Abstract
PH-PLCδ1 [the PH domain (pleckstrin homology domain) of PLCδ1 (phospholipase C-δ1)] is among the best-characterized phosphoinositide-binding domains. PH-PLCδ1 binds with high specificity to the headgroup of PtdIns(4,5)P2, but little is known about its interfacial properties. In the present study, we show that PH-PLCδ1 is also membrane-active and can insert significantly into PtdIns(4,5)P2-containing monolayers at physiological (bilayer-equivalent) surface pressures. However, this membrane activity appears to involve interactions distinct from those that target PH-PLCδ1 to the PtdIns(4,5)P2 headgroup. Whereas the majority of PtdIns(4,5)P2-bound PH-PLCδ1 can be displaced by adding excess of soluble headgroup [Ins(1,4,5)P3], membrane activity of PH-PLCδ1 cannot. PH-PLCδ1 differs from other phosphoinositide-binding domains in that its membrane insertion does not require that the phosphoinositide-binding site be occupied. Significant monolayer insertion remains when the phosphoinositide-binding site is mutated, and PH-PLCδ1 can insert into monolayers that contain no PtdIns(4,5)P2 at all. Our results suggest a model in which reversible membrane binding of PH-PLCδ1, mediated by PtdIns(4,5)P2 or other acidic phospholipids, occurs without membrane insertion. Accumulation of the PH domain at the membrane surface enhances the efficiency of insertion, but does not significantly affect its extent, whereas the presence of phosphatidylethanolamine and cholesterol in the lipid mixture promotes the extent of insertion. This is the first report of membrane activity in an isolated PH domain and has implications for understanding the membrane targeting by this common type of domain.
Keywords: membrane activity, membrane insertion, lipid-binding domain, phosphoinositide, phospholipase C, pleckstrin homology domain (PH domain)
Abbreviations: PC, phosphatidylcholine; PE, phosphatidylethanolamine; PH domain, pleckstrin homology domain; PLCδ1, phospholipase C-δ1; PH-PLCδ1, PH domain of PLCδ1; PS, phosphatidylserine; RU, response unit; SPR, surface plasmon resonance
INTRODUCTION
PH-PLCδ1 [the PH domain (pleckstrin homology domain) of PLCδ1 (phospholipase C-δ1)] was the first PH domain shown to bind with high affinity and stereospecificity to a particular membrane phosphoinositide [PtdIns(4,5)P2] [1,2], by recognizing its headgroup, Ins(1,4,5)P3 [3]. Membrane association of PH domains has been argued to arise from a combination of such direct headgroup binding (which may be specific) and non-specific delocalized electrostatic attraction of the domain to negatively charged membrane surfaces [4–6]. The relative contributions of these two driving forces appear to vary widely for different PH domains, with specific PH domains relying more on headgroup recognition and promiscuous PH domains binding primarily through delocalized electrostatic attraction [7,8].
Recent studies have suggested that the situation may be more complex [8–10]. For example, the PH domains from p130 and PLCδ1 were shown to differ greatly in their membrane-targeting and binding abilities, despite binding to Ins(1,4,5)P3 and monomeric PtdIns(4,5)P2 with almost identical affinities [11]. It was suggested that sequences unique to the C-terminal half of PH-PLCδ1 provide auxiliary interactions with the membrane that are absent for p130. One possible source of these additional interactions is membrane activity, achieved by inserting hydrophobic side chains into the apolar region of the bilayer to strengthen membrane association. Although not previously studied for PH domains, compelling evidence for such membrane insertion/activity has been reported for almost all other phosphoinositide-binding modules [5,12–18]. The crystal structure of PH-PLCδ1 bound to Ins(1,4,5)P3 [3,4] suggested that two hydrophobic side chains in one of the variable loops are well-positioned to penetrate the membrane interface when it docks on to PtdIns(4,5)P2. In addition, a solid-state 13C-NMR study has indicated that the conformation of PH-PLCδ1 is altered upon interaction with PtdIns-(4,5)P2-containing vesicles, and the authors suggested that a part of the PH domain could insert into the membrane [19]. Membrane insertion has been reported for full-length PLCδ1 [20], and may play a role in prolonging the residence time of the enzyme during its action at the plasma membrane.
