purchased from the American Type Culture Collection (Manassas, VA, U.S.A.). The CHO-K1 cell line was grown in GMEM (Glascow's modified Eagle's medium; Invitrogen Life Technologies) supplemented with 10% (v/v) FBS (fetal bovine serum; Hyclone, Logan, UT, U.S.A.), 1×antimycotic/antibiotic solution (Sigma, St. Louis, MO, U.S.A.), 1×G+A (100×stock: 6 mg/ml L-glutamic acid and 6 mg/ml L-asparagine), 1×nucleoside mixture (Invitrogen) and 1×sodium pyruvate (Invitrogen). The NCI-H441 cell line was grown in RPMI 1640 medium (Invitrogen) containing 10% (v/v) heat-inactivated FBS, 1×antimyocotic/antibiotic solution and 1% (w/v) L-glutamine (Sigma). The cells were cultured at 37 °C in a 5% CO2 atmosphere, and passaged weekly.
Transient transfection
Plasmid DNAs were transfected into the cells with the Lipofectamine Plus™ Reagent kit (Invitrogen), when the cells were at 50–90% confluence. At 24 h before transfection, the cells were trypsinized, counted, and then plated into 6-well culture plates until they reached 50–90% confluence, at which time they were used for transfection. At 4 h before transfection, the GMEM/10% FBS was replaced with GMEM. Lipofectamine Plus Reagent was added to 100 μl of GMEM (without serum) containing the SP-B construct (0.3 μg), mixed and incubated for 15 min at room temperature. Then 4 μl of Lipofectamine in 100 μl of GMEM (without serum) was added and incubated for another 15 min at room temperature. The DNA–Lipofectamine complex was added to each well containing fresh GMEM (5 ml). At 4 h after transfection, GMEM/10% FBS was increased to normal volume (10 ml). Transfection was carried out at 37 °C in a 5% CO2 atmosphere for 18 h unless indicated otherwise.
RESULTS
SP-B intron 4 deletion constructs
Constructs containing the SP-B minigene with intron 4 sequences of various lengths were made as illustrated in Figure 1. To study the impact of intron 4 on SP-B mRNA processing, a region spanning 8 bp upstream of the start ATG codon in exon 1 to 433 bp downstream (exon 6) was included in the SP-B minigene. The SP-B minigene was under the control of pCMV in the pcDNA3.1 vector, which is designed for high-level, constitutive expression in a variety of mammalian cell lines.
Five SP-B minigenes from different individuals were characterized and used in the present study. These were pBi4normal (normal-sized intron 4 allele) and four pBi4del (intron 4 deletion) alleles, i.e. pBi4del193 (193 bp deletion), pBi4del211 (211 bp deletion), pBi4del264 (264 bp deletion) and pBi4del340 (340 bp deletion). The sequence alignments of the CA-repeat-rich regions of intron 4 are shown in Figure 2. The deletions are located within a 363 bp sequence of intron 4, i.e. nucleotides 34–396. This 363 bp region is a CA-repeat-rich region in which 65% of the total number of nucleotides are CA. These CAs are clustered in 12 stretches of CA repeats. Another stretch of CA repeats is located within intron 4 at nucleotides 13–23.
Expression of SP-B minigenes in CHO cells
CHO cells were used for the proposed studies. These cells are well suited for such studies because (a) they do not express SP-B, thus eliminating interference by endogenous SP-B expression, and (b) although the last two steps of SP-B precursor protein processing are Type II-cell-specific, SP-B transcription is not known to be cell-specific. Since our end point is SP-B RNA, the CHO cells are advantageous. However, we did perform selected experiments in H441 cells, giving similar results to those obtained with CHO cells (see below).
First we optimized the transfection conditions for the expression of SP-B minigenes in CHO cells. A preliminary experiment showed that no human SP-B mRNA homologue was detected on Northern blots, and no PCR product was amplified by RT-PCR with human SP-B-specific primers from control CHO cells (results not shown), indicating that there would be no interference by sequences similar to that of human SP-B in CHO cells under the experimental conditions used.
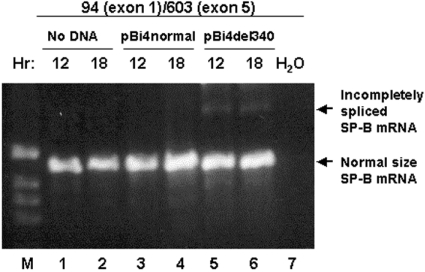
A dose–response experiment with plasmid DNA (0, 0.1, 0.3, 0.5, 0.7, 1.0 and 2.0 μg per well; results not shown) was performed, and a concentration of 0.3 μg of DNA per well was selected for further study. This concentration of plasmid DNA resulted in SP-B mRNA expression that was well below the maximum level. This low concentration of DNA was chosen in order to eliminate the possibility that incompletely spliced RNA was the result of misprocessing due to a high amount of RNA transcribed from the SP-B minigene. A time course of transfection was carried out with pBi4normal and two deletion variants, pBi4del193 and pBi4del264 (Figure 3A). Two bands were observed on the Northern blot, a lower band with high intensity and an upper band with low intensity and varying in size. If SP-B mRNA is completely spliced, the expected molecular size would be approx. 1.1 kb, comprising 686 nt of SP-B sequence (exons 1–6), 335 nt of vectors PCR-XL-Topo and pcDNA3.1, and other sequences such as the 3′ end polyadenylation signal-related sequence and the additional poly(A) tail. The lower band on the Northern blot represents the expected normal size of SP-B mRNA. The upper band of approx. 2 kb is clearly larger than the expected normal SP-B mRNA, and much smaller than 3674 nt that would be the size of the non-spliced SP-B minigene mRNA precursor (3674 nt; 3339 nt of SP-B plus 335 nt of vector sequences). Therefore this is likely to represent an incompletely spliced SP-B RNA.
