Abstract
Inclusion of the PPARα (peroxisome-proliferator-activated receptor α) activator WY 14,643 in the diet of normal mice stimulated the hepatic expression of not only genes of the fatty acid oxidation pathway, but also those of the de novo lipid synthetic pathways. Induction of fatty acid synthase mRNA by WY 14,643 was greater during the light phase of the diurnal cycle, when food intake was low and PPARα expression was high. Hepatic fatty acid pathway flux in vivo showed a similar pattern of increases. The abundance of mRNAs for genes involved in hepatic cholesterol synthesis was also increased by WY 14,643, but was associated with a decrease in cholesterogenic carbon flux. None of these changes were apparent in PPARα-null mice. Mice of both genotypes showed the expected decreases in 3-hydroxy-3-methylglutaryl-CoA reductase mRNA levels and cholesterol synthesis in response to an increase in dietary cholesterol. The increase in fatty acid synthesis due to WY 14,643 was not mediated by increased expression of SREBP-1c (sterol regulatory element binding protein-1c) mRNA, but by an increase in cleavage of the protein to the active form. An accompanying rise in stearoyl-CoA desaturase mRNA expression suggested that the increase in lipogenesis could have resulted from an alteration in membrane fatty acid composition that influenced SREBP activation.
Keywords: cholesterol and fatty acid synthesis, fibrate, gene expression, peroxisome-proliferator-activated receptor α (PPARα), PPARα-null mice, sterol regulatory element binding protein (SREBP)
Abbreviations: ABCA1, ATP-binding cassette transporter A1; ACC, acetyl-CoA carboxylase; Cyp7A1, cholesterol 7α-hydroxylase; FAS, fatty acid synthase; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; LDL, low-density lipoprotein; LXR, liver X receptor; PPAR, peroxisome-proliferator-activated receptor; SCAP, SREBP cleavage-activating protein; SREBP, sterol regulatory element binding protein; SCD-1, stearoyl-CoA desaturase
INTRODUCTION
PPARα (peroxisome-proliferator-activated receptor α) is a member of the nuclear hormone receptor family of transcription factors. It is activated by unsaturated fatty acids and their derivatives, and by pharmacological ligands that include the fibrate group of drugs. PPARα is expressed in the liver, heart, skeletal muscle and brown adipose tissue, where it regulates genes that control mitochondrial and peroxisomal fatty acid oxidation [1]. More recently, a further crucial role for PPARα has been discovered: mediating the effects of leptin in adipose tissue [2].
We have shown previously that PPARα deficiency abolished the normal diurnal variations in the expression of lipogenic genes, such as those encoding FAS (fatty acid synthase), ACC (acetyl-CoA carboxylase) and HMG-CoA (3-hydroxy-3-methylglutaryl-CoA) reductase, in the liver [3]. This disturbance was accompanied by abolition of the corresponding rhythmic changes in the rates of hepatic fatty acid and cholesterol synthesis in vivo. Normally, these diurnal variations are associated with the changing pattern of food intake over the 24 h cycle and are controlled, at least in part, by changes in the plasma insulin concentration. However, the interruption of these natural rhythms in the PPARα-null mice did not result from an abnormal pattern of food intake [3], nor from differences in plasma insulin concentration [4].
The transcription of many lipogenic genes is regulated by members of the SREBP (sterol regulatory element binding protein) family [5]. These sterol-sensitive transcription factors are synthesized as membrane-bound precursor proteins that are cleaved to their mature active form when cellular sterols are depleted. Generally, the SREBP-2 isoform activates genes of the cholesterogenic pathway, whereas the two SREBP-1 isoforms are more active in regulating the synthesis of fatty acids. SREBP-1c is the predominant SREBP-1 isoform in animal tissues, and has been shown [6] to mediate, at least in part, the insulin-dependent regulation of the expression of lipogenic genes by some as yet obscure signalling pathway, which may involve the insulin-sensitive genes INSIG-1 and INSIG-2 [7,8].
Levels of SREBP-1c mRNA were lower in PPARα-null mice than in the wild type [3,9], and remained lower during the changes in insulin concentration that accompanied starvation and refeeding [9]. The concentration of plasma insulin required to sustain a given level of SREBP-1c and FAS mRNA expression, as well as lipogenic carbon flux, was always higher in PPARα-deficient mice than in the wild-type animals [4]. This pattern is suggestive of a degree of insulin-insensitivity in the PPARα-null mice. To investigate this aspect further, we have studied the responses of SREBP-1c and the lipogenic pathway to an increase in PPARα activity resulting from oral administration of a potent PPARα ligand, WY 14,643. Because of the abnormalities of cholesterol biosynthesis that accompanied the disturbed regulation of fatty acid synthesis in the PPARα-null mice [3], we also examined the effects of WY 14,643 on HMG-CoA reductase, SREBP-2 and cholesterogenic carbon flux. In addition, to obtain a wider picture of the effects of fibrate on lipid metabolism, we conducted gene expression analysis using microarrays containing probes for most of the known mouse genes. Throughout, verification of the specificity of PPARα in mediating effects resulting from WY 14,634 was achieved by carrying out identical experiments in PPARα-knockout mice.
MATERIALS AND METHODS
Animals
PPARα-null mice [10] bred on to a SV/129 genetic background were kindly provided by Dr J. Peters and Dr F. J. Gonzalez (National Institutes of Health, Bethesda, MD, U.S.A.). Wild-type SV/129 mice were used as controls. All mice were male, and were used between the ages of 14 and 20 weeks. Animals were maintained in temperature-controlled rooms (22–24 °C) on a 12 h light/12 h dark cycle, and were fed a commercial high-carbohydrate, pelleted diet (Special Diet Service, Witham, Essex, U.K.) containing (by wt) 4.3% fat (0.02% cholesterol), 51.2% carbohydrate (mainly starch), 22.3% protein and 7.7% ash. When required, the diet was supplemented with 2% (w/w) cholesterol (>99% purity; Sigma-Aldrich, Poole, Dorset, U.K.) or 0.1% WY 14,643 (Chemsyn Science Laboratories, Lenexa, KA, U.S.A.) and given for 7 days. Food and, thus, cholesterol consumption was measured on a daily basis, and was similar for each genotype.
Hepatic fatty acid and cholesterol synthesis
Mice were anaesthetized with pentobarbitone and samples of blood were taken from the descending vena cava. The livers were removed and frozen immediately in liquid nitrogen. For assay of the rates of hepatic fatty acid and cholesterol synthesis, mice were injected intraperitoneally with 3H2O (1 mCi) 2 h before they were killed. The specific radioactivity of the plasma water was assayed in samples of blood, and labelled cholesterol and fatty acids were isolated from powdered, frozen livers and assayed as described previously [11,12].