We have analysed the membrane activity and binding of PH-PLCδ1 using mixed-lipid monolayers and bilayers. PH-PLCδ1 appears to be capable of binding to PtdIns(4,5)P2-containing membranes by reversibly associating with the PtdIns(4,5)P2 headgroup in a way that does not lead to detectable membrane insertion. A second mode of PH-PLCδ1 binding to membranes that does involve membrane insertion appears to be independent of PtdIns-(4,5)P2, but is accelerated by its presence. These findings distinguish PH-PLCδ1 from other phosphoinositide-dependent membrane-targeting modules that have been studied so far.
EXPERIMENTAL
Materials
Bovine brain PtdIns4P and PtdIns(4,5)P2, liver PtdIns and the (1,2-dioleoyl) phospholipids PC (phosphatidylcholine), PE (phosphatidylethanolamine) and PS (phosphatidylserine) were obtained from Avanti Polar Lipids (Birmingham, AL, U.S.A.). Cholesterol was obtained from Merck (Darmstadt, Germany) and D- and L-myo-inositol-1,4,5-trisphosphate from Calbiochem (La Jolla, CA, U.S.A.). The PH domain of rat PLCδ1 (residues 11–140) and its triple mutant [K32A (Lys32→Ala), W36N and R38K] were produced and purified as described previously [2,3,21] and stored at −80 °C in 50 mM Mes (pH 6.0), 100 mM NaCl, 1 mM dithiothreitol and 25% (w/v) glycerol. SPR (surface plasmon resonance) studies were performed with freshly produced PH domains. Protein concentration was determined by absorbance A at 278 nm [2].
Monolayer measurements
Monolayer experiments were performed as described in [22] at constant area and 37 °C, using a 1.6 ml Teflon trough, which was 2 cm in diameter and had a monolayer surface area of 3.14 cm2. The subphase contained 20 mM Hepes/NaOH (pH 7.0), 1 mM MgCl2, 2 mM EGTA, 100 mM NaCl and 1 mM dithiothreitol (HCB100).
Analysis of lipid binding by SPR
Membrane binding was monitored by SPR exactly as described in [8] using a Biacore® 3000 (Biacore AB, Uppsala, Sweden) with a L1 sensor chip and HCB100 as the flow buffer (at 25 °C, flow rate of 10 μl/min). Large unilamellar vesicles were prepared and immobilized as described in [23,24]. Flowcell 1 was always coated with pure PC vesicles for real-time background subtraction. PH-PLCδ1 was injected at a range of concentrations (in 50 μl) at 25 °C; dissociation was then monitored for 3 min and a wash with 10 μl of 100 mM NaOH was applied to remove residual bound protein (but not lipid). The equilibrium-binding signal was measured for each injection, and GraphPAD Prism (version 4) was used to fit the curves of binding versus [PH-PLCδ1] to simple one- or two-site models using least-squares regression analysis.
RESULTS
Membrane activity of PH-PLCδ1
To investigate whether PH-PLCδ1 is membrane-active, we spread a mixed-lipid monolayer composed of PC/PE/PS/cholesterol/PtdIns(4,5)P2 (1:1:1:1:1, by mol) at an air/water interface with an initial surface pressure of 25 mN/m and then injected increasing amounts of PH-PLCδ1 into the subphase. As shown in Figure 1, PH-PLCδ1 addition caused a clear increase in the surface pressure of the lipid monolayer. We established that the surface properties of PH-PLCδ1 itself cannot explain this finding by showing that the maximum surface pressure obtainable with the PH domain alone (at 6 μM and in the absence of a lipid monolayer) was just 20.2 mN/m (results not shown). Since this value is significantly lower than the initial surface pressure in our monolayer studies (25 mN/m), the PH-PLCδ1-induced increase in surface pressure observed in Figure 1 must result from the penetration of PH-PLCδ1 into the lipid monolayer. The increase in monolayer surface pressure reached its maximal level at a PH-PLCδ1 concentration of 169 nM, and this saturating amount was then used in all subsequent monolayer experiments.
