Abstract
PARP-1 [poly(ADP-ribose) polymerase-1) is a nuclear enzyme that is involved in several cellular functions, including DNA repair, DNA transcription, carcinogenesis and apoptosis. The activity directed by the PARP-1 gene promoter is mainly dictated through its recognition by the transcription factors Sp1 and Sp3 (where Sp is specificity protein). In the present study, we investigated whether (i) both PARP-1 expression and PARP-1 enzymatic activity are under the influence of cell density in primary cultured cells, and (ii) whether its pattern of expression is co-ordinated with that of Sp1/Sp3 at varying cell densities and upon cell passages. All types of cultured cells expressed PARP-1 in Western blot when grown to sub-confluence. However, a dramatic reduction was observed at post-confluence. Similarly, high levels of Sp1/Sp3 were observed by both Western blot and EMSAs (electrophoretic mobility-shift assays) in sub-confluent, but not post-confluent, cells. Consistent with these results, the promoter of the rPARP-1 (rat PARP-1) gene directed high levels of activity in sub-confluent, but not confluent, cells upon transfection of various CAT (chloramphenicol acetyltransferase)–rPARP-1 promoter constructs into cultured cells. The positive regulatory influence of Sp1 was not solely exerted on the rPARP-1 promoter constructs, as inhibition of endogenous Sp1 expression in HDKs (human dermal keratinocytes) through the transfection of Sp1 RNAi (RNA interference) considerably reduced endogenous hPARP-1 (human PARP-1) expression as well. The reduction in PARP-1 protein expression as cells reached confluence also translated into a corresponding reduction in PARP-1 activity. In addition, expression of both Sp1/Sp3, as well as that of PARP-1, was dramatically reduced as cells were passaged in culture and progressed towards irreversible terminal differentiation. PARP-1 gene expression therefore appears to be co-ordinated with that of Sp1 and Sp3 in primary cultured cells, suggesting that PARP-1 may play some important functions during the proliferative burst that characterizes wound healing.
Keywords: electrophoretic mobility-shift assay (EMSA), poly(ADP-ribose) polymerase-1 (PARP-1), primary cultured cell, promoter, RNA interference (RNAi), specificity protein1 (Sp1)
Abbreviations: AP, activating protein; CAT, chloramphenicol acetyltransferase; DH, Dulbecco's modified Eagle's medium/Ham's F12 (3:1); DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; FBS, foetal bovine serum; HCEC, human corneal epithelial cell; HDK, human dermal keratinocyte; hGH, human growth hormone; HUVEC, human umbilical vein endothelial cell; HVSMC, human vascular smooth muscle cell; NF1, nuclear factor 1; NF-κB, nuclear factor κB; PARP-1, poly(ADP-ribose) polymerase-1; hPARP-1, human PARP-1; rPARP-1, rat PARP-1; RCEC, rabbit corneal epithelial cell; RNAi, RNA interference; RPE, retinal pigmented epithelial; RT, reverse transcriptase; siRNA, small interfering RNA; SL2, Drosophila Schneider line 2; Sp, specificity protein; Sp1m, mutated Sp1 derivative; TCA, trichloroacetic acid
INTRODUCTION
PARP-1 [poly(ADP-ribose) polymerase-1] is the canonical member of the PARP family of enzymes [1]. PARP-1 is a nuclear enzyme, also found in centrosomes [2,3], whose poly(ADP-ribosyl)ation activity is strongly dependent on DNA strand break (reviewed in [4]). Indeed, following binding with DNA breaks, PARP-1 synthesizes negatively charged poly(ADP-ribose) polymers on to acceptor proteins, including histones, transcription factors and PARP-1 itself. Through its ability to post-translationally modify various nuclear proteins, PARP-1 is therefore involved in several important cellular functions, including DNA repair, gene expression, carcinogenesis and apoptosis [4]. The transcriptional regulation of PARP-1 is therefore crucial to the cell's homoeostasis.
Previous studies have shown that the positive transcription factor Sp1 (where Sp is specificity protein) regulates the activity of the rPARP-1 (rat PARP-1) gene promoter by recognizing five upstream Sp1 target sites [5]. Because the rPARP-1 promoter belongs to the TATA-less gene promoter, Sp1 also drives the basal expression via an initiator site [6]. The Sp1-mediated trans-activation of the rPARP-1 promoter was also found to be modulated by the binding of transcription factors that belong to the NF1 (nuclear factor 1) family to the rPARP-1 promoter [7,8]. Studies performed on established cell lines or transformed cells showed that the expression of Sp1 and Sp3, another member of the Sp1 family, is altered by both the state of cell density [9] and the extent of cell differentiation [10]. Expression of Sp1 has been shown to predominate during the G1 phase of the cell cycle and is then subjected to proteasome-dependent degradation before the S phase [10]. Similarly, the PARP-1 mRNA transcript is found throughout the cell cycle, reaching its peak at the G1 phase in both rat [11] and human cells [12]. Although a few studies reported that PARP-1 expression could be modulated by cell density in cancer cells [13,14], no such experiment has ever been conducted on untransformed primary cultured cells. Because they are recognized as being much closer to normal cells than transformed cells or cell lines are, primary cultures are particularly attractive for gene expression studies [15,16].
Over the last several years, we exploited the primary culture of RCECs (rabbit corneal epithelial cells) as a model to study corneal wound healing [9,17,18]. PARP-1 has been shown to regulate the expression of the integrin CD11a through direct interaction with NF-κB (nuclear factor κB) [19], establishing a role for PARP-1 in cell migration during neuronal injury. As cell migration is a major prerequisite for wound healing, it is expected that PARP-1 gene expression will be differently modulated during this process. Wound healing is primarily dictated by changes in the proliferation, migration and adhesion properties of the cells that border the injured area. In turn, these changes depend on the supranormal expression of many structural genes, including those encoding membrane-bound integrins [20]. Transcription of many integrin subunit genes has been shown to rely on the positive regulatory influence of Sp1 [5].
In the present study, we evaluated whether the variations in the expression of the transcription factors Sp1/Sp3 observed when primary cultured cells of various origins are grown at varying densities also result in similar alterations in the level of expression of the PARP-1 gene, and by way of consequence, also in a decreased PARP-1 poly(ADP-ribosyl)ation activity in those cells. We demonstrated that expression of PARP-1 perfectly paralleled that of Sp1/Sp3 in all the primary cultured cell types examined. Expression of both PARP-1 and Sp1/Sp3 predominated in highly prolific cells at sub-confluence and then dramatically decreased as cells reached post-confluence in vitro. Moreover, a similar reduction in PARP-1 expression occurred in RCECs as these cells reached terminal differentiation in vitro.
MATERIALS AND METHODS
Plasmids and oligonucleotides
The plasmids PCR1, PCR2, PCR3, PCR4 and PCR5 have been described previously [5]. All of the plasmids bear the CAT (chloramphenicol acetyltransferase) reporter gene fused to DNA fragments from the rPARP-1 gene promoter extending up to 5′ positions −34 (PCR1), −60 (PCR2), −101(PCR3), −150 (PCR4), −237 (PCR5) respectively, as well as the PCR3 derivative plasmid that bears mutations in the three most proximal Sp1-binding sites (F2, F3 and F4). The Sp1 and Sp3 expression plasmids pPacSp1 and pPacSp3 respectively, which allow for high levels of expression of these proteins in SL2 cells (Drosophila Schneider line 2 cells; ATCC CRL-1963) were obtained from Dr Guntram Suske (Institut für Molekularbiologie und Tumorforschung, Philipps Universität, Marburg, Germany). The LacZ expression plasmid pAC5/V5-His/LacZ was obtained from Invitrogen (Carlsbad, CA, U.S.A.). The double-stranded oligonucleotides bearing the high-affinity binding site for Sp1 (5′-GATCATATCTGCGGGGCGGGGCAGACACAG-3′) [21], the binding site for human HeLa CTF (CCAAT transcription factor)/NF-1 in adenovirus type 2 (Ad2; 5′-GATCTTATTTTGGATTGAAGCCAATATGAG-3′) [22], and a derivative from the Sp1 site identified in the upstream promoter from the α5 gene between positions −81 to −30 that bears mutations (in bold) which prevent its recognition by Sp1 (Sp1m; 5′-GCGGGGAGTTTGGCAAACTAAAAAACGCGTTGAGTCATTCGCCTCTGGGAGG-3′) [9] were all chemically synthesized using a Biosearch 8700 apparatus (Millipore). Preannealed siRNAs (small interfering RNAs) that target three different regions from the human Sp1 gene (catalogue numbers 36737; 116546 and 36912) as well as the Silencer negative control siRNA #1 were all purchased from Ambion Inc. (Austin, TX, U.S.A.).