Gene expression
The frozen livers were ground to a powder under liquid nitrogen, and total RNA was extracted from portions of powder using the RNAzol kit (Biogenesis, Poole, Dorset, U.K.). Samples from four animals were pooled, and cRNA was prepared for hybridization with Affymetrix MU74Av2 microarrays according to the manufacturer's protocols (Affymetrix UK Ltd, High Wycombe, Bucks., U.K.). Data were analysed using the GeneSpring analysis program (Silicon Genetics, Redwood City, CA, U.S.A.), and enzymes were listed that showed a 2-fold or greater increase in expression on WY 16,463 feeding. For more detailed measurements, mRNA was assayed by reverse transcription [13] followed by real-time PCR using an ABI PRISM Sequence Detection System (PE Applied Biosystems, Foster City, CA, U.S.A.) with the primers and probes described previously [3,14]. Reactions were carried out in triplicate in 30 μl of TaqMan Universal PCR Master Mix [3] with β-actin as internal standard. All values were related to a curve generated by a standard liver preparation and were corrected for β-actin mRNA content.
SREBP levels
SREBP proteins were extracted [15] from liver by grinding under liquid nitrogen, followed by thawing in ice-cold lysis buffer containing the protease inhibitors PMSF, leupeptin, pepstatin, chymostatin, aprotinin and N-acetyl-leucinal-leucinal-norleucinal (100 μg of each/ml). Samples (20 μg of protein) of the resultant extracts were then analysed by SDS/PAGE on 4–20% (w/v) polyacrylamide gradient gels. Proteins were transferred to nitrocellulose and SREBP was detected with rabbit anti-SREBP-1 antibody (K10; SC-367; Santa Cruz Biotechnology Inc., Santa Cruz, CA, U.S.A.), using a donkey anti-rabbit horseradish peroxidase-conjugated second antibody followed by ECL® visualization (Amersham Biosciences UK Ltd, Little Chalfont, Bucks., U.K.). This primary antibody recognized pure recombinant nuclear SREBP-1c, and using this antibody we could demonstrate the expected increases in both the precursor and mature forms of SREBP-1 on refeeding after starvation.
Other methods
Plasma and tissue lipid concentrations were assayed as described previously [3]. Tissue protein content was determined by the method of Lowry et al. [16]. The statistical significance of diurnal periodicity was assessed by ANOVA, and the statistical analysis of means was carried out using Student's t test for unpaired data.
Effects of fibrate feeding on gene expression and lipid synthesis
The mRNA extracted from the livers of wild-type and PPARα-null mice fed a diet supplemented with 0.1% WY 14,643 for 7 days was hybridized to Affymetrix microarrays containing probes for known mouse genes. As expected (Table 1), when compared with the chow diet, wild-type animals fed on a diet containing the PPARα-activating drug showed an increase in the expression of mRNAs coding for enzymes involved in the fatty acid activation and oxidation pathways, such as acylcarnitine translocase, acylCoA synthetase (EC 6.2.1.3) and hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35). This confirms data obtained with similar arrays by Yamazaki et al. [17]. In addition, several mRNAs encoding enzymes of de novo lipogenesis were induced, including malate dehydrogenase (EC 1.1.1.40), FAS (EC 2.3.1.85) and SCD-1 (stearoyl-CoA desaturase; EC 1.14.99.5). There were also moderate increases in the expression of genes encoding enzymes involved in the cholesterogenic pathway. Many of these genes were expressed at lower levels in the PPARα-null mice, and were not generally induced in these animals by the inclusion of WY 14,643 in the diet (Table 1).
Table 1. Effects of fibrate feeding on expression of enzymes involved in hepatic lipid metabolism.
PPARα-null and wild-type mice were fed a chow diet either supplemented or not with 0.1% (w/w) WY 14,643 for 7 days. The animals were killed at the mid-point of the light phase of the diurnal cycle. Livers were removed and the RNA extracted and hybridized to Affymetrix MU74Av2 microarrays as described in the Materials and methods section. Data were normalized per experiment and per gene using the autonormalization function in the GeneSpring program.
| mRNA abundance (arbitrary units) | |||||
|---|---|---|---|---|---|
| Enzyme | Accession no. | Wild type | Wild type+WY 14,643 | PPARα-null | PPARα-null+WY 14,643 |
| Fatty acid oxidation and ketogenesis | |||||
| Acyl-CoA synthase | Y14004 | 4.0 | 66.2 | 1.9 | 2.4 |
| Cyp4A10 | AB018421 | 13.6 | 108.9 | 0.9 | 0.6 |
| Acyl-CoA oxidase | AF006688 | 21.3 | 86.2 | 15.1 | 20.5 |
| Hydroxyacyl-CoA dehydrogenase | D29639 | 12.0 | 26.2 | 9.8 | 11.6 |
| Acylcarnitine transferase | AB017112 | 6.6 | 28.6 | 3.4 | 4.1 |
| Multifunctional β-oxidation protein | AJ011864 | 6.0 | 65.1 | 1.7 | 1.7 |
| HMG-CoA lyase | U49878 | 18.7 | 66.6 | 21.4 | 25.7 |
| Carnitine palmitoyltransferase-1 | AF017175 | 12.1 | 32.9 | 11.4 | 16.4 |
| Lipid synthesis | |||||
| FAS | X13135 | 8.5 | 21.7 | 8.1 | 7.2 |
| Malate dehydrogenase | J02652 | 3.7 | 48.6 | 2.0 | 2.8 |
| SCD-1 | M21285 | 12.8 | 104.3 | 7.8 | 12.0 |
| Cholest-7-enol desaturase | AB016248 | 3.4 | 8.8 | 4.7 | 3.2 |
| Squalene synthetase | D29016 | 7.3 | 14.8 | 7.9 | 6.9 |
| HMG-CoA synthase | U12791 | 3.0 | 6.1 | 3.6 | 3.7 |
| Diacylglycerol acyltransferase | AF078752 | 1.0 | 2.9 | 1.3 | 1.5 |
| Glycerol-3-phosphate acyltransferase | U11680 | 6.8 | 24.4 | 5.8 | 6.4 |
| Others | |||||
| Cyp8B1 (sterol 12α-hydroxylase) | AF090317 | 15.8 | 32.5 | 24.6 | 17.0 |
| Fatty acid transport protein | AF072757 | 19.8 | 70.7 | 20.1 | 21.0 |
| Cyp2B39 (arachidonate dehydrogenase) | AF047726 | 5.3 | 18.5 | 3.6 | 5.1 |
| 17β-Hydroxysterol dehydrogenase | X89998 | 8.8 | 35.6 | 8.7 | 7.8 |
| Dodecanoyl-CoA δ-isomerase | Z14050 | 2.3 | 27.8 | 1.3 | 1.4 |
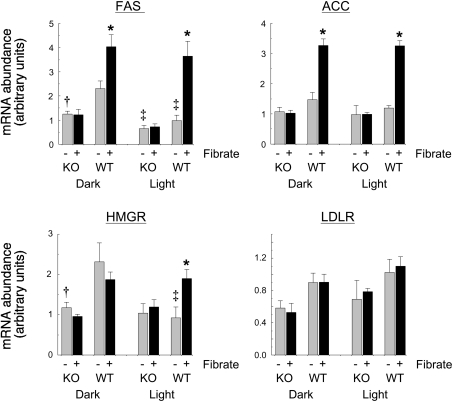
To investigate changes in gene expression more accurately, the hepatic abundance of mRNAs for FAS, ACC and HMG-CoA reductase was determined by quantitative real-time PCR. Because of the pronounced diurnal variations in the abundance of these mRNAs [3], measurements were taken at the mid-point of the dark phase or the mid-point of the light phase of the diurnal cycle (Figure 1). In these experiments, as before [3], values for FAS and HMG-CoA reductase mRNAs in the livers of the wild-type animals were higher during the dark phase of the cycle. Feeding with the fibrate increased the abundance of mRNAs for FAS and ACC in the wild-type mice, but not in the PPARα-null mice. For FAS, the increase was greater in the light than in the dark so that, as for ACC, there was no longer any difference between the phases (Figure 1). WY 14,643 did not affect the hepatic abundance of mRNA for HMG-CoA reductase during the dark phase, but increased it during the light phase, again to the same level as that observed in the dark. There was no effect of the fibrate at either time point in the PPARα-null mice. Fibrate feeding did not affect the hepatic abundance of mRNA for the LDL (low-density lipoprotein) receptor in either the wild-type or the PPARα-null mice (Figure 1).