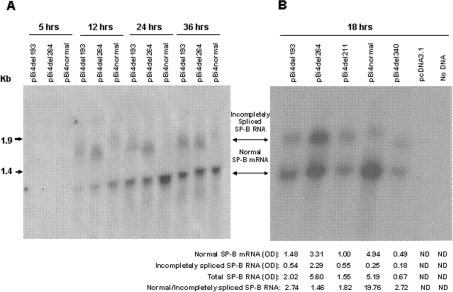
Figure 3. Northern analysis of SP-B minigene mRNA in CHO cells.
(A) Time course (two experiments). (B) Comparison of SP-B variants (three experiments). CHO cells were cultured at 37 °C in a 5% CO2 atmosphere, and fed and passaged weekly. Plasmid DNA was transfected into CHO cells with Lipofectamine Plus™ Reagent kit when the cells were at 50–90% confluence. At the indicated times (A) or at 18 h (B) after transfection, the cells were harvested for total RNA isolation. The probes were 32P-labelled oligos 190 and 603 (see Figure 1). The X-ray film was exposed at −80 °C for 24 h. The density of the bands in (B) was quantified by densitometry, and values are listed under the respective lanes (OD=absorbance). Vector pcDNA3.1 DNA and no DNA were used as negative controls for transfection (ND, not detected).
As shown in Figure 3(A), no visible SP-B band was detected 5 h after transfection, but at 12 h very clear bands were observed. A maximal amount of SP-B transcripts was reached at 24 h for pBi4normal; for pBi4del193 and pBi4del264, the amount of SP-B transcripts was slightly higher at 36 h than at 24 h post-transfection. From 12 to 24 h after transfection, the amount of normal-sized SP-B mRNA was increased markedly with both pBi4normal and pBi4del. However, while incompletely spliced SP-B RNA was absent or barely detectable with pBi4normal, it was detectable with the two pBi4del constructs. This indicated that the observed incompletely spliced SP-B RNA was the result of normal processing and not of misprocessing due to the high amount of SP-B transcripts in CHO cells. At all post-transfection time points (Figure 3A), the pBi4normal allele produced predominantly normal mRNA and only a very small amount of incompletely spliced RNA, while the four pBi4del variants produced lower amounts of normal SP-B mRNA and relatively higher amounts of incompletely spliced SP-B RNA compared with pBi4normal (Figure 3A).
Further comparisons were carried out among pBi4normal and the four pBi4del constructs. Figure 3(B) depicts a Northern blot and densitometric data. To compare the efficiency of mRNA splicing, the ratio of normal to incompletely spliced SP-B RNA was evaluated. The results indicated the following. (1) Significantly lower levels of normal SP-B mRNA were obtained for the deletion variants (except pBi4del264), ranging from 10% to 67% of the value for pBi4normal. The difference in the amount of normal SP-B mRNA among the four pBi4del alleles was up to 6.8-fold (pBi4del264 compared with pBi4del34). (2) The ratio of normal to incompletely spliced RNA was lower in all pBi4del variants compared with pBi4normal, with values for the variants representing only 7–14% of that of the control. The ratio differed among the four pBi4del variants by up to 1.9-fold (pBi4del193 compared with pBi4del264). (3) The total amount of SP-B mRNA (normal plus incompletely spliced) was decreased in three of the four pBi4del variants, to 39% (pBi4del193), 30% (pBi4del211) and 13% (pBi4del340) of that of pBi4normal. Although the total amount of pBi4del264 RNA was similar to that of pBi4normal, the ratio of normal to incompletely spliced SP-B RNA differed (19.76 for pBi4normal compared with 1.46 for pBi4del264), as noted above.
Splicing of SP-B intron 4 in CHO cells
Based on the results described above, we speculated that the SP-B deletion variants alter mRNA splicing, and that abnormal splicing most probably occurs in intron 4. This hypothesis was tested in the following experiments. First, we used pBi4normal to test SP-B mRNA splicing in CHO cells by determining whether all exons in the minigene are appropriately spliced. Different RT-PCR segments were analysed. To further eliminate any possibility of amplification of sequences related to SP-B in the CHO cells, we used vector pcDNA3.1 primer 1221 (upstream from SP-B exon 1) as a sense primer. The antisense primers were the SP-B-specific primers 1219, 190, 171, 603 and 689 for exons 2, 3, 4, 5 and 6 respectively. For the pBi4normal allele (as expected), the correct sizes (completely spliced) of SP-B cDNA segments were amplified: 215 bp for primer pair 1221/1219, 332 bp for 1221/190, 429 bp for 1221/171, 520 bp for 1221/603 and 719 bp for 1221/689 (Figure 4A). Furthermore, the amplification of correct sizes indicated that the PCR products were not due to the amplification of genomic DNA or the SP-B RNA precursor, both of which will include intron sequences. If none of the introns were spliced, the sizes of the above products would have been 0.5, 1.1, 1.5, 2.5 and 2.9 kb respectively. However, a larger band of very low intensity was observed in some PCR products with primer pairs 1221/171 and 1221/603, indicating that very low levels of incompletely spliced mRNA may be present.
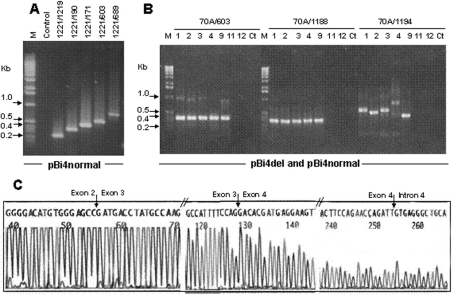
Figure 4. RT-PCR analysis of SP-B minigene mRNA in CHO cells.