Cell culture and media
RCECs were obtained from the central area of freshly dissected rabbit corneas as described in [9]. They were maintained in SHEM (supplemental hormonal epithelium medium) supplemented with 5% (v/v) FBS (foetal bovine serum) and 20 μg/ml gentamicin. HCECs (human corneal epithelial cells) were isolated from the limbal area of normal eyes obtained from 68- and 57-year-old donors following a procedure that we described previously [23] and were maintained in DH [Dulbecco's modified Eagle's medium/Ham's F12 (3:1, v/v)] medium supplemented with 10% (v/v) FBS. HDKs (human dermal keratinocytes) were isolated from a normal adult skin specimen (26-year-old donor) removed during reductive breast surgery and grown on a feeder layer of irradiated mouse Swiss 3T3 fibroblasts in complete keratinocyte medium as described in [24]. HUVECs (human umbilical vein endothelial cells) were obtained by the method of Jaffe et al. [25] and were grown in a medium for endothelial cells [medium M199 supplemented with 20% (v/v) FBS, 2 mM L-glutamine, 50 units/ml heparin, 25 μg/ml endothelial cell growth factor supplement (Sigma), 100 units/ml penicillin G and 25 μg/ml gentamicin]. HUVECs were plated on gelatin-coated tissue culture flasks. Endothelial cell characterization was assessed as described previously [26]. HVSMCs (human vascular smooth muscle cells) were isolated from umbilical veins by the explant method of Ross [27] and were cultured in standard medium [DH medium with 10% (v/v) FBS and antibiotics]. HDKs, HUVECs and HVSMCs were kindly provided by Dr Lucie Germain [Laboratoire d'Organogénèse Expérimentale (LOEX) Hôpital du St-Sacrement du CHA, Québec, Canada]. RPE (retinal pigmented epithelial) cells were isolated from the eyes of a 21-year-old donor as described in [28] and were cultured in DH medium supplemented with 2.5 mM glutamine, 10% (v/v) FBS, 1% penicillin/streptomycin (from a 10000 units/ml stock solution). All cells were grown to sub-confluence (nearly 70% coverage) or post-confluence (100% coverage for 0, 2, 4, 5 or 15 days) under 5% CO2 at 37 °C.
SL2 cells were grown in Schneider insect medium (Sigma–Aldrich) supplemented with 10% (v/v) FBS and 20 μg/ml gentamicin at 27 °C without CO2.
Transfection and CAT assay
RCECs and HDKs were transfected using the polycationic detergent Lipofectamine™ (Gibco BRL, Burlington, Ontario, Canada) as recommended by the manufacturer. Each Lipofectamine™-transfected plate received 1 μg of the test plasmid and 1 μg of the hGH (human growth hormone)-encoding plasmid pXGH5. RPE cells and HCECs were transfected using the calcium phosphate precipitation procedure as described in [29]. Each individual plate received 15 μg of the test plasmid and 5 μg of the plasmid pXGH5 for 4–6 h (RPE cells) or 18 h (HCECs), before being washed with PBS and incubated for an additional 48 h in fresh medium. All cells were plated at sub-confluence (4×105 cells per 35 mm well yielded 50% coverage within 24 h) (SC) and post-confluence [1.5×106 cells per 35 mm well yielded 100% coverage within 24 h; cells were either transfected immediately (C0d) or maintained at full confluence for 2 (C2d), 4 (C4d), 5 (C5d) or 15 (C15d) days, depending on the cell type transfected] before they were transfected as above. siRNA duplexes were also transfected using Lipofectamine™ into HDKs plated at sub-confluence. Based on preliminary results, 0.75 μg from each of the three Sp1 siRNA duplexes (for a total of 2.25 μg), or 2.25 μg of the Silencer negative control siRNA (to give a final concentration of 340 nM), were transfected into each well along with 1 μg of the test plasmid PCR3. Cells were harvested 48 h following the addition of fresh medium as detailed above. Drosophila Schneider cells were transfected according to the calcium phosphate precipitation procedure at a density of 1×106 cells per 60 mm culture plate. CAT activities from transfected SL2 cells were normalized to the amount of β-galactosidase encoded by the plasmid pAC5/V5-His/LacZ and co-transfected along with the CAT recombinant constructs. Each cell-containing plate therefore received 15 μg of the test plasmid, 4 μg of pAC5/V5-His/LacZ and 1 μg of either pPacSp1 or pPacSp3, or both. Levels of CAT activity for all transfected cells were determined as described in [29], normalized to the amount of hGH secreted into the culture medium and assayed using a kit for quantitative measurement of hGH (Immunocorp, Montréal, Québec, Canada). Measurement of the β-galactosidase activity was performed following a standard procedure [30]. Student's t test was performed for comparison of the groups. Differences were considered to be statistically significant at P<0.05. All data are expressed as means±S.D.
Nuclear extracts and EMSA (electrophoretic mobility-shift assay)
Crude nuclear extracts were prepared from all cell types cultured either to sub- or post-confluence, and dialysed against DNase I buffer [50 mM KCl, 4 mM MgCl2, 20 mM K3PO4 (pH 7.4), 1 mM 2-mercaptoethanol and 20% (v/v) glycerol], as described in [9,17]. The Sp1 oligonucleotide was 5′-32P-end-labelled and was used as a probe in EMSA [31]. Approx. 3×104 c.p.m. of labelled DNA was incubated with crude nuclear proteins (5 μg) from each type of cells in the presence of 500 ng of poly(dI-dC)·(dI-dC) (Amersham Biosciences) in buffer D [5 mM Hepes (pH 7.9), 10% (v/v) glycerol, 25 mM KCl, 0.05 mM EDTA, 0.5 mM DTT (dithiothreitol) and 0.125 mM PMSF]. When indicated, unlabelled double-stranded oligonucleotides bearing various DNA target sequences for known transcription factors (Sp1, NF1 and Sp1m) were added as competitors (150- and 500-fold molar excesses) during the assay. DNA–protein complexes were separated by gel electrophoresis through 8% native polyacrylamide gels run against Tris/glycine buffer [9,17]. Gels were dried and autoradiographed at −80 °C to reveal the position of the shifted DNA–protein complexes generated.
SDS/PAGE and Western blot
The protein concentration from the crude nuclear extracts prepared from each primary cultured cells was determined by the Bradford procedure [31a] and was validated precisely through Coomassie Blue staining on SDS/10% polyacrylamide gels. To crude nuclear extracts, 1 vol. of sample buffer [6 M urea, 63 mM Tris/HCl (pH6.8), 10% (v/v) glycerol, 1% (w/v) SDS, 0.00125% (w/v) Bromophenol Blue and 300 mM 2-mercaptoethanol] was added before size-fractionating on a SDS/10% polyacrylamide minigel before being transferred on to a nitrocellulose filter. The blot was then washed twice for 5 min in TS buffer (150 mM NaCl and 10 mM Tris/HCl, pH 7.4) and saturated by incubation for 30 min in TSM buffer (TS buffer with 5% milk and 0.1% Tween 20) as described in [17]. Then, a 1:5000 dilution of polyclonal antibodies raised against the transcription factors Sp1 and Sp3 (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.), or a 1:10000 dilution of a mouse monoclonal antibody (C-2-10 [32]) raised against an epitope in the C-terminal end of the DNA-binding domain of bovine PARP-1 (provided by Dr Guy G. Poirier, Unit of Health and Environment, CHUL Research Center, Sainte-Foy, Québec, Canada) was added to the membrane-containing TSM buffer and incubation proceeded further for 90 min at room temperature (22 °C). The blot was washed in TSM and incubated for an additional 1 h at room temperature in a 1:1000 dilution of a peroxidase-conjugated goat anti-(mouse PARP-1) or a 1:5000 dilution of a peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, Bio/Can Scientific, Mississauga, Ontario, Canada). The membrane was washed in TS buffer before immunoreactive complexes were revealed using a Western blot detection kit (Amersham Biosciences), and autoradiographed. Each Western blot result shown in the present study corresponds to one out of at least three representative experiments.
RT (reverse transcriptase)-PCR analyses
Total RNA was isolated from sub-confluent, or from either 2- or 5-day post-confluent, RCECs using the TRIzol® reagent (Molecular Research Center, Cincinnati, OH, U.S.A.), and reverse-transcribed using the Superscript II Transcriptase kit from Gibco-BRL as described recently [18]. The sequence of the template primers used for the amplification of the PARP-1 transcript were 5′-GTGGCACGGGTCCAGGACCACCAAC-3′ (sense) and 5′-GCCCAAACCTTTGACACTGTGCTTGCCC-3′ (antisense). The oligonucleotide primers used for the amplification of the 18 S rRNA were provided in the Quantum RNA 18 S Internal Standards kit (Ambion). Taq polymerase (Amersham Biosciences) was selected for PCR amplification. Cycle parameters were the same for all primers used (denaturation at 95 °C for 30 s, annealing at 56 °C for 30 s and extension at 72 °C for 30 s) with an identical number of cycles (26, 28, 30, 32, 34 and 36 cycles) for both sets of primers. The PCR products were analysed on a 1.5% agarose gel. The image was scanned using a Visage 110S Bioimage analyser (Millipore) in order to quantify differential gene expression at various cell densities.