Figure 1. Effects of fibrate feeding on the expression of hepatic lipogenic genes.
Wild-type (WT) and PPARα-null (KO) mice were fed a diet with (+) or without (−) 0.1% of the potent fibrate WY 14,643 for 7 days, and were killed at the mid point of the dark phase or the mid point of the light phase of the diurnal cycle. Livers were assayed for their content of mRNAs for FAS, ACC, HMG-CoA reductase (HMGR) and LDL receptor (LDLR). Values are given in arbitrary units relative to a standard preparation and are means±S.E.M. of results from eight mice under each condition in the dark and four mice under each condition in the light. Significant differences: *P<0.05 for effect of fibrate feeding; †P<0.05 for difference between WT and KO; ‡P<0.05 for diurnal variation.
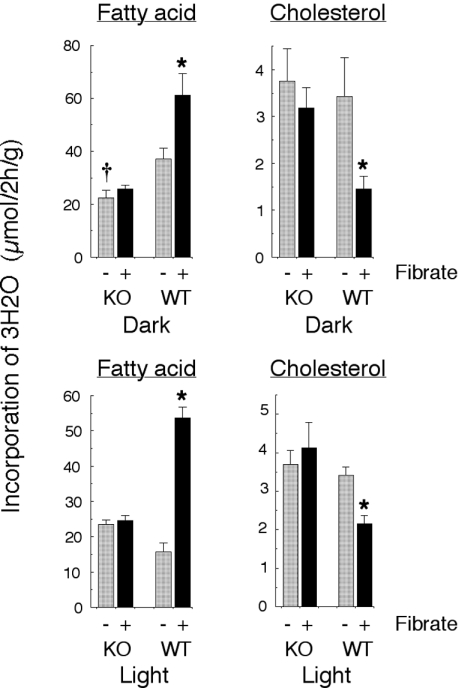
During the dark phase of the cycle, the increase in abundance of mRNAs for FAS and ACC on feeding WY 14,643 to the wild-type mice was reflected in an increase in the incorporation of 3H2O into liver fatty acids (Figure 2). In contrast, incorporation into cholesterol was significantly decreased. A similar pattern was observed during the light phase of the cycle. In this case, the fibrate-mediated increase in fatty acid synthesis in the wild-type mice was potentiated compared with that in the dark phase, which paralleled the changes in FAS and ACC mRNAs. Again, there was an anomalous decrease in the rate of cholesterol synthesis. There were no effects of fibrate feeding on cholesterol or fatty acid synthesis at either time in the livers of the PPARα-null mice (Figure 2).
Figure 2. Effects of fibrate on hepatic lipid synthesis.
Wild-type (WT) and PPARα-null (KO) mice were fed a diet with (+) or without (−) 0.1% WY 14,643 for 7 days, and were killed at the mid point of the dark phase or the mid point of the light phase of the diurnal cycle, 2 h after injection of 1 mCi of 3H2O. Results are shown for the incorporation of 3H2O into liver fatty acids and cholesterol, and values are means±S.E.M. of values from eight mice in the dark and four mice in the light; *P<0.05 for effect of fibrate feeding.
Diurnal changes in hepatic gene expression
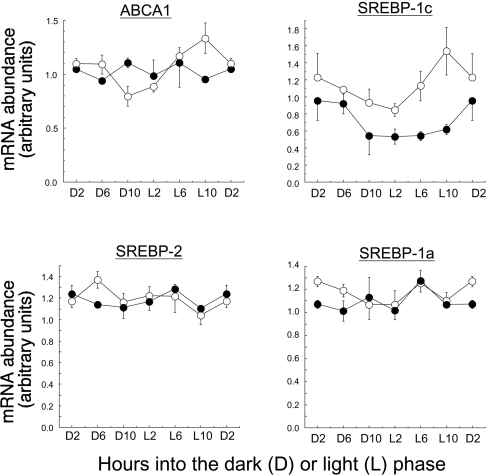
Our results showed that the effects of fibrate feeding on gene expression in the livers of the wild-type mice were generally more pronounced during the light phase of the diurnal cycle, i.e. the time at which PPARα mRNA expression is maximal [3]. FAS and ACC transcription can be regulated by SREBP-1, raising the possibility that this transcription factor could mediate any link between PPARα activation and increased lipogenesis. Measurements of hepatic SREBP-1c mRNA content in wild-type mice (Figure 3) showed a diurnal periodicity (P<0.05), with a pattern broadly similar to that of PPARα mRNA [3]. In the PPARα-null mice, hepatic SREBP-1c mRNA levels were generally lower than in the wild type (Figure 3), and diurnal changes were attenuated or abolished. Interestingly, the expression of the putative cholesterol transporter ABCA1 (ATP-binding cassette transporter A1), which is stimulated by PPARα in human macrophages [18], also showed a diurnal pattern of expression (P<0.05), which was almost superimposable upon that of SREBP-1c and PPARα. Again, this rhythm was abolished in the livers of the PPARα-null mice (Figure 3). There was no diurnal fluctuation in the expression of SREBP-1a or SREBP-2 mRNAs, nor any differences in expression between the genotypes (Figure 3).
Figure 3. Diurnal variations in the content of mRNAs for ABCA1 and the SREBP isoforms in the livers of wild-type (○) and PPARα-null (●) mice.
Results shown are the means±S.E.M. of values from four mice at each time point, given as the number of hours into the dark (D) or light (L) phase of the diurnal cycle. Values are expressed relative to β-actin mRNA, which did not change relative to 18 S RNA during the cycle.