Figure 1. Membrane activity of PH-PLCδ1.
Insertion into lipid monolayers spread at an initial surface pressure of 25 mN/m. Surface pressure increase was measured as a function of PH-PLCδ1 concentration in the subphase (1.6 ml) beneath a mixed-lipid monolayer composed of PC/PE/PS/cholesterol/PtdIns(4,5)P2 (1:1:1:1:1, by mol).
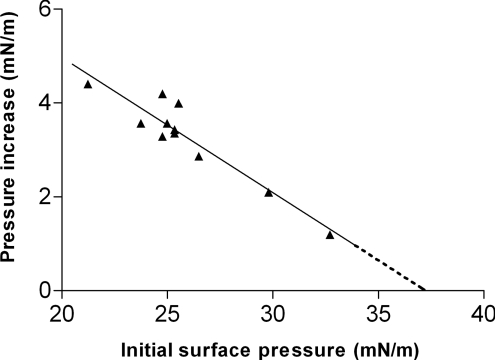
To determine whether the observed membrane activity of PH-PLCδ1 can occur in physiological membranes, we next investigated its ability to insert into PC/PE/PS/cholesterol/PtdIns(4,5)P2 (1:1:1:1:1) monolayers at a series of different initial surface pressures. Higher initial monolayer surface pressures correlate with higher lipid packing densities and hinder protein insertion. This effect is observed as a progressive reduction in the surface pressure increase that is induced by PH-PLCδ1 as the initial surface pressure is increased (Figure 2). Extrapolation of these data yields the exclusion pressure, defined as the initial surface pressure beyond which the protein can no longer insert into the monolayer. The data presented in Figure 2 yield an exclusion pressure estimate for PH-PLCδ1 in the PtdIns(4,5)P2-containing mixed-lipid monolayers of 37.2 mN/m. Physiological bilayers are estimated to have surface pressures of 30–35 mN/m [25,26]. Since the PH-PLCδ1 exclusion pressure significantly exceeds all estimates of the surface pressures in biological membranes, we suggest that PH-PLCδ1 is capable of inserting into biomembranes at physiological surface pressures, and this is corroborated by SPR analysis of immobilized mixed-lipid membrane bilayers (see below).
Figure 2. Dependence of PH-PLCδ1 insertion on initial monolayer surface pressure.
The increase in monolayer surface pressure induced by the addition of PH-PLCδ1 (169 nM) was monitored as a function of the initial surface pressure. Mixed-lipid monolayers composed of PtdIns(4,5)P2 (20 mol%) in PC/PE/PS/cholesterol (1:1:1:1) were used, and the surface pressure increase was measured after equilibrium was reached. Linear regression analysis gave an x-axis intercept, the monolayer exclusion pressure, of 37.2 mN/m (r2=0.88).
Specificity and lipid dependence of membrane activity
We next tested the lipid specificity of monolayer insertion by injecting PH-PLCδ1 into the subphase beneath a series of mixed-lipid monolayers composed respectively of PC/PE/PS/cholesterol (1:1:1:1) plus (a) 20% (mol/mol) PtdIns(4,5)P2; (b) 20% PtdIns4P; (c) 20% (mol/mol) PS (to give a final 40% PS) and (d) a neutral mixture of PC/PE/cholesterol (1:1:1). As shown in Figure 3(A), significant surface pressure increases were observed in every case. The rate and extent (after 30 min) of PH-PLCδ1 insertion were greatest for the PtdIns(4,5)P2-containing monolayer (curve a), but only decreased slightly (extent by approx. 5% and rate by approx. 22%) when PtdIns(4,5)P2 was replaced by PtdIns4P (curve b). In the monolayer containing 40% PS (curve c), the extent of insertion after 30 min was decreased by only 25% compared with that seen for the PtdIns(4,5)P2-containing monolayer, and the rate of insertion was further decreased (compare curve c with a). In the absence of any acidic phospholipids (using a 1:1:1 mixture of PC/PE/cholesterol), the rate of PH-PLCδ1 insertion into the lipid monolayer was substantially decreased (curve d), but the final extent of insertion in the experiment was still only approx. 30% smaller than that seen with 20% PtdIns(4,5)P2+20% PS. Thus, although the most rapid monolayer insertion by PH-PLCδ1 was seen when PtdIns(4,5)P2 was present, these results show that this phosphoinositide is not required for insertion. In fact, significant monolayer insertion of PH-PLCδ1 can occur in the absence of any anionic phospholipid at all. The presence of these lipids [or the presence of PtdIns(4,5)P2] appears primarily to influence the rate of PH-PLCδ1 insertion.