Total RNA (1 μg) from transfected CHO cells was reverse transcribed by M-MLV reverse transcriptase, with a poly(T) primer. The reverse transcription product (1 μl) was used as a template for PCR amplification. (A) PCR amplification of different regions of SP-B mRNA from cells transfected with pBi4normal. The PCR primer pair for each lane is given above the gel. (B) PCR amplification of SP-B mRNA from cells transfected with recombinant plasmid DNAs: lane 1, pBi4del193; lane 2, pBi4del264; lane 3, pBi4del211; lane 4, pBi4normal; lane 9, pBi4del340; lane 11, vector pcDNA3.1 (control); lane 12, no plasmid DNA; lane M, 1 kb DNA ladder; lane Ct, control for PCR with distilled water. The PCR primers are given above the gel. (C) Sequence analysis of RT-PCR products amplified with primers 70A and 1194 from pBi4del193. The PCR products were purified and sequenced with primer 70A. DNA sequencing was done with an Applied Biosystems ABI 377 DNA sequencer. The junctions of exons 2 and 3, exons 3 and 4, and exon 4 and intron 4, are indicated by arrows.
Next we analysed the SP-B transcripts of the four pBi4del variants. When only exon sequences were used as PCR primers (primer 70A located in exon 2 and primer 603 in exon 5) (Figure 4B), a very strong band was observed in all the pBi4del variants at the expected size of 325 bp. In addition, a low-intensity but very clear high-molecular-mass band with a size that varied among the pBi4del variants was also observed. This upper band was considered to be incompletely spliced RNA. With the pBi4normal allele, this upper band was barely detectable (lane 4 for 70A/603, Figure 4B), consistent with the results of Figure 4(A), while with the four pBi4del variants the upper band was clearly visible.
To examine whether intron 4 sequence was present in the incompletely spliced SP-B RNA, we forced preferential PCR amplification of the incompletely spliced RNA by using intron 4 primers (1188 and 1194) as antisense primers and an exon 2 primer (70A) as sense primer (Figure 4B). Significant amounts of PCR products were detectable. The PCR products were the same size for all variants (including pBi4normal) when the primer (1188) was located upstream of the CA-repeat-rich region of intron 4 (i.e. 34 nt from the exon 4/intron 4 splice junction). However, the size of the amplified fragments differed among the variants when the intron 4 antisense primer 1194 was used. This primer is located downstream of the intron 4 CA-repeat-rich region. The difference in size was similar to the length of intron 4 in each construct (Figure 4B). The size of the pBi4normal product (Figure 4B, 70A/1194, lane 4) was approx. 0.74 kb (as expected), larger than any of the pBi4del products.
The RT-PCR results confirmed the findings of the Northern analysis: (1) the presence of an incompletely spliced SP-B RNA; and (2) a much lower ratio of normal to incompletely spliced SP-B RNA for pBi4del alleles than for pBi4normal. Moreover, the incompletely spliced SP-B RNA contained intron 4 sequence.
The existence of intron 4 sequence in SP-B mRNA was confirmed by direct sequencing of RT-PCR products. The PCR products of primers 70A and 1194 from pBi4del193 were isolated, purified and sequenced (Figure 4C). As indicated by arrows, exon 2 sequence was followed by exon 3 sequence and exon 3 sequence by exon 4, with no intron sequences in between. However, exon 4 sequence was followed by intron 4 sequence, instead of exon 5, indicating that SP-B intron 4 sequence was present in the incompletely spliced SP-B RNA.
Selected experiments in NCI-H441 cells
The SP-B gene is expressed in epithelial alveolar type II and Clara cells of the lung. However, although the final two protein processing steps are cell specific [33,34], nothing is known regarding the cell specificity of steps prior to protein processing. To ensure that cell specificity is not involved in mRNA processing, which is our end point, we carried out a selected experiment in H441 cells, a human adenocarcinoma cell line that expresses SP-B. As shown in Figure 5, a significant amount of endogenous SP-B mRNA was amplified from exons 1–5 in untransfected H441 cells (lanes 1 and 2), and in H441 cells transfected with the pBi4normal (lanes 3 and 4) or with pBi4del340 (lanes 5 and 6). Similar to what we found in transfected CHO cells (Figure 4B), an incompletely spliced SP-B mRNA (upper band, arrow) was observed in pBi4del340-transfected H441 cells (lanes 5 and 6) but not in pBi4normal-transfected (lanes 3 and 4) or non-transfected (lanes 1 and 2) H441 cells. Considering that the focus of the present study is not on SP-B protein processing, and that similar observations of incompletely spliced SP-B mRNA were made in both H441 and CHO cells, the CHO cells were chosen for further study, to avoid any interference of endogenous SP-B mRNA in H441 cells.
Figure 5. RT-PCR analysis of SP-B mRNA in H441 cells.
Total RNA (1 μg) from non-transfected (lanes 1 and 2) and transfected (lanes 3–6) H441 cells was reverse transcribed by M-MLV with a poly(T) primer. Samples of 1 μl of the reverse transcription products were used for PCR with exon 1 (no. 94) and exon 5 (no. 603) primers. PCR products at 12 and 18 h from the start of the experiment (lanes 1 and 2) or following transfection with pBi4normal (lanes 3 and 4) or with pBi4del340 (lanes 5 and 6) are shown. A band of high intensity is shown in all sample lanes (arrow; normal-sized SP-B mRNA) and a band of low intensity is present in lanes 5 and 6 (arrow; incompletely spliced SP-B mRNA). M, 1 kb DNA ladder; H2O, no reverse transcription product was present in the reaction.