Fibrate feeding does not increase SREBP-1c mRNA levels, but enhances cleavage of the immature protein
To discover if the effects of fibrate on the expression of hepatic lipogenic genes could be mediated by changes in SREBP expression, mRNA levels for the three SREBP isoforms were measured in the livers of wild-type and PPARα-null mice fed WY 14,643. There was no effect of fibrate on the abundance of mRNAs for any of the SREBP isoforms in either the wild-type or the PPARα-null animals (Table 2). Transcription of the SREBP-1c gene is known to be stimulated by the oxysterol-activated factor LXRα (liver X receptor α) [19,20], which also increases the transcription of ABCA1 [21–23] and Cyp7A1 (cholesterol 7α-hydroxylase) [24,25]. However, fibrate feeding did not increase the mRNA levels for either of these proteins, nor for LXRα itself, in the wild-type livers (Table 2), and there was no effect of PPARα deficiency. In fact, the mRNA level for Cyp7A1 was significantly reduced by fibrate feeding, but only in the wild-type animals. This may have resulted from a decrease in the hepatic cholesterol concentration (see below). In contrast, the mRNA level for a known PPARα-responsive gene, acyl-CoA oxidase, was increased over 5-fold by fibrate feeding in wild-type mice, and was significantly reduced by PPARα deficiency (Table 2).
Table 2. Effects of fibrate feeding on hepatic mRNA levels.
Wild-type and PPARα-null mice were fed a diet with or without 0.1% WY 14,643 for 7 days, and were killed at the mid point of the dark phase or the mid point of the light phase of the diurnal cycle. Livers were assayed for their content of mRNAs for the genes shown by real-time PCR. Values from the dark and light phases were not different and were combined. They are given in arbitrary units relative to a standard preparation and are means±S.E.M. of results from the numbers of mice shown in parentheses. Significant differences: *P<0.05 for effect of fibrate feeding; †P<0.05 for difference between wild type and PPARα-null.
| mRNA abundance (arbitrary units) | ||||
|---|---|---|---|---|
| Protein | Wild-type | Wild-type+WY 14,643 | PPARα-null | PPARα-null+WY 14,643 |
| SREBP-2 (11) | 0.94±0.08 | 1.13±0.08 | 0.84±0.09 | 0.75±0.03 |
| SREBP-1a (8) | 0.76±0.05 | 0.68±0.03 | 0.59±0.08 | 0.63±0.05 |
| SREBP-1c (11) | 1.15±0.08 | 1.23±0.11 | 0.76±0.05† | 0.78±0.06 |
| INSIG-2a (8) | 0.31±0.03 | 0.36±0.07 | 0.39±0.02 | 0.30±0.04 |
| ABCA1 (8) | 0.80±0.08 | 0.81±0.04 | 0.71±0.05 | 0.78±0.06 |
| Cyp7A1 (8) | 1.36±0.19 | 0.63±0.05* | 1.11±0.12 | 1.20±0.14 |
| LXRα (8) | 0.70±0.05 | 0.61±0.05 | 0.66±0.07 | 0.68±0.05 |
| PPARα (8) | 0.99±0.05 | 0.90±0.07 | − | − |
| PPARγ (6) | 1.23±0.15 | 1.63±0.13 | 1.84±0.31 | 2.36±0.30 |
| Acyl-CoA oxidase (8) | 0.89±0.07 | 5.21±0.67* | 0.58±0.07† | 0.69±0.04 |
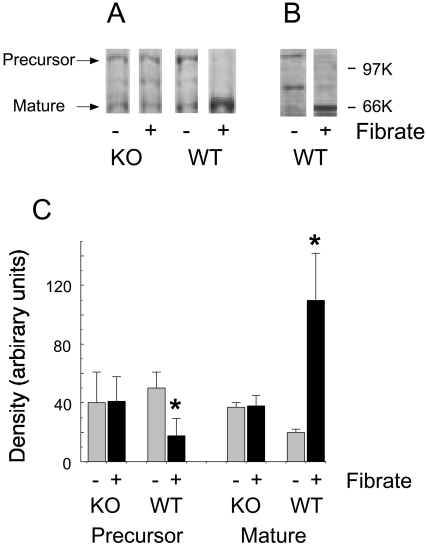
The absence of any increase in SREBP-1c mRNA expression following fibrate feeding suggested that the observed stimulation of fatty acid synthesis could have resulted from an effect of fibrate on SREBP cleavage. To test this proposition, we subjected liver extracts to Western blotting. The blots showed that fibrate feeding decreased the amount of SREBP-1 precursor protein in the livers of wild-type mice and increased the amount of the mature, active form (Figures 4A and 4B). These changes were not observed in the livers of the PPARα-null mice (Figure 4A). Results from a number of independent experiments showed a statistically significant decrease in the precursor form of SREBP-1 on WY 14,643 feeding and a significant increase in the mature, active form (Figure 4C).
Figure 4. Response of SREBP-1 cleavage to fibrate administration.
Wild-type (WT) and PPARα-null (KO) mice were fed a chow-based diet alone (−) or a diet supplemented (+) with 0.1% (w/w) WY 14,643 for 7 days. The mice were killed at the mid-point of the dark phase of the diurnal cycle, and the livers were frozen in liquid nitrogen. Proteins were separated by SDS/PAGE and transferred to nitrocellulose. The mature and precursor forms of SREBP-1 were detected using a polyclonal antibody as the primary antibody. (A) Blots from whole livers from WT and KO mice; (B) blots from different whole WT livers; (C) relative intensities (means±S.E.M.) of the precursor and mature forms of SREBP-1 obtained by quantitative scanning of the blots (four mice in each group). *P<0.01 for effect of fibrate feeding.
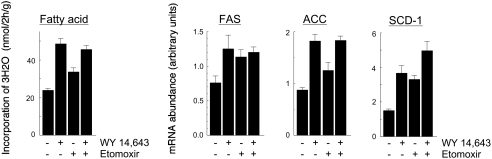
The increase in lipogenesis following WY 14,643 feeding could have been related to the well known ability of fibrates to stimulate the oxidation of fatty acids. However, addition to the diet of etomoxir, which inhibits the fatty acid oxidation [26], did not abolish the effect of WY 14,643 on the incorporation of 3H2O into fatty acids, and nor did it abolish the increases in mRNA levels for FAS, ACC or SCD-1 (Figure 5). Nevertheless, the stimulatory effect of WY 14,643 was less pronounced if etomoxir were present in the diet than if it were absent.
Figure 5. Effects of etomoxir on fatty acid synthesis.
Wild-type mice were fed a diet with (+) or without (−) 0.1% (w/w) WY 14,643 or 0.02% (w/w) etomoxir for 7 days, and were killed at the mid-point of the light phase of the diurnal cycle, 2 h after injection of 1 mCi of 3H2O. Results are shown for the incorporation of 3H2O into liver fatty acids, and for the hepatic content of mRNAs for FAS, ACC, and SCD-1. Values are means±S.E.M. from five mice under each condition. All effects of WY 14,643 and etomoxir alone were statistically significant (P<0.05).