Figure 3. Dependence of PH-PLCδ1 membrane activity on lipid type.
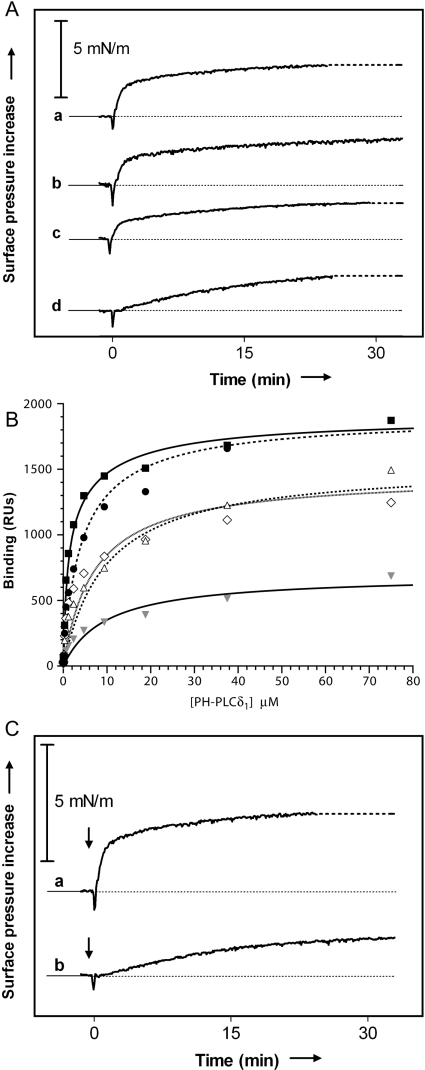
(A) PH-PLCδ1 (169 nM) was injected beneath monolayers of different lipid compositions at an initial surface pressure of 25 mN/m. Curve a, PtdIns(4,5)P2 (20 mol%) in PC/PE/PS/cholesterol (1:1:1:1); curve b, PtdIns4P (20 mol%) in PC/PE/PS/cholesterol (1:1:1:1); curve c, PS (20 mol%) in PC/PE/PS/cholesterol (1:1:1:1); and curve d, PC/PE/cholesterol (1:1:1). Typical results are shown (n>2). (B) SPR analysis of PH-PLCδ1 binding to mixed-lipid membranes. PH-PLCδ1 was injected at a range of concentrations on to immobilized membranes composed of PC/PE/PS/cholesterol (1:1:1:1)+20 mol% of PtdIns(4,5)P2 (■), PtdIns4P (●), PtdIns (△), PS (◇) or PC (grey triangle). Results shown are from representative experiments, and the best-fit curves plotted represent an average of fits to at least three independent experiments. Curves were fit to a two-site binding model in which one site (constant in all fits) represented binding to the background lipid mixture (with KD=9.7 μM and Rmax=690 RU), and the other site represented simple 1:1 binding to PtdIns(4,5)P2 (KD=0.8 μM), PtdIns4P (KD=2.8 μM) etc. (C) Surface pressure increase as a function of time after the addition (at t=0) of PH-PLCδ1 (169 nM) for monolayers containing 20% PtdIns(4,5)P2 in a lipid background of PC/PE/PS/cholesterol (1:1:1:1) (curve a) or pure PC (curve b) (initial surface pressure, 25 mN/m).