PARP assay
PARP activity was measured as TCA (trichloroacetic acid)-precipitable radioactivity incorporated from [adenylate-32P]NAD+ [33]. Crude nuclear extract samples (25 μg in 70 μl) were incubated at 25 °C for 30 min in a standard assay mixture (152 μl) containing 100 mM Tris/HCl, pH 8.0, 10 mM MgCl2, 10% (v/v) glycerol, 1.5 mM DTT, 10 μg/ml activated DNA (DNase I-treated) and 200 μM NAD+. The reaction was started by adding test samples and stopped by adding ice-cold 20% (w/v) TCA. Samples were filtered on GF/C filters (Millipore) saturated previously with 25% (w/v) TCA, 0.02 M sodium pyrophosphate and 1 mM NAD+. Filters were washed twice with 10 ml of 100% ethanol, dried briefly under vacuum, and 32P incorporation into TCA-precipitated proteins was measured. PARP specific activity was expressed as units/mg of total proteins. One unit is defined as the amount of enzyme required to convert 1 nmol of NAD+/min under standard conditions [33]. Each value corresponds to the average of five separate measurements.
RESULTS
The transcriptional activity directed by the rPARP-1 promoter is modulated by cell density
Only a few studies reported a reduction of PARP-1 gene expression linked to an increase in cell density [13,14]. However, these experiments were conducted on transformed cancer cells and at the time of submission of the present paper, no results were available that detailed the influence that cell density might play on PARP-1 gene expression in primary cultured cells. We successfully used primary cultures of RCECs grown to various densities as an in vitro model to study expression of membrane-bound integrins during corneal wound healing [9,17,18]. We therefore exploited this cell culture model to examine whether PARP-1 expression is influenced by cell density.
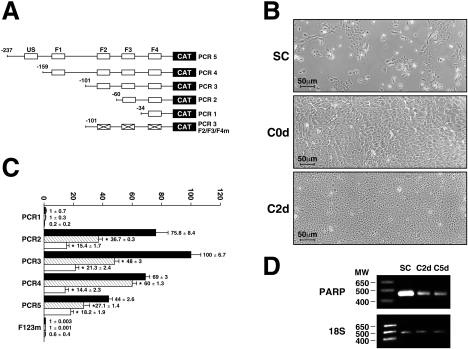
Recombinant plasmids bearing the CAT gene fused to various segments from the rPARP-1 promoter that include one or more of the five target sites for Sp1 (US1, F1–F4; Figure 1A) were transfected into RCECs either grown to sub-confluence or maintained to complete confluence for various periods of time [either 0 h (C0d) or 2 days (C2d) at full confluence]. Sub-confluent RCECs are sparsely dispersed and cover approx. 50–70% of the culture plate (Figure 1B). As they just reach confluence, RCECs are covering the entire culture surface, but are still actively dividing (C0d; Figure 1B), whereas, at 2 days post-confluence, cells were smaller and much more packed (C2d; Figure 1B). The rPARP-1 promoter construct PCR1 that only bears the most proximal Sp1 site (F4) was totally inefficient in yielding any CAT activity, as this single site was shown previously to be insufficient by itself to ensure basal rPARP-1 promoter activity [5,6]. On the other end, extending the rPARP-1 promoter to include the F3 Sp1 site (in PCR2) yielded a dramatic increase in CAT activity upon transfection of sub-confluent RCECs (Figure 1C). The promoter activity increased further by approx. 20% when the F2 site was added. Any further extension of the promoter resulted in a progressive reduction in CAT activity to approx. 45% of that directed by PCR3 in sub-confluent cells (for PCR5). All plasmids yielded a reduced rPARP-1 promoter activity when transfected in confluent cells (C0d). The reduction in promoter function was even greater when RCECs were maintained for 2 days at post-confluence (up to 5-fold reduction when compared with the results from sub-confluent cells). This reduction in rPARP-1 promoter activity was not restricted to the transfected rPARP-1 promoter plasmids, as transcription of the endogenous rabbit PARP-1 gene was also affected by cell density. Indeed, semi-quantitative RT-PCR experiments conducted using total RNA from RCECs grown to either sub-confluence or full confluence for 2 or 5 days revealed a high level of PARP-1 expression at sub-confluence, which then decreased as cells were maintained at confluence for various periods of time (Figure 1D). Normalization of the PARP-1 PCR product over that of the 18 S rRNA and obtained from three separate experiments conducted on three different batches of total RNAs revealed a ratio of 2.12±0.24 at sub-confluence, which then progressively decreased to 0.62±0.09 and 0.15±0.08 at both 2 days and 5 days post-confluence respectively. We therefore conclude that transcription of the PARP-1 gene progressively decreases as RCECs progress to quiescence in vitro.
Figure 1. rPARP-1 promoter activity in sub-confluent and confluent RCECs.
Plasmids bearing various lengths from the rPARP-1 promoter (PCR1, PCR2, PCR3, PCR4 and PCR5) were transfected into RCECs at varying cell densities. (A) Schematic representation of the constructs transfected. The position of the Sp1 sites (US1, F1, F2, F3 and F4 [5]) is provided, along with the 5′ end points (−34 to −237 respectively). ‘X’ indicates mutation of the F2, F3 and F4 Sp1 sites in the PCR3F2/F3/F4m plasmid. (B) Phase-contrast images of RCECs grown to sub-confluence (SC), or to confluence for either 0 (C0d) or 2 days (C2d). Magnification, ×200. Scale bar, 50 μm. (C) CAT activities from RCECs transfected with the plasmids shown in (A) at either sub-confluence (black bars) or confluence (hatched bars: RCECs maintained at confluence for 0 days; white bars: RCECs maintained at post-confluence for 2 days). *CAT activities from transfected RCECs grown to post-confluence that are statistically different from those measured in sub-confluent cells (P<0.005; Student's paired t test). Results are means±S.D. (D) RT-PCR analysis of the rabbit endogenous PARP-1 mRNA transcript. Total RNA from sub-confluent (SC) and both 2-day (C2d) and 5-day (C5d) post-confluent RCECs were reverse-transcribed and PCR co-amplified using the PARP-1 and 18 S rRNA specific primers. The position of both the amplified 171 bp PARP-1 and 489 bp 18 S fragments is indicated, along with that of the most relevant molecular mass markers (MW, sizes in kDa). Results shown are from PCR amplification cycle 28.
Regulatory influence of Sp1 and Sp3 on the rPARP-1 promoter
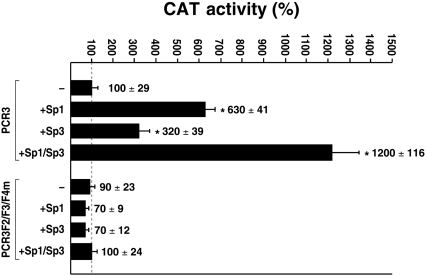
Three target sites for Sp1 were identified through DNase I foot-printing in the basal rPARP-1 promoter contained on the PCR3 recombinant construct [5,34]. However, as the rPARP-1 promoter is highly GC-rich, the possibility remained that sequences in the vicinity of the F2–F4 Sp1 site might also act as low-affinity target sites for this transcription factor. Besides, the contribution of Sp3 to the basal transcription directed by the rPARP-1 promoter has not been the subject of any study to date. Sp3, a member from the Sp family that is widely expressed in most type of cells and usually functions as a transcriptional activator, also possesses the ability to bind to Sp1 target sites. In order to evaluate the respective contribution of both Sp1 and Sp3 on the activity directed by the rPARP-1 promoter, co-transfection experiments were conducted into SL2 cells, which express neither of these transcription factors. Both the plasmid PCR3 and its mutative derivative PCR3F2/F3/F4m that bear mutations in each of the three promoter-proximal Sp1 sites (F2–F4) were co-transfected into SL2 cells either alone or with the recombinant plasmids pPacSp1 and pPacSp3. These constructs bear either the Sp1 or Sp3 cDNA under the control of the Drosophila actin gene promoter and therefore ensure expression of high levels of both these proteins in SL2 cells. As shown in Figure 2, PCR3 directed only a low level of CAT activity when transfected alone in SL2 cells. However, its activity increased 6.3- and 3.2-fold when co-transfected with either pPacSp1 (+Sp1) or pPacSp3 (+Sp3) respectively. Sp1 and Sp3 acted in an additive manner on rPARP-1 promoter activity, as co-transfection of both the Sp1 and Sp3 expression plasmids together (+Sp1/Sp3) resulted in a CAT activity (12-fold increase) that is close to the sum of the regulatory influences exerted by Sp1 and Sp3 when transfected individually with PCR3. Neither the Sp1 nor the Sp3 expression plasmid could improve the activity directed by the mutated derivative PCR3F2/F3/F4m, a clear indication that both Sp1 and Sp3 exert their positive regulatory influences through their interaction with the F2–F4 sites and that no other low-affinity target sites for these transcription factors are present along the rPARP-1 basal promoter to which they could bind.