The effects on SREBP-1c cleavage and lipogenesis could also have been related to changes in membrane fatty acid composition. The ratio of C18:1 to C18:0 fatty acids in hepatic membranes increased (P<0.05) from 0.60±0.02 to 1.18±0.06 (mean±S.E.M. for four mice) on feeding wild-type mice with fibrate. This effect was absent in the PPARα-null mice, which exhibited ratios of 0.38±0.02 in the absence and 0.45±0.03 in the presence of dietary fibrate.
PPARα deficiency does not affect the hepatic response to cholesterol feeding
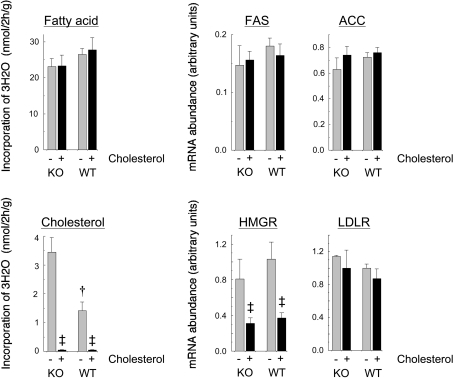
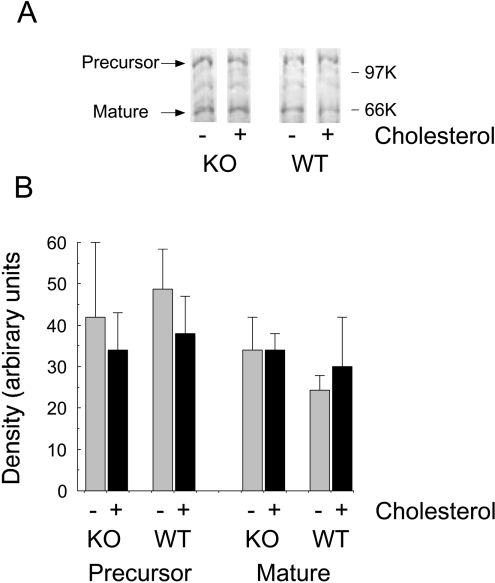
The lack of response of cholesterogenesis to fibrate feeding in the livers of the PPARα-null mice (Figure 2) suggested that the normal regulation of cholesterol synthesis might be impaired under conditions of PPARα deficiency. To examine this possibility, we fed the mice a diet containing 2% (w/w) cholesterol for 7 days. Figure 6 shows that the rate of cholesterogenesis was greatly diminished following cholesterol feeding in mice of both genotypes, and that the expression of HMG-CoA reductase mRNA was decreased by a similar amount in the wild-type and PPARα-null mice. There was no effect on LDL receptor mRNA expression. Cholesterol feeding also had no effect on pathway flux into fatty acids, or on the expression of ACC or FAS mRNAs in either genotype. Notwithstanding the constancy of these parameters of de novo lipogenic activity, cholesterol feeding resulted in a large increase in the expression of SREBP-1c mRNA in the livers of mice of both genotypes (Table 3). However, this cholesterol-mediated increase in the abundance of SREBP-1c mRNA failed to promote a similar increase in the concentration of the mature transcription factor, as measured by immunoblotting (Figure 7). No increases were observed in the expression of SREBP-1a or SREBP-2 mRNAs (Table 3). Indeed, that of SREBP-2 decreased with cholesterol feeding. In both genotypes, the mRNAs for the LXRα-activated genes Cyp7a1 and Abca1 increased in response to cholesterol feeding, although there were no effects on the level of LXRα mRNA. Interestingly, PPARγ expression was stimulated by cholesterol feeding in both the wild type and PPARα-null mice.
Figure 6. Effects of a high-cholesterol diet on cholesterogenesis.
Wild-type (WT) and PPARα-null (KO) mice were fed a diet with (+) or without (−) 2% (w/w) cholesterol for 7 days, and were killed at the mid point of the light phase of the diurnal cycle, 2 h after injection of 1 mCi of 3H2O. Results are shown for the incorporation of 3H2O into liver fatty acids and cholesterol, and for the hepatic content of mRNAs for FAS, ACC, HMG-CoA reductase (HMGR) and LDL receptor (LDLR). Values are the means±S.E.M. from six mice under each condition. Significant differences: ‡P<0.05 for effect of cholesterol feeding; †P<0.05 for difference between WT and KO.
Table 3. Effects of cholesterol feeding on hepatic mRNA levels.
Wild-type and PPARα-null mice were fed a diet with or without 2% (w/w) cholesterol for 7 days, and were killed at the mid point of the dark phase of the diurnal cycle. Livers were assayed for their content of mRNA for the genes shown by real-time PCR. Values are given in arbitrary units relative to a standard preparation, and are means±S.E.M. of results from six mice. Significant differences: *P<0.05 for effect of cholesterol feeding; †P<0.05 for difference between wild-type and PPARα-null.
| mRNA abundance (arbitrary units) | ||||
|---|---|---|---|---|
| Protein | Wild-type | Wild-type+cholesterol | PPARα-null | PPARα-null+cholesterol |
| SREBP-2 (6) | 1.04±0.03 | 0.88±0.05* | 1.15±0.06 | 0.79±0.03* |
| SREBP-1a (6) | 1.09±0.06 | 1.27±0.05 | 0.94±0.08 | 1.13±0.08 |
| SREBP-1c (6) | 1.28±0.12 | 2.22±0.16* | 0.88±0.08† | 2.62±0.24* |
| INSIG-2a (6) | 0.75±0.04 | 0.71±0.07 | 0.82±0.07 | 0.83±0.03 |
| ABCA1 (6) | 0.91±0.05 | 1.23±0.05* | 0.90±0.03 | 1.21±0.06* |
| Cyp7A1 (6) | 0.96±0.24 | 3.80±0.68* | 1.92±1.02 | 4.12±1.44 |
| LXRα (6) | 0.95±0.14 | 0.96±0.08 | 0.86±0.13 | 0.94±0.09 |
| PPARα (6) | 0.92±0.07 | 1.04±0.08 | − | − |
| PPARγ (6) | 0.88±0.14 | 2.02±0.32* | 3.64±0.76† | 7.10±0.61* |
Figure 7. Response of SREBP-1 cleavage to dietary cholesterol.
Wild-type (WT) and PPARα-null (KO) mice were fed a diet with (+) or without (−) 2% (w/w) cholesterol for 7 days, and were killed at the mid point of the light phase of the diurnal cycle. The mature and precursor forms of SREBP-1 were detected as described in the legend to Figure 4. (A) Blots from whole livers; (B) relative intensities (means±S.E.M.) of the precursor and mature forms of SREBP-1 obtained by quantitative scanning of the blots (four mice in each group).
Effects of fibrate and cholesterol on plasma and liver lipid levels
The overall effects of fibrate or cholesterol feeding on lipid concentrations in the mice are shown in Table 4. Neither of the treatments had any significant effect on the plasma non-esterified fatty acid concentration. However, fibrate produced a 45% decrease in plasma triacylglycerol concentration in the wild-type mice, without affecting the liver triacylglycerol concentration. It also lowered the concentration of cholesterol in the liver, with a slight, but not significant (P=0.10), reduction in the plasma cholesterol level. There were no effects of fibrate on lipid levels in the plasma or livers of the PPARα-null mice. Dietary cholesterol, as expected, gave rise to significant increases in the concentrations of cholesterol in the liver and plasma of both genotypes. Surprisingly, there was also a significant increase in hepatic triacylglycerol, but not in plasma triacylglycerol, in both types of mice.