Parallel to these studies, we also used SPR for a direct analysis of PH-PLCδ1 binding to membranes with lipid compositions identical with those used in our monolayer studies. As shown in Figure 3(B, filled grey triangles), PH-PLCδ1 bound significantly to membranes composed of PC/PE/PS/cholesterol (2:1:1:1). The resulting binding curves saturated at 690 RU (response units) and gave an apparent dissociation constant KD=9.7 μM. In membranes composed of PC/PE/PS/cholesterol (1:1:2:1) or PC/PE/PS/cholesterol/PI (1:1:1:1:1), in which the percentage of acidic lipids is increased by 2-fold (Figure 3B, open diamonds and triangles), the binding capacity of the sensor chip-immobilized membrane was approximately doubled (maximal response Rmax∼1400 RU), but the apparent affinity was unchanged (KD=5–10 μM in each case). As anticipated, adding PtdIns(4,5)P2 or PtdIns4P to 20% PS had a much greater influence. Curves for PH-PLCδ1 binding to the phosphoinositide-containing membranes appeared to represent a two-site combination of a simple (1:1) PtdIns(4,5)P2 (or PtdIns4P) binding and a lower affinity binding to the background lipid mix. The best fit to the PtdIns(4,5)P2 data suggests that PH-PLCδ1 binds PtdIns(4,5)P2 with a KD of 0.8 μM (Rmax=1207 RU) and the background lipid with KD=9.7 μM and Rmax=690 RU (using values fit for the background lipid alone). At saturation, PtdIns(4,5)P2 recruitment would thus be responsible for approx. 64% of the bound PH-PLCδ1. For PtdIns4P, the best-fit KD value for the higher affinity is 3.5-fold weaker (at 2.8 μM; Rmax=1214), in agreement with previous studies [1,2]. The relative affinity of PH-PLCδ1 for the different mixed-lipid membranes thus correlates well with the rate of its insertion into the corresponding monolayers (Figure 3A).
These studies argue that PtdIns(4,5)P2 strongly promotes PH-PLCδ1 binding to membranes, but that its presence is not critical for the observed membrane activity of PH-PLCδ1. Rather, the overall nature of the membrane or monolayer surface may be a more important determinant of PH-PLCδ1 membrane activity. To investigate this possibility further, we analysed PH-PLCδ1 insertion into monolayers that contained identical amounts of PtdIns-(4,5)P2, but in two quite different background contexts: one of pure PC, and another similar to cellular membranes (PC/PE/PS/cholesterol in the ratio 1:1:1:1). As shown in Figure 3(C), both the extent (after 30 min) and the rate of PH-PLCδ1 insertion were significantly decreased for monolayers containing PtdIns(4,5)P2 in a pure PC background when compared with the PC/PE/PS/cholesterol (1:1:1:1) background. This result suggests that the membrane activity of PH-PLCδ1 may be largely independent of its phosphoinositide binding and that the presence of PE, PS or cholesterol is an important determinant of the maximum extent and efficiency of PH-PLCδ1 insertion into PtdIns(4,5)P2-containing monolayers.