Figure 2. Transfection in Sp1/Sp3-deficient SL2 cells.
The plasmid PCR3 and its mutated derivative PCR3 F2/F3/F4m were transfected either alone (−) or in combination with the Sp1 (+Sp1) or Sp3 (+Sp3) expression plasmids pPacSp1 and pPacSp3 respectively, or both (+Sp1/Sp3) into SL2 cells. Cells were harvested, and CAT activity (expressed as fold activity relative to the level directed by the PCR3 promoter construct alone) was determined and normalized. Asterisks (*) indicate CAT activities that are statistically different from those measured in SL2 cells transfected with PCR3 in the absence of co-transfected Sp1 or Sp3 expression plasmids (P<0.005; Student's paired t test). Results are means±S.D.
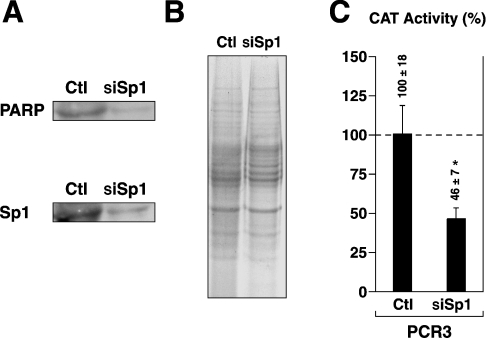
As Sp1 was shown to positively influence PARP-1 promoter activity through the use of indirect approaches, we then used RNAi (RNA interference) to assess whether inhibition of endogenous Sp1 will also alter the expression of endogenous PARP-1. The results illustrated in Figure 3(A) show that transfection of a combination of three siRNA duplexes (siSp1) directed against three different segments of the Sp1 gene decreased both Sp1 and PARP-1 proteins by approx. 84% and approx. 86% respectively in nuclear extracts from HDKs. The reduced Sp1 and PARP-1 proteins in siSp1-transfected cells did not result from corresponding variations in the amount of proteins loaded on the gel as Coomassie-Blue staining of SDS/gel-fractionated nuclear proteins from both the negative control (Ctl; cells transfected with the Silencer negative control #1) and siSp1-transfected HDKs (siSp1) was found to be identical (Figure 3B). As for endogenous PARP-1, reducing Sp1 protein expression through RNAi also resulted in a significant reduction (approx. 54%) in the activity of the rPARP-1 promoter upon co-transfection of HDKs with both pCR3 and siSp1 duplexes (Figure 3C). We therefore conclude that, as with the rPARP-1 promoter bearing constructs, Sp1 also positively regulates expression of endogenous PARP-1 gene in primary cultured HDKs.
Figure 3. Down-regulation of endogenous hPARP-1 via Sp1 RNAi.
(A) Nuclear extracts (10 μg) from mid-confluent HDKs transfected either with the Silencer negative control #1 (Ctl) or with a combination of three different Sp1 siRNA duplexes (siSp1) were examined in Western blot using antisera raised either against PARP-1 or Sp1. (B) Nuclear proteins (20 μg) from both the Silencer negative control (Ctl) and Sp1 (siSp1) siRNA transfected HDKs used in (A) were loaded on an SDS/12% polyacrylamide gel and stained with Coomassie Blue for comparison purposes. (C) The plasmid PCR3 was transfected either alone (Ctl) or with the Sp1 siRNA duplexes (siSp1) into mid-confluent HDKs. Cells were harvested, and CAT activities were determined. *CAT activities from transfected HDKs that are statistically different from those measured for PCR3 in the absence of added siRNA (P<0.005; Student's paired t test). Results are means±S.D.
The influence of cell density on Sp1/Sp3 and PARP-1 expression is ubiquitous
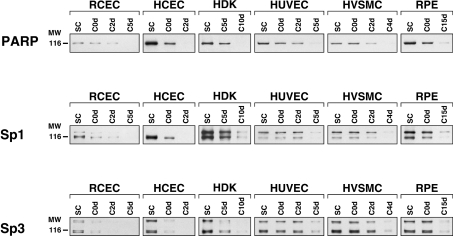
As the rPARP-1 promoter activity is primarily dictated through the recognition of the F2–F4 promoter target sites by Sp1 and Sp3 and that expression of both proteins was shown recently to decrease with increasing cell density [9], we examined whether the cell-density-dependent extinction of Sp1/Sp3 also occurs in primary cultured cells other than RCECs. Nuclear proteins were therefore prepared from a variety of primary cultured cells (HCECs, HDKs, HUVECs, HVSMCs and RPE cells), grown at either sub-confluence (SC) or post-confluence for various periods of time (C0d, C2d, C5d, C10d and C15d). An extended period of time at post-confluence (up to 15 days) was required, as some types of cells, such as the HDKs and RPE cells, require maintenance at full confluence for more than 10 days before they reach growth arrest (quiescence) when cultured in vitro.
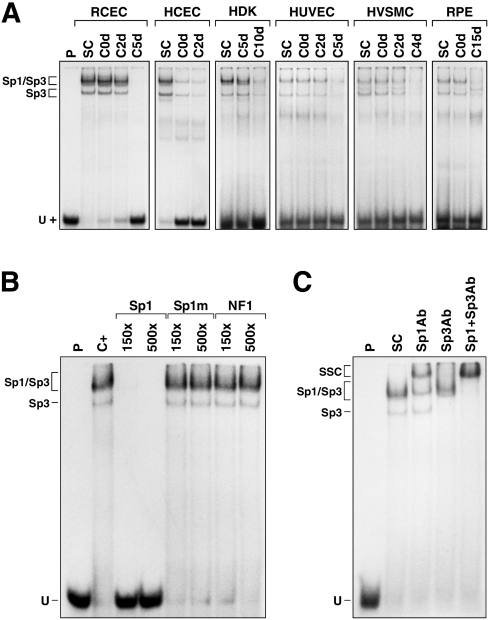
Incubation of the nuclear extracts with a labelled probe bearing the high-affinity binding site for Sp1 revealed the expression of Sp1 in all the cell types grown to sub-confluence (SC; Figure 4A). However, and as observed for RCECs, expression of both Sp1 and Sp3 is progressively lost in all types of cells as they reach quiescence (Figure 4A). The identity of the proteins yielding the shifted DNA–protein complexes as being Sp1 and Sp3 was confirmed by competitions in EMSAs (Figure 4B). Indeed, only the unlabelled oligonucleotide bearing the intact Sp1 target site (Sp1), but neither its mutated derivative (Sp1m), nor the oligomer bearing the site for the unrelated transcription factor NF1 (NF1), could prevent the formation of the shifted complexes observed in EMSA. As Figure 4(C) indicates, addition of polyclonal antibodies directed against either Sp1 or Sp3 prevented to various extents the formation of the shifted DNA–protein signals and yielded SSCs (supershifted complexes) resulting from the binding of antibodies against either the Sp1 or the Sp3 protein present in these complexes (Figure 4C). No shifted complexes could be observed when both antibodies were added to the reaction mixture before their analysis by EMSAs (Sp1+Sp3Ab).
Figure 4. EMSA analyses of Sp1/Sp3 in cells primary cultured at varying densities.