Table 4. Effects of fibrate and cholesterol feeding on plasma and liver lipid levels.
Wild-type and PPARα-null mice were fed a diet with or without 0.1% WY 14,643 or 2% cholesterol for 7 days. Plasma and livers were assayed for the lipids shown. Values are means±S.E.M. of results from 26 mice with no addition, 16 mice fed with fibrate and 12 mice fed with cholesterol. Significant differences: *P<0.05 for effect of fibrate feeding; †P<0.05 for difference between wild-type and PPARα-null; ‡P<0.05 for effect of cholesterol. NEFA, non-esterified fatty acids; TAG, triacylglycerol.
| Plasma | Liver | ||||||
|---|---|---|---|---|---|---|---|
| Animals and treatment | NEFA (μmol/l) | Total cholesterol (mmol/l) | Ester (%) | TAG (μmol/l) | Total cholesterol (μmol/g) | Ester (%) | TAG (μmol/g) |
| Wild-type | 337±23 | 2.82±0.11 | 68.7 | 844±52 | 10.03±0.99 | 21.6 | 15.3±2.2 |
| Wild-type+WY 14,643 | 405±32 | 2.54±0.12 | 66.6 | 476±39* | 7.41±0.49* | 16.7 | 16.6±2.1 |
| Wild-type+cholesterol | 317±26 | 3.62±0.29‡ | 70.6 | 617±91‡ | 20.96±1.16‡ | 41.2 | 31.93±4.1‡ |
| PPARα-null | 353±36 | 3.26±0.10† | 69.4 | 721±41 | 11.69±0.94 | 18.1 | 20.4±3.5 |
| PPARα-null+WY 14,643 | 378±63 | 2.95±0.08 | 66.7 | 793±79 | 9.91±1.21 | 20.1 | 22.6±5.2 |
| PPARα-null+cholesterol | 323±27 | 3.92±0.25‡ | 71.1 | 649±77 | 24.63±2.38‡ | 46.9 | 45.9±9.1‡ |
DISCUSSION
The results presented here indicate that fibrate feeding activates a portfolio of genes involved in lipid synthesis in the liver (Table 1). Furthermore, the extent of the effects on ACC and FAS was greater during the light phase of the diurnal cycle, when food intake is relatively low and PPARα is maximally expressed [3]. The physiological significance of these changes at the transcriptional level was apparent from the increase in hepatic fatty acid synthesis, as determined by the incorporation of 3H2O in vivo. These effects were mediated by PPARα, since they were not observed in PPARα-null animals. The lack of response to fibrate in the knock-out mice resembles the unresponsiveness of lipogenic genes and fatty acid synthetic rate in this genotype to the increase in food intake that occurs during the dark phase of the diurnal cycle [3]. The observed effects of PPARα activation in the wild-type but not in the PPARα-null mice support the proposal that PPARα is involved in the transduction of dietary signals to regulate lipid metabolism in the liver.
There is no evidence for a direct effect of PPARα on the promoter regions of the FAS and ACC genes, so it is likely that the observed changes are an indirect result of PPARα activation. A promising candidate target for these effects is SREBP-1c, since the mature form of this transcription factor is known to activate fatty acid synthesis by increasing transcription of the lipogenic genes [5,27,28]. However, the actions of fibrate were effected without any change in the abundance of SREBP-1c mRNA (Table 2). Conversely, cholesterol feeding, which gave rise to a considerable increase in SREBP-1c abundance (Table 3), probably as a result of sterol-mediated activation of LXRα [29], produced no change either in the expression of FAS and ACC mRNAs or in the rate of lipogenic pathway flux. It would appear, therefore, that there is no invariable relationship between the expression of SREBP-1c mRNA and downstream effects on lipogenesis. These examples highlight the difficulty in precisely predicting the metabolic consequences of physiological changes in vivo simply on the basis of changes in gene expression alone.
SREBP activity may, of course, be regulated at the post-translational as well as at the transcriptional level by alterations in the rate of proteolytic cleavage of the membrane-bound precursor form to give the mature transcription factor. The stimulatory effect of fibrates on the hepatic concentration of the mature form of SREBP-1 in the wild-type mice (Figure 4) provides a likely explanation for the increases in the lipogenic parameters observed. This notion is supported by the lack of effect of fibrate feeding on SREBP-1 maturation in the livers of the PPARα-null mice. On the other hand, cholesterol feeding, which increased SREBP-1c mRNA expression, had no effect on the concentration of the mature form of the transcription factor in mice of either genotype. This would explain the impotence of dietary cholesterol in effecting changes in hepatic de novo lipogenesis in the mouse (Figure 6), as was found many years ago to be the case in the dog [30]. It is likely that an LXR-mediated increase in SREBP-1c transcription resulting from the increase in dietary cholesterol represents a compensatory response to maintain a constant rate of mature SREBP-1c production in the face of an incipient constraint on cleavage imposed by an increased sterol content of intracellular membranes.
The observed relationship between fibrate feeding, lipogenesis and SREBP-1 maturation in the wild-type mice raises the question of the mechanisms by which these parameters are linked. It is well known that fibrate-mediated activation of PPARα increases the rate of fatty acid oxidation [1]. In the present work, this is supported by the increased expression of acyl-CoA oxidase (Table 2), the increased expression of genes for enzymes involved in fatty acid activation, oxidation and ketogenesis, such as acyl-CoA synthase, Cyp4A10, hydroxyacyl-CoA dehydrogenase and HMG-CoA synthase (Table 1), and the decreased plasma concentration of triacylglycerol (Table 4). None of these effects was observed in the PPARα-null mice. However, the lack of effect of etomoxir indicates that the stimulation of lipogenesis by fibrate is not merely a compensatory response to the increase in fatty acid oxidation.