Membrane insertion of PH-PLCδ1 does not require an intact PtdIns(4,5)P2-binding site
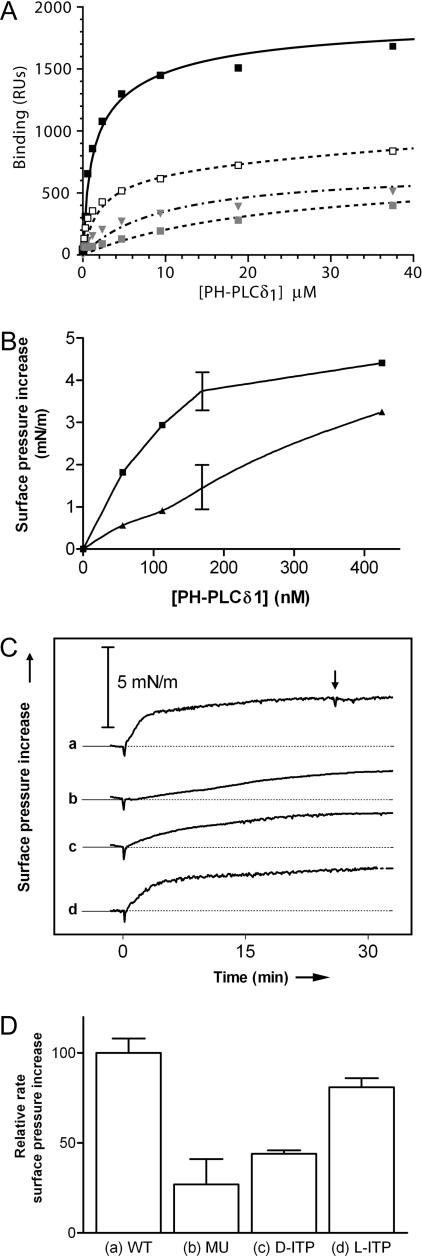
Since the presence of PtdIns(4,5)P2 is not required for monolayer insertion by PH-PLCδ1 (although it does increase the rate of insertion, as shown in Figure 3A), we predicted that mutating the PtdIns(4,5)P2-binding site should not abolish PH-PLCδ1 insertion (although it should decrease its efficiency). To test this, we used a mutated form of PH-PLCδ1 with substitutions (K32A, W36N and R38K) of three amino acids that are critical for phosphoinositide binding [6,21]. We used SPR to demonstrate that these mutations greatly decrease the affinity of PH-PLCδ1 for PtdIns(4,5)P2 (Figure 4A). The best fit to the binding curve suggested a very low affinity contribution from residual PtdIns(4,5)P2 binding on top of a slightly increased affinity for background lipid (by 2–3-fold). We have previously shown that this set of mutations prevents PH-PLCδ1 from inhibiting PtdIns(4,5)P2-dependent processes [21]. Monolayer studies presented in Figure 4(B) showed that the membrane activity of the mutated PH domain towards PC/PE/PS/cholesterol/PtdIns(4,5)P2 (1:1:1:1:1) monolayers is decreased when it is added at low protein concentrations (<400 nM; see also curve b in Figure 4C). However, the binding site mutations do not greatly decrease the maximum extent of monolayer insertion by PH-PLCδ1 observed at protein concentrations above 400 nM. Thus, disrupting the PtdIns(4,5)P2-binding site does not prevent PH domain insertion into the lipid monolayer.
Figure 4. Dependence of PH-PLCδ1 binding and membrane activity on occupancy of the phosphoinositide-binding site.
(A) In SPR studies, a triple mutation in the phosphoinositide-binding site (□) or addition of 100 μM D-myo-Ins(1,4,5)P3 (grey square) significantly decreased binding to membranes comprising 20 mol% PtdIns(4,5)P2 in a PC/PE/PS/cholesterol (1:1:1:1) background, as compared with control (wild-type PH-PLCδ1; ■). Both of these manipulations caused binding resembling that seen for wild-type PH-PLCδ1 with a PC/PE/PS/cholesterol (2:1:1:1) mixture (grey triangle). (B) To assess membrane activity, lipid monolayers containing 20 mol% of PtdIns(4,5)P2 in PC/PE/PS/cholesterol (1:1:1:1:1) were spread at 25 mN/m. Surface pressure increase as a function of protein concentration is plotted for wild-type PH-PLCδ1 (■) and the binding-defective mutated form of PH-PLCδ1 (▲). (C) Surface pressure increase induced by the injection of 169 nM of the following: (a) wild-type PH-PLCδ1; (b) mutated PH-PLCδ1; (c) wild-type PH-PLCδ1+10 μM D-myo-Ins(1,4,5)P3; (d) wild-type PH-PLCδ1+10 μM L-myo-Ins(1,4,5)P3 (initial surface pressure of 25 mN/m). The vertical arrow in curve a indicates the time at which 10 μM D-myo-Ins(1,4,5)P3 was added after insertion had been allowed to proceed. Data from (C) are quantified in (D) where the surface pressure increase in the first 5 min after protein injection is presented as a measure of the initial rate of insertion (n=2, error bars indicate range).