(A) Sp1/Sp3 expression in primary cultured cells. The Sp1 double-stranded oligonucleotide was 5′ end-labelled and incubated with nuclear proteins (5 μg) from cells (RCECs, HCECs, HDKs, HUVECs, HVSMCs and RPE cells) primary cultured to either sub- (SC) or post-confluence for 0, 2, 4, 5, 10 or 15 days (C0d, C2d, C4d, C5d, C10d and C15d respectively). Formation of DNA–protein complexes was then monitored by EMSA on an 8% native gel. The positions of both the Sp1 and Sp3 DNA–protein complexes are shown, as well as that of the free probe (U). P, labelled probe alone. (B) Competition experiment in EMSA. The Sp1 probe was incubated with nuclear proteins (10 μg) from sub-confluent RCECs in the presence of either no (C+) or 150- and 500-fold molar excesses of unlabelled competitor oligonucleotides (Sp1, Sp1m and NF-1). Formation of DNA–protein complexes was then monitored by EMSA on an 8% native gel. (C) Supershift experiments in EMSA. Proteins (10 μg) from sub-confluent RCECs were incubated with the Sp1 probe either alone (SC), or with 1 μl of a polyclonal antibody directed against Sp1 (Sp1Ab) or Sp3 (Sp3Ab) and added either individually or in combination (Sp1+Sp3Ab). Formation of both the Sp1 and Sp3 complexes, as well as their corresponding supershifted complexes (SSC), is indicated.
The influence of cell density on endogenous PARP-1 and both Sp1 and Sp3 expression was next monitored at the protein level through Western blot analyses. As shown on Figure 5, high levels of PARP-1 expression could be observed while cells are highly prolific at sub-confluence (SC) for all types of cultured cells, and then dramatically decreased as they reached growth arrest at quiescence. The cell-density-dependent pattern of PARP-1 extinction was totally identical with that observed for both Sp1 and Sp3 under similar culture conditions (Figure 5).
Figure 5. Western blot analyses of PARP-1, Sp1 and Sp3 expression in cells primary cultured at varying densities.
Nuclear extracts (10 μg) from primary cultured cells of different histological origins (RCECs, HCECs, HDKs, HUVECs, HVSMCs and RPE cells) and grown to either sub- (SC) or post-confluence for 0, 2, 4, 5 10 or 15 days (C0d, C2d, C4d, C5d, C10d and C15d respectively) were examined by Western blot using a monoclonal antibody directed against PARP-1 and antisera raised against Sp1 or Sp3. The position of the 116 kDa β-galactosidase protein used as a molecular mass marker (MW) is indicated.
As expression of PARP-1 appears to be intimately linked to that of both Sp1 and Sp3, we then verified whether these cell-density-dependent alterations in the level of Sp1/Sp3 would also translate into similar alterations in the transcriptional activity of the rPARP-1 promoter in cell types other than just RCECs. The PCR3 construct was therefore transfected into either sub- or 2-day post-confluent cultures of RCECs, HCECs, HDKs and RPE cells, and CAT activity was determined and compared. A clear reduction in the CAT activity directed by PCR3 (which ranged from 2- to 4-fold) was observed for all types of cells (Figure 6), although none of them reached complete growth arrest after being maintained for only 2 days at post-confluence.
Figure 6. rPARP-1 promoter activity in sub- and post-confluent primary cultured cells.
The plasmid PCR3 was transfected into both sub-confluent (black bars) and 2-days post-confluent (hatched bars) primary cultures of RCECs, HCECs, HDKs and RPE cells, and CAT activities were determined and normalized. Asterisks (*) indicate CAT activities at post-confluence that are statistically different from those measured in sub-confluent cells (P<0.005; Student's paired t test). Results are means±S.D.
Cellular poly(ADP-ribosyl)ation activity decreases with increasing cell density
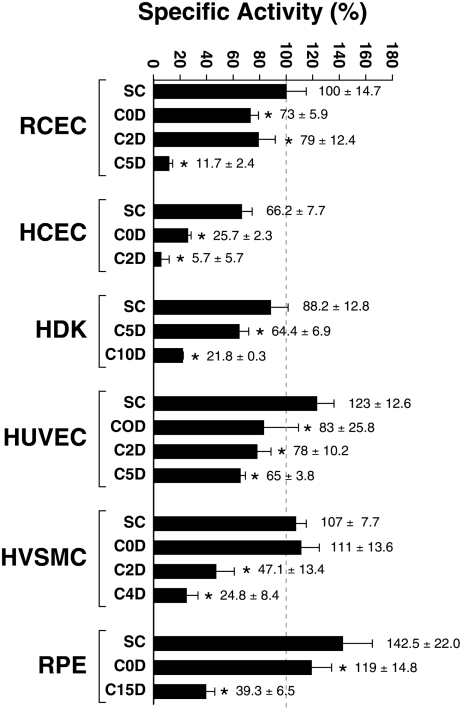
As a decrease in both the transcription directed by the PARP-1 gene and the amount of PARP-1 protein might not necessarily translate into a similar reduction in its ability to catalyse the transfer of ADP-ribose units to acceptor proteins, we sought to examine the cellular poly(ADP-ribosyl)ation activity through a standard PARP assay that measures the incorporation of [adenylate-32P]NAD+ into acceptor proteins [33]. Interestingly, basal unstimulated cellular PARP activity was very similar at sub-confluence for all types of primary cultured cells (the PARP activity was measured as units of PARP per mg of total nuclear proteins and was expressed as percentage specific activity relative to the level measured in sub-confluent RCECs, which corresponded to 100%) (Figure 7). However, as all the different types of cells progressed toward quiescence, the cellular poly(ADP-ribosyl)-ation activity was strongly impaired, reaching levels up to 12 times lower (for HCECs) at quiescence (Figure 7). In most cases, the reduction in the cellular poly(ADP-ribosyl)ation activity correlated perfectly with a similar reduction in PARP-1 protein expression (Figure 5) at the corresponding cell densities. This is in direct line with earlier reports stating the major contribution of PARP-1 to the overall poly(ADP-ribosyl)ation reactions in a cell [35].
Figure 7. Cellular poly(ADP-ribosyl)ation activity in cells cultured at varying densities.
The PARP enzymatic activity was measured in primary cultured cells (RCECs, HCECs, HDKs, HUVECs, HVSMCs and RPE cells) grown to sub-confluence (SC) or to post-confluence for 0, 2, 4, 5, 10 and 15 days (C0D, C2D, C4D, C5D, C10D and C15D respectively). PARP activity was measured as units of PARP per mg of total nuclear proteins and is expressed as percentage specific activity relative to the level measured in sub-confluent RCECs (which corresponded to 100%). Asterisks (*) indicate PARP activities at post-confluence that are statistically different from those measured in sub-confluent cells for each specific cell type (P<0.005; Student's paired t test). Results are means±S.D.
Expression of both PARP-1 and Sp1 is under the influence of cell differentiation
As primary cultured RCECs terminally differentiate early upon cell passages (they usually sustain up to five passages in culture), we next examined how rPARP-1 promoter activity, and endogenous PARP-1 protein expression, was affected by cell differentiation in vitro. RCECs were therefore obtained from the central area of freshly dissected rabbit corneas and were sequentially passaged up to passage 5 (P5). Cells from each passage were then used for both transfection analyses and preparation of crude nuclear extracts.
RCECs produced large colonies of small cells in the primary culture (Figure 8A; P0). Although they preserved their morphological properties for a few passages (up to P2), these were progressively lost as they progressed beyond P2, the colonies being much smaller and essentially made up of large cells which could hardly be subcultured to the following passages (Figure 8A; P3 and P4). Many RCEC subcultures could not be maintained beyond P5 (Figure 8A; P5), at which point the culture exhibited a terminally differentiated phenotype mainly consisting of large non-dividing cells.
Figure 8. Influence of cell passages on PARP-1 and Sp1/Sp3 expression.
(A) Phase-contrast images of RCECs at the primary culture (P0) or at each cell passage (P1–P5). Magnification, ×200. Scale bar, 50 μm. (B) Nuclear proteins were prepared from sub-confluent RCECs serially passaged in culture up to P5 (P0–P5) and used in Western blot analyses. Expression of both PARP-1 and Sp1 proteins was monitored using the corresponding monoclonal (C-2-10 for PARP-1) or polyclonal (for Sp1) antibodies. The positions of the 116 kDa and 85 kDa molecular-mass markers (MW; sizes in kDa) are indicated. Nuclear proteins prepared from either untreated or VP16-treated HL60 cells were also included as controls for the expression of the mature 113 kDa PARP-1 protein or its 89 kDa apoptotic cleavage derivative respectively. (C) The plasmid PCR3 was transfected in sub-confluent (SC) and 2-days post-confluent (C) RCECs at P1, P2 and P3. Cells were harvested, and CAT activities were measured and expressed as percentage CAT activity relative to the level directed by sub-confluent RCECs transfected at P1 (considered as 100%). Asterisks (*) indicate CAT activities at both P2 and P3 that are statistically different from those measured in P1 cells (P<0.005; Student's paired t test). Results are means±S.D.