Search for ESEs (exonic splicing enhancers) in the deletion sequences
To determine whether cis-acting elements known to play a role in splicing are present in intron 4 of the SP-B gene, we searched the intron 4 sequence for sequence motifs of ESEs. The deleted sequences in the constructs under study included regions from 88 bp downstream of exon 4 to 385 bp upstream of exon 5. These deletions did not eliminate usage of exon 4/intron 4 and intron 4/exon 5 splice junctions, as indicated by the presence of normal-sized (i.e. correctly spliced) products. However, our results indicated that although the content of normally spliced RNA was relatively high in the deletion variants (albeit lower than in pBi4normal), the content of incompletely spliced SP-B RNA was increased for the deletion variants compared with pBi4normal. Furthermore, although the deleted regions may contain ESEs and/or ESSs (exonic splicing silencers) that may be necessary for appropriate splicing [39–41], the observations favour the hypothesis that ESEs are more important in this process. ESSs exhibit negative effects on RNA splicing. Recent reports have shown that ESSs inhibit exon 7 splicing in the SMN2 (survival of motor neuron 2) gene [42,43] and inhibit splicing of the upstream intron of tat exon 2 of HIV-1 RNA [44]. If putative ESSs in the deleted SP-B intron 4 sequences were playing a role, following elimination of cis-inhibitory factors, we would have expected a considerably higher amount of normally spliced RNA and a lower amount of incompletely spliced RNA.
Four groups of ESE consensus motifs described previously [39] were found to be present in the deleted sequences. A total of 30 potential ESEs within an approx. 372 bp deletion region (Figure 2) were identified. These included (1) ten (nos. 4, 8, 9, 11, 12, 15, 18, 19, 24 and 30) motifs of SF2/ASF (splicing factor 2/alternative splicing factor) [consensus sequence C(GA)A-(GUC)C(G)A(CU)C(G)G(U)A(UG); the bold letters indicate high frequencies]; (2) ten (nos. 1, 7, 10, 13, 14, 20, 21, 25, 26 and 29) motifs of SC35 (splicing component 35) [consensus sequence G(ACU)G(AU)C(UAG)C(U)C(AUG)C(GAU)U(CAG)G(A)]; (3) five (nos. 3, 6, 17, 23, and 28) motifs of SRP40 (serine/arginine-rich protein 40) [consensus sequence U(CA)G(A)-C(UA)G(A)U(G)C(AG)]; and (4) five (nos. 2, 5, 16, 22 and 27) motifs of SRP55 [consensus sequence U(CA)C(UG)A(UCG)-C(AGU)A(UC)G(CA)G(C)]. The loss of these intron 4 ESEs in deletion variants may compromise normal splicing and increase the amount of incompletely spliced SP-B RNA.
Detection of incompletely spliced SP-B RNAs containing intron 4 sequences in human lung tissues
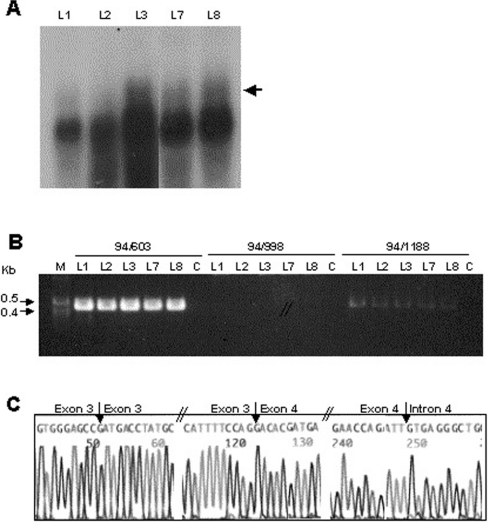
We next examined whether incompletely spliced SP-B RNAs containing intron 4 sequences observed in CHO cells could also be identified in human lung tissues. To this end, we performed a Northern blot analysis to detect SP-B mRNA in lung tissues. We used SP-B antisense RNA probes (955 nt; covering exons 1–9) to increase the sensitivity of the Northern blot analysis, because, based on the observations from the transient transfection experiments (Figure 3), we anticipated a low amount of incompletely spliced SP-B RNA to be present in normal lung tissue. A band with very high intensity was observed after only a 2 min exposure of the blot to X-ray film at −80 °C (Figure 6A). An SP-B RNA band of lower intensity and of higher molecular mass was also observed in these lung tissues, although this band was not clearly defined (arrow).
Figure 6. Detection of incompletely spliced SP-B RNA containing intron 4 sequence from normal human lung.
Total RNA was isolated from normal human lung tissues L1, L2, L3, L7 and L8 as described in the Experimental section. (A) Northern analysis. Total RNA (10 μg) from human lung tissue was separated on a 1% (w/v) agarose gel containing formaldehyde (0.22 M), and transferred on to a GeneSceen Plus membrane. The blot was fixed under UV light, and probed with [32P]UTP-labelled SP-B antisense RNA (exons 1–9), in Ultrahyb solution according to the manufacturer's instructions. The blot was exposed to Kodak film (XAR 5) at −80 °C for 2 min. The experiment was repeated two times. The arrow indicates incompletely spliced SP-B RNA. (B) RT-PCR. Total RNA (1 μg) from each normal human lung tissue was used for in vitro RT by M-MLV reverse transcriptase, and a poly(T) primer. Samples of 1 μl of reverse transcription product were used as a template for PCR amplification with sense primer 94 in exon 1 and antisense primer 603 in exon 5, primer 998 in intron 3, or primer 1188 in intron 4, as indicated above the gels. M, 1 kb DNA ladder; C, control (distilled water instead of RNA was used for reverse transcription). The experiment was repeated three times. (C) Partial sequences derived from PCR products of primer pair 94/1188. The PCR products of 94/1188 from normal human lung tissue L7 were separated, isolated, purified from a 1.2% (w/v) agarose gel, and sequenced directly with an Applied Biosystems ABI 377 DNA sequencer. The sequences shown are from the 9th nucleotide upstream of the 3′ end of exon 2 to the 12th nucleotide downstream of the 5′ end of exon 3; the 10th nucleotide upstream of the 3′ end of exon 3 to the 10th nucleotide downstream of the 5′ end of exon 4; and the 10th nucleotide upstream of the 3′ end of exon 4 to the 10th nucleotide downstream of the 5′ end of intron 4. The arrows indicate the exon–exon and exon–intron junction positions.