One of the most marked effects of fibrate feeding was on the expression of SCD-1 (Table 1), which would be predicted to increase the degree of unsaturation of cellular fatty acids. Indeed, we observed a doubling of the ratio of C18:1 to C18:0 fatty acids in hepatic membranes on feeding wild-type mice with fibrate. In invertebrates at least, changes in fatty acid composition can regulate SREBP cleavage and are an important means of maintaining the physicochemical homoeostasis of intracellular membrane structures [31,32]. Polyunsaturated fatty acids and WY 14,643 have been shown to reduce SREBP-1 maturation and lipogenesis in HEK cells [33] and rat liver [34,35]. This also seems to apply to oleate, although the data are less consistent. In contrast, the experiments described here with WY 16,463, and recently by Miyazaki et al. [36] with dietary oleate, showed a stimulation of lipogenesis. However, a high dietary oleate concentration of 20% (w/w) was necessary to achieve this stimulation, and it was suggested that oleate produced within the membrane by SCD-1 could be a more potent influence on the maturation of SREBP-1 than oleate supplied in the diet [36]. It is difficult to compare general feeding experiments with those in which changes in desaturation by SCD-1 could influence specific regions of the membrane to keep the structure and cholesterol content optimal. In this respect it may be relevant that long-term fibrate feeding greatly reduced hepatic cholesterol levels. While this would not be expected to affect SREBP-1c cleavage directly (Figure 7), the difference in membrane composition could result in a different effect of fatty acids than in cells replete with cholesterol. It has been shown that oleate produced by SCD-1 is necessary for the fructose-mediated induction of lipogenic genes [36]. By analogy, the increase in SCD-1 activity observed here could also be responsible for the effects of fibrate feeding on lipogenic genes. It would lead to an alteration in membrane composition which would affect the conformation of the transport protein SCAP (SREBP cleavage-activating protein) and allow the transfer of SREBP-1c from the endoplasmic reticulum to the Golgi for cleavage. Transport involves the release of the SCAP from its membrane anchor INSIG, but there was no evidence that the effects of fibrate or cholesterol were mediated by changes in the abundance of the liver-specific INSIG-2a (Tables 2 and 3).
The overall response to fibrate feeding in the livers of the wild-type mice included a decrease in cholesterogenic pathway flux (Figure 2), which was presumably responsible, at least in part, for the observed decrease in hepatic cholesterol content (Table 4). The absence of these effects in the PPARα-null mice suggested the involvement of PPARα. However, the knockout animals responded normally to dietary cholesterol (Figure 6), indicating that PPARα deficiency per se did not give rise to any defect in the cholesterol-feedback machinery. It is more likely that the decreased cholesterogenic flux arose from a reduced availability of pre-reductase precursors, rather than from a decrease in HMG-CoA reductase itself, whose mRNA expression actually rose in response to fibrate during the light phase of the diurnal cycle (Figure 1). It has been shown previously [12,37,38] that variations in the demand for acetyl-CoA for fatty acid synthesis can influence carbon flux into cholesterol, so it is likely that the stimulation of fatty acid synthesis by PPARα activation was responsible for the accompanying decrease in flux of the common precursor, acetyl-CoA, through the cholesterol pathway. The resulting reduction in hepatic cholesterol concentration provoked a compensatory response by HMG-CoA reductase mRNA (Figure 1), in a mechanism that probably involved enhanced cleavage of SREBP-2.
Since Srebp-1c gene transcription is dependent to some extent upon activation of LXRα by cholesterol-derived oxysterols [20,21], the relatively low levels of SREBP-1c mRNA in the livers of the PPARα-null mice (Table 2) could have resulted from the reduced hepatic concentration of non-esterified cholesterol that has been observed in these animals [3]. That LXRα could still be activated by cholesterol in the PPARα-null mice was shown by the increase in the expression of its target genes Srebp-1c, Cyp7a1 and Abca1 following cholesterol feeding (Table 3). Activation of Srebp-1c transcription leads to an increased expression of glycerophosphate acyltransferase [39], one of its target genes, which probably accounted for the increase in triacylglycerol content in the livers of the cholesterol-fed mice (Table 4).
In summary, the experiments described here indicate that PPARα activation regulates fatty acid and cholesterol synthesis in a response which may compensate for increased fatty acid oxidation. Provision of ligands for LXRα can also influence the transcription of lipogenic genes under physiological conditions, but is not responsible for the effects resulting from the activation of PPARα by fibrate, which are mediated by an increase in the rate of cleavage of SREBP-1c to its active form. Overall, the changes observed following fibrate and cholesterol feeding indicate that the liver acts to compensate for the effects of external influences by maintaining the homoeostasis of essential membrane lipids and preventing the possibility of deleterious changes in membrane physicochemical behaviour.
Acknowledgments
This work was supported by a programme grant (G. F. G.) and direct funding (B. L. K.) from the Medical Research Council (U.K.), and a project grant (D. H.) from the British Heart Foundation.
References
- 1.Schoonjans K., Staels B., Auwerx J. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J. Lipid Res. 1996;37:907–925. [PubMed] [Google Scholar]
- 2.Lee Y., Yu X., Mangelsdorf D. J., Wang M.-Y., Richardson C., Witters L. A., Unger R. H. PPARα is necessary for the lipopenic action of hyperleptinemia on white adipose and liver tissue. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11848–11853. doi: 10.1073/pnas.182420899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel D. D., Knight B. L., Wiggins D., Humphreys S. M., Gibbons G. F. Disturbances in the normal regulation of SREBP-sensitive genes in peroxisome proliferator-activated receptor alpha (PPARα) deficient mice. J. Lipid Res. 2001;42:328–337. [PubMed] [Google Scholar]
- 4.Sugden M. C., Bulmer K., Gibbons G. F., Knight B. L., Holness M. J. Peroxisome-proliferator-activated receptor-α (PPARα) deficiency leads to dysregulation of hepatic lipid and carbohydrate metabolism by fatty acids and insulin. Biochem. J. 2002;364:361–368. doi: 10.1042/BJ20011699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horton J. D., Goldstein J. L., Brown M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foufelle F., Ferre P. New perspectives in the regulation of hepatic glycolytic and lipogenic genes by insulin and glucose: a role for the transcription factor sterol regulatory element binding protein-1c. Biochem. J. 2002;366:377–391. doi: 10.1042/BJ20020430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., Brown M. S. Crucial step in cholesterol homeostasis: Sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 8.Yabe D., Brown M. S., Goldstein J. L. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibbons G. F., Patel D. D., Wiggins D., Knight B. L. The functional efficiency of lipogenic and cholesterogenic gene expression in normal mice and in mice lacking the peroxisomal proliferator-activated receptor-α (PPAR-α) Adv. Enzyme Regul. 2002;42:227–247. doi: 10.1016/s0065-2571(01)00033-4. [DOI] [PubMed] [Google Scholar]