Nuclear extracts from RCECs grown at each passage were then used to monitor the expression of both PARP-1 and Sp1 by Western blot (Figure 8B). Expression of PARP-1 diminished progressively up to P3 and then remained stable until cells terminally differentiated (at P5). The strong reduction in PARP-1 expression did not result from RCECs being committed into apoptosis, as the 89 kDa PARP-1 apoptotic fragment yielded from the cleavage of mature PARP-1 by caspase 3 was absent from the extracts collected at each passage [compare with the nuclear proteins from HL60 cells exposed or not to the cell-death-inducer VP16 and used as controls in the assay (right-hand panel of Figure 8B)]. Expression of Sp1 was found to decrease with cell passages, following a pattern of extinction identical with that seen for the PARP-1 protein (Figure 8B). As expression of both PARP-1 and Sp1 decreased with cell passages, we then examined whether transcription directed by the rPARP-1 promoter was similarly affected by cell differentiation in vitro. PCR3 was therefore transfected into both sub-confluent and 2-day post-confluent RCECs at P1, P2 and P3 (RCECs could not be transfected efficiently beyond P3). High levels of CAT activities were observed in both sub- and 2-day post-confluent RCECs at P1. However, a dramatic reduction (10- and 14-fold reduction in sub-confluent and 2-day post-confluent cells respectively) in rPARP-1 promoter activity resulted from transfection of RCECs at P2, which fell even further at P3 (17- and 22-fold reduction in sub-confluent and 2-day post-confluent cells respectively, relative to the levels measured at P1). Despite the dramatic influence of cell passages on the rPARP-1 promoter activity, post-confluent cells always expressed the rPARP-1 promoter to a level lower than that measured at sub-confluence for each specific passage (2.3-, 3.3- and 3.0-fold reduction at P1, P2 and P3 respectively). We therefore conclude that, although basal unstimulated PARP-1 expression is strongly abrogated by differentiation through cell passaging, it remains, however, under the influence of cell density.
DISCUSSION
PARP-1 possesses many distinctive functions other than just helping or enhancing DNA repair, for which it has been initially the subject of many intensive studies (for a review, see [36]). At the transcription regulation level, PARP-1 plays at least two major functions: (i) it possesses the ability to post-translationally modify histones and therefore promotes the decondensation of higher-order chromatin structures, and (ii) it actively participates as a component of the enhancer/promoter regulatory complexes. The present study was aimed at investigating whether PARP-1 expression is influenced by cell proliferation and differentiation, two properties that are required for proper repair of wounded tissues, in primary cultured cells of various histological origins. We hypothesized that expression of PARP-1 might play a role during the proliferative phase that characterizes wound healing of the cornea, as well as other mammalian tissues, by preserving the integrity of the genomic DNA through its DNA repair function. Alternatively, by favouring unwinding of active chromatin through its action on histone proteins, PARP-1 may also facilitate the expression of genes whose encoded products are required for cell adhesion and migration of the leading edge during tissue repair. We demonstrated that primary cultured cells that progress toward terminal differentiation express levels of PARP-1 activity many times lower than those observed in undifferentiated highly prolific cells. This reduction of PARP-1 expression probably results from the co-ordinated extinction of both Sp1 and Sp3 in these cells, as the basal level of expression for these transcription factors decreased considerably at both the protein- and DNA-binding levels when all primary cultured cells reached growth arrest at post-confluence.
We have shown previously that transcription of the PARP-1 gene is primarily dictated by the recognition of its upstream promoter by the positive transcription factor Sp1 [5,8,34]. However, these results were obtained indirectly through the use of recombinant constructs bearing the rPARP-1 promoter fused to the CAT reporter gene, or through DNase I footprinting using recombinant preparations of Sp1. By exploiting inhibition of endogenous Sp1 expression through RNAi [37], we demonstrated that expression of endogenous hPARP-1 is indeed under the positive influence of this transcription factor in primary cultured HDKs.
Sp1 was reported recently by us to play a critical function during the proliferative phase required to restore an intact epithelial layer in a cell culture model recapitulating some aspects of the corneal wound healing [9,38]. The reduction of Sp1/Sp3 expression observed at post-confluence was expected to result in a similar reduction in promoter activity, as transcription directed by the rPARP-1 promoter was shown to be heavily dependent on the regulatory influence of these nuclear proteins [5,8,34]. Indeed, a comparison of the rPARP-1 promoter strength in highly prolific sub-confluent RCECs with that measured in 2-day post-confluent cells revealed a consistent reduction in promoter function in all types of post-confluent cells transfected (RCECs, HCECs, HDKs and RPE cells). The influence of cell density on the rPARP-1 promoter activity was not specific to the rPARP-1–CAT recombinant plasmids used, as a similar reduction of the endogenous PARP-1 transcripts was also observed by semi-quantitative RT-PCR. The relationship of PARP-1 extinction with cell differentiation was best appreciated through cell passaging of RCECs in vitro. Indeed, the progression of RCECs through irreversible terminal differentiation by serial passages also correlated with a dramatic, co-ordinated reduction of both Sp1 (and consequently of the rPARP-1 promoter activity as well) and PARP-1 proteins in these cells. Expression of Sp1 has been shown to predominate during the G1 phase of the cell cycle and is then subjected to proteasome-dependent degradation before the S phase, a process that is thought to be dependent on the level of Sp1 phosphorylation [10]. Consistent with this cell-cycle pattern of Sp1 expression, PARP-1 expression was also reported to increase during the G1 phase and then remained stable during the S phase of the cell cycle [13].
Much evidence points towards a major function for PARP-1 in tissue damage [39]. Tissue insults lead to DNA damage, which can arise from the formation of NO derivatives, such as peroxynitrite [39]. As a consequence of such massive DNA damage, PARP-1 becomes overactivated and may lead to an important depletion in its substrate NAD+. In response to the NAD+ depletion, the cell's attempt to resynthesize this substrate leads to a depletion of ATP and triggers the cell to die from energy loss. This allows for the elimination of cells that are too damaged to progress towards the many steps (cell adhesion, migration and proliferation) that characterize the wounding process. However, one alternative way through which PARP-1 may influence wound healing without the need for the cell to progress towards apoptosis is through alteration of transcription factors that regulate genes whose encoded products are required for cell adhesion and migration, such as membrane-bound integrins. Indeed, gene disruption or pharmacological inactivation of PARP-1 has been reported to reduce the cytokine-mediated expression of ICAM-1 (intercellular adhesion molecule-1), P-selectin and E-selectin, as well as MAdCAM-1 (mucosal addressin cell-adhesion molecule-1) in human umbilical vein endothelial cells [40]. PARP-1 has been reported to modulate the expression of the integrin CD11a in the migration of microglial cells following brain injury [41]. PARP-1 may do so either by directly interacting with transcription factors, as has been demonstrated for YY-1 (Yin Yang 1), AP-2 (where AP is activating protein), B-MYB, Oct-1 (octamer binding protein-1), TEF-1 (transcriptional enhancer factor-1) and NF-κB (reviewed in [36]), or through their poly(ADP-ribosyl)ation, as seen for p53, fos, NF-κB, and both RNA polymerases I and II [36]. Although PARP-1 has been reported most often to interfere with the positive regulatory influences that are mediated by these transcription factors, some evidence suggests that it may also act as a co-activator or enhancer factor and thereby promote gene transcription, as has been shown with NF-κB and AP-1 [42,43]. Target sites for some of these transcription factors (AP-1, AP-2, B-MYB and NF-κB) were identified in many integrin gene promoters. Both AP-1 and AP-2 are of particular interest, as binding sites for these transcription factors have been identified in the promoter of the α4, α5 and α6 integrin gene subunits [44–46], whose expression was reported to be increased during corneal wound healing [9,18,47]. The transcription factor PAX-6, a member of the Pax family that is required for proper development of many eye structures, including the cornea, the lens and the retina, is also worth mentioning, as its expression has been demonstrated to be under the influence of PARP-1, which participates as a component of protein complexes bound to the EP enhancer of the Pax-6 P0 promoter [48]. Interestingly, Pax-6 expression is increased at the migrating edge as the epithelium resurfaces the cornea after injury [49] and may contribute to corneal wound healing by modulating the expression of Pax-6-responsive genes which comprise those encoding the integrin subunits β1, α4 and α5 [18,50,51] whose encoded proteins are required for cell adhesion and migration. That Pax-6 also controls the expression of matrix metalloproteinases (for instance, MMP-9) is not surprising, as remodelling of the extracellular matrix beneath the leading edge of the healing corneal epithelium is required during its migration over the wounded area [49,51]. It is interesting to point out that activation of the Sp1 DNA-binding activity by TNF-α (tumour necrosis factor α) or LPS (lipopolysaccharide) requires PARP-1 activity, as Sp1 activation was found to be lower in PARP-1−/− relative to the level measured in PARP-1+/+ glial cells [52]. However, no clear evidence that PARP-1 may either interact with Sp1 directly or use it as a substrate for poly(ADP-ribosyl)ation has been reported to date.