We next used RT-PCR to determine whether the SP-B RNA from normal human lung contains SP-B intron 4 sequences. As shown in Figure 6(B), a 458 bp fragment from exons 1–5 was amplified with PCR primers 94 (exon 1) and 603 (exon 5). Based on the size, these PCR products were amplified from completely spliced SP-B cDNA without sequences of intron 1 (300 bp), intron 2 (501 bp) or intron 3 (364 bp). However, if the antisense primer was the intron 4 primer 1188, a clear PCR product of lower intensity was also amplified. The size of the PCR product (435 bp) was the sum of exons 1–4 (401 bp) plus intron 4 sequence (34 bp) between exon 4 and primer 1188. No PCR products were detected with the primers 94 (exon 1) and 998 (intron 3). The products of primers 94/1188 are likely to reflect incompletely spliced SP-B RNA.
PCR products obtained with primer pair 94/1188 from lung tissue L7 were purified and sequenced. The partial sequences are shown in Figure 6(C). The sequencing primer was 70A in exon 2. After exon 2, the sequence was that of exon 3, and exon 3 was followed by exon 4. There was no sequence of introns 2 or 3. However, after exon 4 the sequence was that of intron 4. We concluded that incompletely spliced SP-B RNAs with intron 4 sequence exist in normal human lung.
RT-PCR analysis of SP-B mRNA in normal and cancerous lung tissue
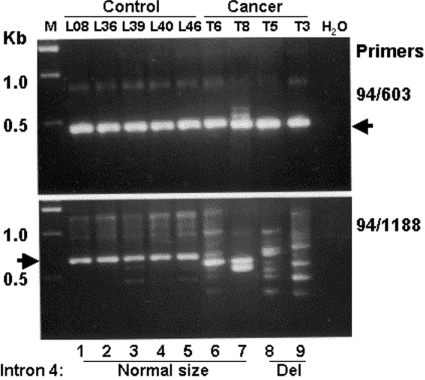
To gain insight into the potential role of intron 4 deletion in human lung disease, we used tissues from four patients with squamous cell carcinoma of known SP-B intron 4 genotype. Tissue from normal healthy lungs obtained via The Gift of Life Donor Program was used as controls. The results (Figure 7) indicate that when exon primers 94 (exon 1) and 603 (exon 5) were used, a band of high intensity was observed in all samples (upper panel, arrow). However, when an exon 1 primer (94) and an intron 4 primer (1188) were used, a variety of patterns were observed.
Figure 7. RT-PCR analysis of SP-B mRNA in normal and cancerous lung tissues.
Total RNA was isolated from normal healthy lung tissue (lanes 1–5) and from lung tissue of four patients (T6, T8, T5 and T3) with squamous cell carcinoma (lanes 6–9). The RNA was reverse transcribed by M-MLV reverse transcriptase with a poly(T) primer, and 1 μl of reverse transcription product was used as a template for PCR. The primer pairs used are indicated on the right. Primers 94 and 603 (upper panel) are located in exons 1 and 5 respectively; primers 94 and 1188 (lower panel) are located in exon 1 and intron 4 respectively. All five control normal lung tissue samples and two of the four patient lung tissue samples (lanes 6 and 7) had a normal-sized intron 4, whereas tissue samples from the other two patients had a deletion within intron 4 (lanes 8 and 9). M, 1 kb DNA ladder; H2O, no DNA was added in the sequencing reaction.
The control samples (lower panel, lanes 1–5) with normal-sized intron 4 exhibited one major band with high intensity (arrow). The tissues from cancer patients T6 and T8 with a normal-sized intron 4 showed one (T6) or two (T8) lower-sized bands (lanes 6 and 7). However, the patterns in cancerous tissues from patients (T5 and T3) with intron 4 deletions were more complex, with a large number of bands of various sizes. These data indicate that when primers in intron 4 are used, (1) the SP-B RT-PCR products from normal healthy lungs are similar (Figure 7, lower panel, lanes 1–5), but differ significantly from those from patients with squamous cell carcinoma (lower panel, lanes 6–9); and (2) the pattern of the RT-PCR bands in the cancer tissues is more complex in tissues from patients with intron 4 deletion (lanes 8–9).
DISCUSSION
Size length variants of intron 4 in the human SP-B gene have been associated with several pulmonary diseases [16,17,22,23]. In the present paper, we show that (1) deletion variants of intron 4 affect SP-B gene expression and mRNA splicing in CHO cells, (2) incompletely spliced SP-B RNA molecules are detectable in human lung tissue, and (3) intron 4 deletion variants are associated with a complex RT-PCR pattern in cancerous lung tissue. In CHO cells, the content of normal-sized SP-B mRNA and the ratio of normal to incompletely spliced SP-B RNA were markedly reduced on expression of all deletion variants under study compared with the pBi4normal allele, and the incompletely spliced SP-B RNA contained intron 4 sequence. Low levels of incompletely spliced SP-B RNA containing intron 4 sequence were also observed in normal human lung.
Previous studies have indicated that conserved cis-acting elements, the 5′- and 3′-splice sites and the branch site are not sufficient to define exon/intron boundaries for mRNA splicing. Exonic and intronic splicing cis-acting elements, such as ESEs and ESSs, have been shown to play a role in splicing [39–41]. A total of 30 ESE motifs were detected in the deleted intron 4 sequences of the deletion constructs studied in the present work (Figure 2), These included four groups (SF2/ASF, SC35, SRP40 and SRP55) of elements. Although the positions and the numbers of ESE motifs present in the four deletion variants differed, there were 11 motifs, from no. 13 to no. 23, located in the region between 189 bp downstream of exon 4 and 515 bp upstream of exon 5 that were deleted in all four deletion variants.