In a complementary set of experiments, we analysed the effect of adding excess D-myo-Ins(1,4,5)P3 on PH-PLCδ1 membrane activity. D-myo-Ins(1,4,5)P3 binds to the PH-PLCδ1 phosphoinositide-binding site approx. eight times more strongly than does membrane-embedded PtdIns(4,5)P2, and D-myo-Ins(1,4,5)P3 efficiently competes PH-PLCδ1 away from PtdIns(4,5)P2-containing membranes [2,7,27]. SPR studies show that the presence of 100 μM D-myo-Ins(1,4,5)P3 decreases wild-type PH-PLCδ1 binding to PtdIns(4,5)P2-containing membranes from the high-affinity interaction described above (filled black squares in Figures 3B and 4A) to a binding curve that resembles binding of unliganded PH-PLCδ1 to background lipid, but with decreased affinity (KD∼24 μM, Rmax∼650 RU; Figure 4A, filled grey squares). Singh and Murray [4] have reported that binding of D-myo-Ins(1,4,5)P3 to PH-PLCδ1 neutralizes much of the charge on its positively charged face, which in turn will decrease its non-specific electrostatic attraction for negatively charged membranes, and this may explain the fact that D-myo-Ins(1,4,5)P3 binding decreases its membrane affinity to below background.
In monolayer studies, adding 10 μM D-myo-Ins(1,4,5)P3 to the subphase together with PH-PLCδ1 also considerably decreased the rate of monolayer insertion by the PH domain, but cut the extent of its insertion after 30 min by only approx. 35% (curve c in Figure 4C, quantified in Figure 4D). As a specificity control, we showed that L-myo-Ins(1,4,5)P3 (which binds PH-PLCδ1 42 times more weakly than the D-isomer [2]) has little influence on PH-PLCδ1 insertion (d in Figures 4C–4D).
Thus mutating the PtdIns(4,5)P2-binding site of PH-PLCδ1, or occupying it with excess Ins(1,4,5)P3, appears to negate the enhancing effect of PtdIns(4,5)P2 on monolayer insertion by this PH domain. However, these manipulations [or omitting PtdIns(4,5)P2 from the lipid mixture] do not abolish monolayer insertion and appear to have little effect on its extent. The membrane activity of PH-PLCδ1 therefore appears to be independent of its phosphoinositide binding, although phosphoinositide binding increases the rate at which it occurs.
Monolayer insertion of PH-PLCδ1 is not reversible
To investigate further what causes the surface pressure increase observed in Figure 1, we took advantage of the long-standing observation that D-myo-Ins(1,4,5)P3 displaces most of the bound PH-PLCδ1 from PtdIns(4,5)P2-containing membranes in vitro [7] and in vivo [27]. If recruitment of PH-PLCδ1 to PtdIns(4,5)P2-containing membranes leads to its insertion into the membrane, we would expect the increase in surface pressure to be reversed when D-myo-Ins(1,4,5)P3 is added. Surprisingly, this was not the case. Although addition of D-myo-Ins(1,4,5)P3 during the insertion phase clearly decreased the efficiency of insertion (Figure 4C, curve c), it could not reverse the process if added after allowing insertion to proceed [Figure 4C, curve a: D-myo-Ins(1,4,5)P3 was added at the time indicated by the vertical arrow]. In contrast, direct binding studies indicate that excess D-myo-Ins(1,4,5)P3 can displace ≥ 80% of PH-PLCδ1 from similar membranes [7]. The SPR sensorgram shown in Figure 5 (solid trace) illustrates that most of the binding of PH-PLCδ1 to PtdIns(4,5)P2-containing membranes is rapidly reversible, although dissociation is not complete, and it is possible that the ‘residual’ (∼200 RU or 20%) binding might represent PH-PLCδ1 that has inserted into the membrane irreversibly (but is dissociable with 0.1 M NaOH). Adding an excess 100 μM D-myo-Ins(1,4,5)P3 in SPR experiments during the association phase greatly impaired binding of 2 μM PH-PLCδ1 to PC/PE/PS/cholesterol/PtdIns(4,5)P2 membranes (Figure 5, broken line). However, if 2 μM PH-PLCδ1 was allowed to bind the membrane, and D-myo-Ins(1,4,5)P3 was added (to 100 μM) after the dissociation phase (Figure 5, vertical arrow), it had no effect at all on the ‘residual’ PH-PLCδ1 binding. These observations mirror those seen in the monolayer studies in Figure 4(C, curve a).