Wound healing of damaged tissues is a particularly complex process that remains poorly understood at the molecular level, despite the tremendous clinical advancements reached over the last decade. The results presented in the present study raised a possible, yet undefined, function for PARP-1 in wound healing, and have added to the increasing diversity of the cellular tasks that this enzyme can achieve to dictate the fate of any given cell.
Acknowledgments
We thank Claudia Fugère and Karina Laflamme [Laboratoire d'Organogénèse Expérimentale (LOEX), Hôpital du St-Sacrement du CHA, Québec, Canada] for their help with the preparation of HDKs, HUVECs, and HVSMCs primary cultures. We are also grateful to Steve Gagnon for technical assistance with the PARP enzymatic assays. S. D. and S. L. G. are Scholars from the Fonds de la Recherche en Santé du Québec (FRSQ). K. Z. and A. R. were supported by Studentships from the Réseau de Recherche en Santé de la Vision of the FRSQ and the National Sciences and Engineering Research Council of Canada (NSERC) respectively. This work was supported by a grant from the NSERC (grant OGP0138624) to S. L. G.
References
- 1.Ame J. C., Spenlehauer C., de Murcia G. The PARP superfamily. BioEssays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 2.Augustin A., Spenlehauer C., Dumond H., Menissier-De Murcia J., Piel M., Schmit A. C., Apiou F., Vonesch J. L., Kock M., Bornens M., De Murcia G. PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J. Cell Sci. 2003;15:1551–1562. doi: 10.1242/jcs.00341. [DOI] [PubMed] [Google Scholar]
- 3.Kanai M., Tong W. M., Sugihara E., Wang Z. Q., Fukasawa K., Miwa M. Involvement of poly(ADP-ribose) polymerase 1 and poly(ADP-ribosyl)ation in regulation of centrosome function. Mol. Cell. Biol. 2003;23:2451–2462. doi: 10.1128/MCB.23.7.2451-2462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Amours D., Desnoyers S., D'Silva I., Poirier G. G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 5.Bergeron M. J., Leclerc S., Laniel M. A., Poirier G. G., Guérin S. L. Transcriptional regulation of the rat poly(ADP-ribose) polymerase gene by Sp1. Eur. J. Biochem. 1997;250:342–353. doi: 10.1111/j.1432-1033.1997.0342a.x. [DOI] [PubMed] [Google Scholar]
- 6.Laniel M. A., Poirier G. G., Guérin S. L. A conserved initiator element on the mammalian poly(ADP-ribose) polymerase-1 promoters, in combination with flanking core elements, is necessary to obtain high transcriptional activity. Biochim. Biophys. Acta. 2004;1679:37–46. doi: 10.1016/j.bbaexp.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Laniel M. A., Beliveau A., Guérin S. L. Electrophoretic mobility shift assays for the analysis of DNA–protein interactions. Methods Mol. Biol. 2001;148:13–30. doi: 10.1385/1-59259-208-2:013. [DOI] [PubMed] [Google Scholar]
- 8.Laniel M. A., Bergeron M. J., Poirier G. G., Guérin S. L. A nuclear factor other than Sp1 binds the GC-rich promoter of the gene encoding rat poly(ADP-ribose) polymerase in vitro. Biochem. Cell Biol. 1997;75:427–434. [PubMed] [Google Scholar]
- 9.Gingras M. E., Larouche K., Larouche N., Leclerc S., Salesse C., Guérin S. L. Regulation of the integrin subunit α5 gene promoter by the transcription factors Sp1/Sp3 is influenced by the cell density in rabbit corneal epithelial cells. Invest. Ophthalmol. Visual Sci. 2003;44:3742–3755. doi: 10.1167/iovs.03-0191. [DOI] [PubMed] [Google Scholar]
- 10.Grinstein E., Jundt F., Weinert I., Wernet P., Royer H. D. Sp1 as G1 cell cycle phase specific transcription factor in epithelial cells. Oncogene. 2002;21:1485–1492. doi: 10.1038/sj.onc.1205211. [DOI] [PubMed] [Google Scholar]
- 11.Thibodeau J., Gradwohl G., Dumas C., Clairoux-Moreau S., Brunet G., Penning C., Poirier G. G., Moreau P. Cloning of rodent cDNA coding the poly(ADP-ribose) polymerase catalytic domain and analysis of mRNA levels during the cell cycle. Biochem. Cell Biol. 1989;67:653–660. doi: 10.1139/o89-097. [DOI] [PubMed] [Google Scholar]
- 12.Menegazzi M., Gerosa F., Tommasi M., Uchida K., Miwa M., Sugimura T., Suzuki H., Gelosa F. Induction of poly(ADP-ribose) polymerase gene expression in lectin-stimulated human T lymphocytes is dependent on protein synthesis. Biochem. Biophys. Res. Commun. 1988;156:995–999. doi: 10.1016/s0006-291x(88)80942-2. [DOI] [PubMed] [Google Scholar]
- 13.Leduc Y., Lawrence J. J., De Murcia G., Poirier G. G. Cell cycle regulation of poly(ADP-ribose) synthetase in FR3T3 cells. Biochim. Biophys. Acta. 1988;968:275–282. doi: 10.1016/0167-4889(88)90018-3. [DOI] [PubMed] [Google Scholar]
- 14.Ludwig A., Behnke B., Holtlund J., Hilz H. Immunoquantitation and size determination of intrinsic poly(ADP-ribose) polymerase from acid precipitates: an analysis of the in vivo status in mammalian species and in lower eukaryotes. J. Biol. Chem. 1988;263:6993–6999. [PubMed] [Google Scholar]
- 15.Gao J. G., Mazella J., Tseng L. Activation of the human IGFBP-1 gene promoter by progestin and relaxin in primary culture of human endometrial stromal cells. Mol. Cell. Endocrinol. 1994;104:39–46. doi: 10.1016/0303-7207(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 16.Park H. M., Okumura J., Muramatsu T. Modulation of transcriptional activity of the chicken ovalbumin gene promoter in primary cultures of chicken oviduct cells: effects of putative regulatory elements in the 5′-flanking region. Biochem. Mol. Biol. Int. 1995;36:811–816. [PubMed] [Google Scholar]
- 17.Larouche K., Leclerc S., Salesse C., Guérin S. L. Expression of the α5 integrin subunit gene promoter is positively regulated by the extracellular matrix component fibronectin through the transcription factor Sp1 in corneal epithelial cells in vitro. J. Biol. Chem. 2000;275:39182–39192. doi: 10.1074/jbc.M002945200. [DOI] [PubMed] [Google Scholar]
- 18.Zaniolo K., Leclerc S., Cvekl A., Vallieres L., Bazin R., Larouche K., Guérin S. L. Expression of the alpha4 integrin subunit gene promoter is modulated by the transcription factor Pax-6 in corneal epithelial cells. Invest. Ophthalmol. Visual Sci. 2004;45:1692–1704. doi: 10.1167/iovs.03-0908. [DOI] [PubMed] [Google Scholar]
- 19.Ullrich O., Diestel A., Eyupoglu I. Y., Nitsch R. Regulation of microglial expression of integrins by poly(ADP-ribose) polymerase-1. Nat. Cell Biol. 2001;3:1035–1042. doi: 10.1038/ncb1201-1035. [DOI] [PubMed] [Google Scholar]
- 20.Lu L., Reinach P. S., Kao W. W. Corneal epithelial wound healing. Exp. Biol. Med. 2001;226:653–664. doi: 10.1177/153537020222600711. [DOI] [PubMed] [Google Scholar]
- 21.Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- 22.de Vries E., van Driel W., van den Heuvel S. J., van der Vliet P. C. Contactpoint analysis of the HeLa nuclear factor I recognition site reveals symmetrical binding at one side of the DNA helix. EMBO J. 1987;6:161–168. doi: 10.1002/j.1460-2075.1987.tb04734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Germain L., Auger F. A., Grandbois E., Guignard R., Giasson M., Boisjoly H., Guérin S. L. Reconstructed human cornea produced in vitro by tissue engineering. Pathobiology. 1999;67:140–147. doi: 10.1159/000028064. [DOI] [PubMed] [Google Scholar]
- 24.Germain L., Rouabhia M., Guignard R., Carrier L., Bouvard V., Auger F. A. Improvement of human keratinocyte isolation and culture using thermolysin. Burns. 1993;19:99–104. doi: 10.1016/0305-4179(93)90028-7. [DOI] [PubMed] [Google Scholar]