The present study identified a reduction in the ratio of normal to incompletely spliced SP-B RNA on expression of the deletion variants compared with the normal size intron 4, and also identified the presence of intron 4 sequences in the incompletely spliced mRNA. These observations indicated that one or more of the 11 ESEs may play a role in intron 4 splicing. Of interest is the observation that disruption of a consensus ESE in SCNM1 (sodium channel modifier 1) in mice resulted in a decrease in the amount of correctly spliced sodium channel mRNA below the threshold for survival [45]. However, because the total content of SP-B RNA was decreased with the three deletion variants, but with the pBi4del264 variant was similar to that of pBi4normal, it is possible that transcription elements within intron 4 also play a role in determining SP-B RNA levels. In the CA repeat region of intron 4, there are several putative transcription factor binding sites [e.g. for HNF-3 (hepatocyte nuclear factor-3), C/EBPβ (CCAAT/enhancer-binding protein), AML1 (acute myeloid leukaemia 1) factors, and others] that are located within the deleted region of all four deletion constructs studied. Although HNF-3 (also present in CHO cells) has been implicated in the stimulation of SP-B promoter activity [46], it is currently unknown whether elimination from intron 4 of the HNF-3 binding site or of binding sites for other transcription factors compromises SP-B expression.
The observations that both deletions and insertions of intron 4 are associated with lung diseases, and that deletions result in increased amounts of incompletely spliced SP-B RNA, indicate that the length of intron 4 may contribute to lung diseases by affecting SP-B RNA processing. For a long time, biologists have argued that alternating purines and pyrimidines could form an alternative DNA structure that may affect gene transcription. A polymorphic dinucleotide repeat in the rat nucleolin gene can form a Z-DNA structure and inhibit gene transcription [11]. Deletion of a CA repeat in intron 1 of HSD11B2 led to a significant decrease (40%) in the expression of this gene in human cortical collecting duct cells, and its influence on expression was dependent upon the location and orientation of the CA repeat [14]. Length variants of CA repeat polymorphisms have been shown to affect gene expression, and a decrease in HSD11B2 gene expression is associated with a shorter CA repeat length [13]. In SP-B intron 4, deletion of between 193 and 340 nucleotides, a region that contains a high content of CA repeats, is a major change, and could affect SP-B DNA structure and mRNA secondary structure.
For SP-B, intron 4 variants may affect the functional protein not only quantitatively (reducing the amount of a normal-sized mRNA), but also qualitatively (altering gene function). Incompletely spliced SP-B RNA containing intron 4 sequences may generate a stop codon and thus produce a truncated protein, or shift the reading frame after exon 4 and thus change the amino acid sequence of the protein. In addition to shifting of the reading frame, the addition of extra amino acids may compromise SP-B function. It is also possible that an incompletely spliced SP-B RNA molecule containing a stop codon can initiate the NMD (nonsense-mediated mRNA decay) pathway to rapidly degrade the aberrant transcripts [47]. Therefore aberrant transcripts may not be readily detectable, and may not be translated into aberrant protein molecules. However, in the present study, the amount of incompletely spliced SP-B RNA was increased on expression of the deletion variants. Although it is not known whether these incompletely spliced transcripts are translated, a decline in normal SP-B mRNA content and an increase in incompletely spliced SP-B RNA may lead to insufficient amounts of functional SP-B protein and/or to a dysfunctional SP-B protein. Both of these possibilities may fail to support normal lung function. This may be one mechanism through which SP-B contributes to disease susceptibility. The observation that lung tumour tissues with an SP-B intron 4 deletion genotype were associated with an increased number of seemingly abnormally spliced SP-B mRNAs supports the notion that SP-B intron 4 abnormalities may increase disease risk or disease severity. Further support for this possibility is provided by genetic associations of SP-B intron 4 length variants with several lung diseases. These include RDS [16,17], ARDS [20], cancer [23], COPD [22] and most recently BPD [21], where the frequency of intron 4 deletion variants was increased in infants with BPD when compared with those at risk who did not develop BPD [21]. Thus intron 4 deletion variants in the presence of unknown insults (as may be the case in cancer) may enhance abnormal splicing. Further study is necessary to determine the impact of the alternatively spliced SP-B RNA on the quantity and/or function of SP-B, as well as on mechanisms involved in the pathogenesis of lung disease.
In summary, using transient transfections of a variety of constructs of intron 4 variants, we have shown that the length of intron 4 of the human SP-B gene influences splicing of the exon 4/intron 4 junction. We also showed that (1) inappropriately spliced SP-B RNA containing intron 4 sequences is detectable in normal lung tissue, and (2) a larger amount of incompletely spliced SP-B mRNA was evident in cancerous tissues with an intron 4 deletion genotype.
Acknowledgments
We thank Jelena Pavlovic, Brana Janic and Susan DiAngelo for reading the manuscript. This work is supported by NIH grants R37HL34788, ALA RG-066-N. We gratefully acknowledge the Gift of Life Donor Program (Philadelphia, PA, U.S.A.) and the generosity of the organ donor families for allowing these organs that are not suitable for transplantation to be utilized to advance the understanding of human disease.