- 10.Lee S. S., Pineau T., Drago J., Lee E. J., Owens J. W., Kroetz D. L., Fernandez S. P., Westphal H., Gonzalez F. J. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 1995;15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.deVasconcelos P. R. L., Kettlewell M. G. W., Gibbons G. F., Williamson D. H. Increased rates of hepatic cholesterogenesis and fatty acid synthesis in septic rats in vivo: evidence for the possible involvement of insulin. Clin. Sci. 1989;76:205–211. doi: 10.1042/cs0760205. [DOI] [PubMed] [Google Scholar]
- 12.Gibbons G. F., Pullinger C. R., Munday M. R., Williamson D. H. Regulation of cholesterol synthesis in the liver and mammary gland of the lactating rat. Biochem. J. 1983;212:843–848. doi: 10.1042/bj2120843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J., Frisch E. F., Maniatis T. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 14.Knight B. L., Patel D. D., Humphreys S. M., Wiggins D., Gibbons G. F. Inhibition of cholesterol absorption associated with a PPARα-dependent increase in ABC binding cassette transporter A1 in mice. J. Lipid Res. 2003;44:2049–2058. doi: 10.1194/jlr.M300042-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Sheng Z., Otani H., Brown M. S., Goldstein J. L. Independent regulation of sterol regulatory element-binding proteins 1 and 2 in hamster liver. Proc. Natl. Acad. Sci. U.S.A. 1995;92:935–938. doi: 10.1073/pnas.92.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 17.Yamazaki K., Kuromitsu J., Tanaka I. Microarray analysis of gene expression changes in mouse liver induced by peroxisome proliferator-activated receptor α agonsts. Biochem. Biophys. Res. Commun. 2002;290:1114–1122. doi: 10.1006/bbrc.2001.6319. [DOI] [PubMed] [Google Scholar]
- 18.Chinetti G., Lestavel S., Bocher V., Remaley A. T., Neve B., Torra I. P., Teissier E., Minnich A., Jaye M., Duverger N., et al. PPAR-α and PPAR-γ activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nat. Med. 2001;7:53–58. doi: 10.1038/83348. [DOI] [PubMed] [Google Scholar]
- 19.Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J.-M. A., Shimomura I., Shan B., Brown M. S., Goldstein J. L., Mangelsdorf D. J. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultz J. R., Tu H., Luk A., Repa J. J., Medina J. C., Li L., Schwender S., Wang S., Thoolen M., Mangelsdorf D. J., et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costet P., Luo Y., Wang N., Tall A. R. Sterol-dependent transactivation of the ABC1 promoter by the liver X receptor/retinoid X receptor. J. Biol. Chem. 2000;275:28240–28245. doi: 10.1074/jbc.M003337200. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz K., Lawn R. M., Wade D. P. ABC1 gene expression and ApoA-I-mediated cholesterol efflux are regulated by LXR. Biochem. Biophys. Res. Commun. 2000;274:794–802. doi: 10.1006/bbrc.2000.3243. [DOI] [PubMed] [Google Scholar]
- 23.Repa J. J., Turley S. D., Lobaccaro J. A., Medina J., Li L., Lustig K., Shan B., Heyman R. A., Dietschy J. M., Mangelsdorf D. J. Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science. 2000;289:1524–1529. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- 24.Lehmann J. M., Kliewer S. A., Moore L. B., Smith O. T., Oliver B. B., Su J. L., Sundseth S. S., Winegar D. A., Blanchard D. E., Spencer T. A., Willson T. M. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J. Biol. Chem. 1997;272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- 25.Peet D. J., Turley S. D., Ma W., Janowski B. A., Lobaccaro J. M., Hammer R. E., Mangelsdorf D. J. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 26.Hegardt F. G., Serra D., Asins G. Influence of Etomoxir on the expression of several genes in liver, testis and heart. Gen. Pharmacol. 1995;26:897–904. doi: 10.1016/0306-3623(94)00281-q. [DOI] [PubMed] [Google Scholar]
- 27.Shimomura I., Shimano H., Korn B. S., Bashmakov Y., Horton J. D. Nuclear sterol regulatory element-binding proteins activate genes responsible for the entire program of unsaturated fatty acid biosynthesis in transgenic mouse liver. J. Biol. Chem. 1998;273:35299–35306. doi: 10.1074/jbc.273.52.35299. [DOI] [PubMed] [Google Scholar]
- 28.Osborne T. F. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J. Biol. Chem. 2000;275:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- 29.Saucier S. E., Kandutsch A. A., Gayen A. K., Swahn D. K., Spencer T. A. Oxysterol regulators of 3-hydroxy-3-methylglutaryl-CoA reductase in liver. Effect of dietary cholesterol. J. Biol. Chem. 1989;264:6863–6869. [PubMed] [Google Scholar]
- 30.Gould R. G., Taylor C. B., Hagerman J. S., Warner I., Campbell D. J. Cholesterol metabolism. I. Effect of dietary cholesterol on the synthesis of cholesterol in dog tissues in vitro. J. Biol. Chem. 1953;201:519–528. [PubMed] [Google Scholar]
- 31.Dobrosotskaya I. Y., Seegmiller A. C., Brown M. S., Goldstein J. L., Rawson R. B. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science. 2002;296:879–883. doi: 10.1126/science.1071124. [DOI] [PubMed] [Google Scholar]
- 32.Seegmiller A. C., Dobrosotskaya I., Goldstein J. L., Ho Y. K., Brown M. S., Rawson R. B. The SREBP pathway in Drosophila: Regulation by palmitate, not sterols. Dev. Cell. 2002;2:229–238. doi: 10.1016/s1534-5807(01)00119-8. [DOI] [PubMed] [Google Scholar]
- 33.Hannah V. C., Ou J. F., Luong A., Goldstein J. L., Brown M. S. Unsaturated fatty acids down-regulate SREBP isoforms 1a and 1c by two mechanisms in HEK-293 cells. J. Biol. Chem. 2001;276:4365–4372. doi: 10.1074/jbc.M007273200. [DOI] [PubMed] [Google Scholar]
- 34.Xu J., Nakamura M. T., Cho H. P., Clarke S. D. Sterol regulatory element binding protein-1 expression is suppressed by dietary polyunsaturated fatty acids. J. Biol. Chem. 1999;274:23577–23583. doi: 10.1074/jbc.274.33.23577. [DOI] [PubMed] [Google Scholar]
- 35.Xu J., Cho H., O'Malley S., Park J. H. Y., Clarke S. D. Dietary polyunsaturated fats regulate rat liver sterol regulatory element binding proteins-1 and -2 in three distinct stages. J. Nutr. 2002;132:3333–3339. doi: 10.1093/jn/132.11.3333. [DOI] [PubMed] [Google Scholar]
- 36.Miyazaki M., Dobrzyn A., Man W. C., Chu K., Sampath H., Kim H.-J., Ntambi J. M. Stearoyl-CoA desaturase gene 1 expression is necessary for fructose-mediated induction of lipogenic gene expression by sterol regulatory element-binding protein-1c-dependent and -independent mechanisms. J. Biol. Chem. 2004;279:25164–25171. doi: 10.1074/jbc.M402781200. [DOI] [PubMed] [Google Scholar]
- 37.Pullinger C. R., Gibbons G. F. The role of substrate supply in the regulation of cholesterol biosynthesis in rat hepatocytes. Biochem. J. 1983;210:625–632. doi: 10.1042/bj2100625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibbons G. F., Bjornsson O. G., Pullinger C. R. Evidence that changes in hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase activity are required partly to maintain a constant rate of sterol synthesis. J. Biol. Chem. 1984;259:14399–14405. [PubMed] [Google Scholar]
- 39.Ericsson J., Jackson S. M., Kim J. B., Spiegelman B. M., Edwards P. A. Identification of glycerol-3-phosphate acyltransferase as an adipocyte determination and differentiation factor 1- and sterol regulatory element-binding protein-responsive gene. J. Biol. Chem. 1997;272:7298–7305. doi: 10.1074/jbc.272.11.7298. [DOI] [PubMed] [Google Scholar]