- 25.Jaffe E. A., Nachman R. L., Becker C. G., Minick C. R. Culture of human endothelial cells derived from umbilical veins: identification by morphologic and immunologic criteria. J. Clin. Invest. 1973;52:2745–2756. doi: 10.1172/JCI107470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voyta J. C., Via D. P., Butterfield C. E., Zetter B. R. Identification and isolation of endothelial cells based on their increased uptake of acetylated-low density lipoprotein. J. Cell Biol. 1984;99:2034–2040. doi: 10.1083/jcb.99.6.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross R. The smooth muscle cell. II. Growth of smooth muscle in culture and formation of elastic fibers. J. Cell Biol. 1971;50:172–186. doi: 10.1083/jcb.50.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Proulx S., Guérin S. L., Salesse C. Effect of quiescence on integrin α5β1 expression in human retinal pigment epithelium. Mol. Vision. 2003;9:473–481. [PubMed] [Google Scholar]
- 29.Pothier F., Ouellet M., Julien J. P., Guérin S. L. An improved CAT assay for promoter analysis in either transgenic mice or tissue culture cells. DNA Cell Biol. 1992;11:83–90. doi: 10.1089/dna.1992.11.83. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J., Fritsch E. F., Maniatis T. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 31.Laniel M. A., Poirier G. G., Guérin S. L. Nuclear factor 1 interferes with Sp1 binding through a composite element on the rat poly(ADP-ribose) polymerase promoter to modulate its activity in vitro. J. Biol. Chem. 2001;276:20766–20773. doi: 10.1074/jbc.M010360200. [DOI] [PubMed] [Google Scholar]
- 31a.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 32.Duriez P. J., Desnoyers S., Hoflack J. C., Shah G. M., Morelle B., Bourassa S., Poirier G. G., Talbot B. Characterization of anti-peptide antibodies directed towards the automodification domain and apoptotic fragment of poly(ADP-ribose) polymerase. Biochim. Biophys. Acta. 1997;1334:65–72. doi: 10.1016/s0304-4165(96)00077-3. [DOI] [PubMed] [Google Scholar]
- 33.Shah G. M., Poirier D., Duchaine C., Brochu G., Desnoyers S., Lagueux J., Verreault A., Hoflack J. C., Kirkland J. B., Poirier G. G. Methods for biochemical study of poly(ADP-ribose) metabolism in vitro and in vivo. Anal. Biochem. 1995;227:1–13. doi: 10.1006/abio.1995.1245. [DOI] [PubMed] [Google Scholar]
- 34.Potvin F., Roy R. J., Poirier G. G., Guérin S. L. The US-1 element from the gene encoding rat poly(ADP-ribose) polymerase binds the transcription factor Sp1. Eur. J. Biochem. 1993;215:73–80. doi: 10.1111/j.1432-1033.1993.tb18008.x. [DOI] [PubMed] [Google Scholar]
- 35.Shieh W. M., Ame J. C., Wilson M. V., Wang Z. Q., Koh D. W., Jacobson M. K., Jacobson E. L. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 36.Bouchard V. J., Rouleau M., Poirier G. G. PARP-1, a determinant of cell survival in response to DNA damage. Exp. Hematol. 2003;31:446–454. doi: 10.1016/s0301-472x(03)00083-3. [DOI] [PubMed] [Google Scholar]
- 37.Abdelrahim M., Samudio I., Smith R., 3rd, Burghardt R., Safe S. Small inhibitory RNA duplexes for Sp1 mRNA block basal and estrogen-induced gene expression and cell cycle progression in MCF-7 breast cancer cells. J. Biol. Chem. 2002;277:28815–28822. doi: 10.1074/jbc.M203828200. [DOI] [PubMed] [Google Scholar]
- 38.Gaudreault M., Carrier P., Larouche K., Leclerc S., Giasson M., Germain L., Guérin S. L. Influence of sp1/sp3 expression on corneal epithelial cells proliferation and differentiation properties in reconstructed tissues. Invest. Ophthalmol. Visual Sci. 2003;44:1447–1457. doi: 10.1167/iovs.02-0707. [DOI] [PubMed] [Google Scholar]
- 39.Szabo C. Role of poly(ADP-ribose)synthetase in inflammation. Eur. J. Pharmacol. 1998;350:1–19. doi: 10.1016/s0014-2999(98)00249-0. [DOI] [PubMed] [Google Scholar]
- 40.Oshima T., Pavlick K. P., Laroux F. S., Verma S. K., Jordan P., Grisham M. B., Williams L., Alexander J. S. Regulation and distribution of MAdCAM-1 in endothelial cells in vitro. Am. J. Physiol. Cell Physiol. 2001;281:C1096–C1105. doi: 10.1152/ajpcell.2001.281.4.C1096. [DOI] [PubMed] [Google Scholar]
- 41.Diestel A., Aktas O., Hackel D., Hake I., Meier S., Raine C. S., Nitsch R., Zipp F., Ullrich O. Activation of microglial poly(ADP-ribose)-polymerase-1 by cholesterol breakdown products during neuroinflammation: a link between demyelination and neuronal damage. J. Exp. Med. 2003;198:1729–1740. doi: 10.1084/jem.20030975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliver F. J., Menissier-de Murcia J., Nacci C., Decker P., Andriantsitohaina R., Muller S., de la Rubia G., Stoclet J. C., de Murcia G. Resistance to endotoxic shock as a consequence of defective NF-κB activation in poly(ADP-ribose) polymerase-1 deficient mice. EMBO J. 1999;18:4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zingarelli B., Hake P. W., O'Connor M., Denenberg A., Wong H. R., Kong S., Aronow B. J. Differential regulation of activator protein-1 and heat shock factor-1 in myocardial ischemia and reperfusion injury: role of poly(ADP-ribose) polymerase-1. Am. J. Physiol. Heart. Circ. Physiol. 2004;286:H1408–H1415. doi: 10.1152/ajpheart.00953.2003. [DOI] [PubMed] [Google Scholar]
- 44.De Meirsman C., Schollen E., Jaspers M., Ongena K., Matthijs G., Marynen P., Cassiman J. J. Cloning and characterization of the promoter region of the murine α-4 integrin subunit. DNA Cell Biol. 1994;13:743–754. doi: 10.1089/dna.1994.13.743. [DOI] [PubMed] [Google Scholar]
- 45.Lin C. S., Chen Y., Huynh T., Kramer R. Identification of the human α6 integrin gene promoter. DNA Cell Biol. 1997;16:929–937. doi: 10.1089/dna.1997.16.929. [DOI] [PubMed] [Google Scholar]
- 46.Schollen E., De Meirsman C., Matthijs G., Cassiman J. J. A regulatory element in the 5′UTR directs cell-specific expression of the mouse α4 gene. Biochem. Biophys. Res. Commun. 1995;211:115–122. doi: 10.1006/bbrc.1995.1785. [DOI] [PubMed] [Google Scholar]
- 47.Stepp M. A., Zhu L., Cranfill R. Changes in β4 integrin expression and localization in vivo in response to corneal epithelial injury. Invest. Ophthalmol. Visual Sci. 1996;37:1593–1601. [PubMed] [Google Scholar]
- 48.Plaza S., Aumercier M., Bailly M., Dozier C., Saule S. Involvement of poly (ADP-ribose)-polymerase in the Pax-6 gene regulation in neuroretina. Oncogene. 1999;18:1041–1051. doi: 10.1038/sj.onc.1202406. [DOI] [PubMed] [Google Scholar]
- 49.Sivak J. M., Mohan R., Rinehart W. B., Xu P. X., Maas R. L., Fini M. E. Pax-6 expression and activity are induced in the reepithelializing cornea and control activity of the transcriptional promoter for matrix metalloproteinase gelatinase B. Dev. Biol. 2000;222:41–54. doi: 10.1006/dbio.2000.9694. [DOI] [PubMed] [Google Scholar]
- 50.Chauhan B. K., Reed N. A., Yang Y., Cermak L., Reneker L., Duncan M. K., Cvekl A. A comparative cDNA microarray analysis reveals a spectrum of genes regulated by Pax6 in mouse lens. Genes Cells. 2002;7:1267–1283. doi: 10.1046/j.1365-2443.2002.00602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sivak J. M., West-Mays J. A., Yee A., Williams T., Fini M. E. Transcription factors Pax6 and AP-2α interact to coordinate corneal epithelial repair by controlling expression of matrix metalloproteinase gelatinase B. Mol. Cell. Biol. 2004;24:245–257. doi: 10.1128/MCB.24.1.245-257.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ha H. C., Hester L. D., Snyder S. H. Poly(ADP-ribose) polymerase-1 dependence of stress-induced transcription factors and associated gene expression in glia. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3270–3275. doi: 10.1073/pnas.052712399. [DOI] [PMC free article] [PubMed] [Google Scholar]