References
- 1.Subramanian S., Mishra R. K., Singh L. Genome-wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biol. 2003;4:R13. doi: 10.1186/gb-2003-4-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu C. W., Chen G. D., Jiang K. C., Li A. F., Chi C. W., Lo S. S., Chen J. Y. A genome-wide study of microsatellite instability in advanced gastric carcinoma. Cancer. 2001;92:92–101. doi: 10.1002/1097-0142(20010701)92:1<92::aid-cncr1296>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 3.Enomoto A., Esumi M., Yamashita K., Takagi K., Takano S., Iwai S. Abnormal nucleotide repeat sequence in the TGF-betaRII gene in hepatocellular carcinoma and in uninvolved liver tissue. J. Pathol. 2001;195:349–354. doi: 10.1002/path.963. [DOI] [PubMed] [Google Scholar]
- 4.Merlo A., Mabry M., Gabrielson E., Vollmer R., Baylin S. B., Sidransky D. Frequent microsatellite instability in primary small cell lung cancer. Cancer Res. 1994;54:2098–2101. [PubMed] [Google Scholar]
- 5.Cario E., Podolsky D. K. Differential alteration in intestinal epithelial cell expression of toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect. Immun. 2000;68:7010–7017. doi: 10.1128/iai.68.12.7010-7017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993. pp. 971–983. [DOI] [PubMed]
- 7.Djian P., Hancock J. M., Chana H. S. Codon repeats in genes associated with human diseases: fewer repeats in the genes of nonhuman primates and nucleotide substitutions concentrated at the sites of reiteration. Proc. Natl. Acad. Sci. U.S.A. 1996;93:417–421. doi: 10.1073/pnas.93.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomita N., Fujita R., Kurihara D., Shindo H., Wells R. D., Shimizu M. Effects of triplet repeat sequences on nucleosome positioning and gene expression in yeast minichromosomes. Nucleic Acids Res. Suppl. 2002;2:231–232. doi: 10.1093/nass/2.1.231. [DOI] [PubMed] [Google Scholar]
- 9.Liu L., Panangala V. S., Dybvig K. Trinucleotide GAA repeats dictate pMGA gene expression in Mycoplasma gallisepticum by affecting spacing between flanking regions. J. Bacteriol. 2002;184:1335–1339. doi: 10.1128/JB.184.5.1335-1339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L., Dybvig K., Panangala V. S., van Santen V. L., French C. T. GAA trinucleotide repeat region regulates M9/pMGA gene expression in Mycoplasma gallisepticum. Infect. Immun. 2000;68:871–876. doi: 10.1128/iai.68.2.871-876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rothenburg S., Koch-Nolte F., Rich A., Haag F. A polymorphic dinucleotide repeat in the rat nucleolin gene forms Z-DNA and inhibits promoter activity. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8985–8990. doi: 10.1073/pnas.121176998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Streelman J. T., Kocher T. D. Microsatellite variation associated with prolactin expression and growth of salt-challenged tilapia. Physiol. Genomics. 2002;9:1–4. doi: 10.1152/physiolgenomics.00105.2001. [DOI] [PubMed] [Google Scholar]
- 13.Agarwal A. K., Giacchetti G., Lavery G., Nikkila H., Palermo M., Ricketts M., McTernan C., Bianchi G., Manunta P., Strazzullo P., et al. CA-repeat polymorphism in intron 1 of HSD11B2: effects on gene expression and salt sensitivity. Hypertension. 2000;36:187–194. doi: 10.1161/01.hyp.36.2.187. [DOI] [PubMed] [Google Scholar]
- 14.Agarwal A. K. Transcriptional influence of two poly purine-pyrimidine tracts located in the HSD11B2 (11beta-hydroxysteroid dehydrogenase type 2) gene. Endocr. Res. 2001;27:1–9. doi: 10.1081/erc-100107163. [DOI] [PubMed] [Google Scholar]
- 15.Gebhardt F., Zanker K. S., Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J. Biol. Chem. 1999;274:13176–13180. doi: 10.1074/jbc.274.19.13176. [DOI] [PubMed] [Google Scholar]
- 16.Floros J., Veletza S. V., Kotikalapudi P., Krizkova L., Karinch A. M., Friedman C., Buchter S., Marks K. Dinucleotide repeats in the human surfactant protein-B gene and respiratory-distress syndrome. Biochem. J. 1995;305:583–590. doi: 10.1042/bj3050583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kala P., Ten Have T., Nielsen H., Dunn M., Floros J. Association of pulmonary surfactant protein A (SP-A) gene and respiratory distress syndrome: interaction with SP-B. Pediatr. Res. 1998;43:169–177. doi: 10.1203/00006450-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 18.Floros J., Kala P. Surfactant proteins: molecular genetics of neonatal pulmonary diseases. Annu. Rev. Physiol. 1998;60:365–384. doi: 10.1146/annurev.physiol.60.1.365. [DOI] [PubMed] [Google Scholar]
- 19.Floros J., Lin Z. Genetic variability of surfactant protein-B and respiratory distress diseases. Medscape General Medicine 1. 1999 1999 ( http://www.medscape.com/viewarticle/408742?src=search) [Google Scholar]
- 20.Max M., Pison U., Floros J. Frequency of SP-B and SP-A1 gene polymorphisms in the acute respiratory syndrome (ARDS) Appl. Cardiopulm. Physiol. 1996;6:111–118. [Google Scholar]
- 21.Rova M., Haataja R., Marttila R., Ollikainen V., Tammela O., Hallman M. Data mining and multiparameter analysis of lung surfactant protein genes in bronchopulmonary dysplasia. Hum. Mol. Genet. 2004;13:1095–1104. doi: 10.1093/hmg/ddh132. [DOI] [PubMed] [Google Scholar]
- 22.Seifart C., Plagens A., Brodje D., Muller B., Von Wichert P., Floros J. Surfactant protein B intron 4 variation in German patients with COPD and acute respiratory failure. Dis. Markers. 2002;18:129–136. doi: 10.1155/2002/194075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seifart C., Seifart U., Plagens A., Wolf M., von Wichert P. Surfactant protein B gene variations enhance susceptibility to squamous cell carcinoma of the lung in German patients. Br. J. Cancer. 2002;87:212–217. doi: 10.1038/sj.bjc.6600353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.deMello D. E., Lin Z. Pulmonary alveolar proteinosis: a review. Pediatr. Pathol. Mol. Med. 2001;20:413–432. [PubMed] [Google Scholar]
- 25.Tokieda K., Whitsett J. A., Clark J. C., Weaver T. E., Ikeda K., McConnell K. B., Jobe A. H., Ikegami M., Iwamoto H. S. Pulmonary dysfunction in neonatal SP-B-deficient mice. Am. J. Physiol. 1997;273:L875–L882. doi: 10.1152/ajplung.1997.273.4.L875. [DOI] [PubMed] [Google Scholar]
- 26.Clark J. C., Weaver T. E., Iwamoto H. S., Ikegami M., Jobe A. H., Hull W. M., Whitsett J. A. Decreased lung compliance and air trapping in heterozygous SP-B-deficient mice. Am. J. Respir. Cell Mol. Biol. 1997;16:46–52. doi: 10.1165/ajrcmb.16.1.8998078. [DOI] [PubMed] [Google Scholar]
- 27.Clark J. C., Wert S. E., Bachurski C. J., Stahlman M. T., Stripp B. R., Weaver T. E., Whitsett J. A. Targeted disruption of the surfactant protein B gene disrupts surfactant homeostasis, causing respiratory failure in newborn mice. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7794–7798. doi: 10.1073/pnas.92.17.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tokieda K., Ikegami M., Wert S. E., Baatz J. E., Zou Y., Whitsett J. A. Surfactant protein B corrects oxygen-induced pulmonary dysfunction in heterozygous surfactant protein B-deficient mice. Pediatr. Res. 1999;46:708–714. doi: 10.1203/00006450-199912000-00014. [DOI] [PubMed] [Google Scholar]
- 29.Epaud R., Ikegami M., Whitsett J. A., Jobe A. H., Weaver T. E., Akinbi H. T. Surfactant protein B inhibits endotoxin-induced lung inflammation. Am. J. Respir. Cell Mol. Biol. 2003;28:373–378. doi: 10.1165/rcmb.2002-0071OC. [DOI] [PubMed] [Google Scholar]
- 30.Pilot-Matias T. J., Kister S. E., Fox J. L., Kropp K., Glasser S. W., Whitsett J. A. Structure and organization of the gene encoding human pulmonary surfactant proteolipid SP-B. DNA. 1989;8:75–86. doi: 10.1089/dna.1.1989.8.75. [DOI] [PubMed] [Google Scholar]
- 31.Vamvakopoulos N. C., Modi W. S., Floros J. Mapping the human pulmonary surfactant-associated protein B gene (SFTP3) to chromosome 2p12→p11.2. Cytogenet. Cell Genet. 1995;68:8–10. doi: 10.1159/000133878. [DOI] [PubMed] [Google Scholar]
- 32.Jacobs K. A., Phelps D. S., Steinbrink R., Fisch J., Kriz R., Mitsock L., Dougherty J. P., Taeusch H. W., Floros J. Isolation of a cDNA clone encoding a high molecular weight precursor to a 6-kDa pulmonary surfactant-associated protein. J. Biol. Chem. 1987;262:9808–9811. [PubMed] [Google Scholar]
- 33.Weaver T. E. Synthesis, processing and secretion of surfactant proteins B and C. Biochim. Biophys. Acta. 1998;1408:173–179. doi: 10.1016/s0925-4439(98)00066-0. [DOI] [PubMed] [Google Scholar]
- 34.Guttentag S. H., Beers M. F., Bieler B. M., Ballard P. L. Surfactant protein B processing in human fetal lung. Am. J. Physiol. 1998;275:L559–L566. doi: 10.1152/ajplung.1998.275.3.L559. [DOI] [PubMed] [Google Scholar]
- 35.Hawgood S., Derrick M., Poulain F. Structure and properties of surfactant protein B. Biochim. Biophys. Acta. 1998;1408:150–160. doi: 10.1016/s0925-4439(98)00064-7. [DOI] [PubMed] [Google Scholar]
- 36.Lin Z., Floros J. Heterogeneous allele expression of pulmonary SP-D gene in rat large intestine and other tissues. Physiol. Genomics. 2002;11:235–243. doi: 10.1152/physiolgenomics.00061.2002. [DOI] [PubMed] [Google Scholar]
- 37.Lin Z., deMello D. E., Batanian J. R., Khammash H. M., DiAngelo S., Luo J., Floros J. Aberrant SP-B mRNA in lung tissue of patients with congenital alveolar proteinosis (CAP) Clin. Genet. 2000;57:359–369. doi: 10.1034/j.1399-0004.2000.570506.x. [DOI] [PubMed] [Google Scholar]
- 38.Lin Z., deMello D. E., Wallot M., Floros J. An SP-B gene mutation responsible for SP-B deficiency in fatal congenital alveolar proteinosis: evidence for a mutation hotspot in exon 4. Mol. Genet. Metab. 1998;64:25–35. doi: 10.1006/mgme.1998.2702. [DOI] [PubMed] [Google Scholar]
- 39.Cartegni L., Chew S. L., Krainer A. R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 40.Xu Q., Modrek B., Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez A. J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 42.Kashima T., Manley J. L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 43.Singh N. N., Androphy E. J., Singh R. N. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem. Biophys. Res. Commun. 2004;315:381–388. doi: 10.1016/j.bbrc.2004.01.067. [DOI] [PubMed] [Google Scholar]
- 44.Zahler A. M., Damgaard C. K., Kjems J., Caputi M. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J. Biol. Chem. 2004;279:10077–10084. doi: 10.1074/jbc.M312743200. [DOI] [PubMed] [Google Scholar]
- 45.Buchner D. A., Trudeau M., Meisler M. H. SCNM1, a putative RNA splicing factor that modifies disease severity in mice. Science. 2003;301:967–969. doi: 10.1126/science.1086187. [DOI] [PubMed] [Google Scholar]
- 46.Li C., Zhu N. L., Tan R. C., Ballard P. L., Derynck R., Minoo P. Transforming growth factor-beta inhibits pulmonary surfactant protein B gene transcription through SMAD3 interactions with NKX2.1 and HNF-3 transcription factors. J. Biol. Chem. 2002;277:38399–38408. doi: 10.1074/jbc.M203188200. [DOI] [PubMed] [Google Scholar]
- 47.Singh G., Lykke-Andersen J. New insights into the formation of active nonsense-mediated decay complexes. Trends Biochem. Sci. 2003;28:464–466. doi: 10.1016/S0968-0004(03)00176-2. [DOI] [PubMed] [Google Scholar]