Figure 5. Influence of added D-myo-Ins(1,4,5)P3 on PH-PLCδ1 binding to PtdIns(4,5)P2-containing membranes.
PH-PLCδ1 at 2 μM was injected on to a mixed PC/PE/PS/cholesterol/PtdIns(4,5)P2 (1:1:1:1:1) membrane either without (solid line) or with (broken line) 100 μM D-myo-Ins(1,4,5)P3 during the association phase. Contact time for this particular injection was only half that used for equilibrium binding experiments, so steady-state binding was not yet fully attained. The vertical arrow indicates the addition of D-myo-Ins(1,4,5)P3 (to 100 μM) after the dissociation phase.
Taken together, these studies show that monolayer insertion by PH-PLCδ1 is accelerated by (but is not dependent on) phosphoinositides, and also suggest that monolayer insertion by PH-PLCδ1 is irreversible. Importantly, these findings also argue that the reversible PtdIns(4,5)P2-dependent recruitment of PH-PLCδ1 to membranes seen in vitro and in vivo occurs without detectable membrane insertion. The membrane activity of PH-PLCδ1 seems to be an independent property that may play a role in longer-lived membrane association.
DISCUSSION
The most straightforward interpretation of our data is that PH-PLCδ1 has two modes of membrane binding. The PH domain appears to bind the surface of PtdIns(4,5)P2-containing membranes in a freely reversible manner by specifically recognizing the phosphoinositide headgroup, and this mode of membrane association does not involve detectable membrane penetration. The lack of apparent membrane activity associated with this mode of interaction suggests that the crystal structure of a PH-PLCδ1–Ins(1,4,5)P3 complex [3] provides a faithful representation of how the PH domain recognizes phosphoinositides at the membrane surface.
Our findings reveal a contrast between PtdIns(4,5)P2-dependent membrane recruitment of PH-PLCδ1 and membrane targeting of other phosphoinositide-binding domains. Recent studies of FYVE (Fab1p, YOTB, Vac1p and EEA1) [14–16], PX (Phox homology) [18] and ENTH (epsin N-terminal homology) [28] domains have all indicated that the respective phosphoinositide-binding sites must be occupied for maximal membrane insertion by each domain. In each of these cases, it has been argued that binding to the phosphoinositide headgroup induces conformational changes in the domain that are required for its subsequent penetration of the membrane. Our results with PH-PLCδ1 paint quite a different picture. Whether its phosphoinositide-binding site is mutated, is occupied by Ins(1,4,5)P3, is occupied by PtdIns(4,5)P2 or is simply unoccupied, the maximum extent of monolayer insertion by PH-PLCδ1 appears to be similar. Binding to PtdIns(4,5)P2 is therefore not required for maximum monolayer insertion by PH-PLCδ1, although it does appear to enhance the rate of monolayer insertion (to reach that maximum). Thus PH-PLCδ1 appears to have intrinsic (uninduced) membrane activity that is not seen with other phosphoinositide-binding modules. Since most of the PH domains do not bind strongly to phosphoinositides [8], whereas (at least in yeast) all PX and FYVE domains specifically recognize PtdIns3P, this distinction may have significant functional implications for this domain class.
It remains unclear which parts of PH-PLCδ1 are responsible for its observed membrane activity. The fact that the reversible PtdIns(4,5)P2-dependent membrane association of PH-PLCδ1 does not involve membrane penetration argues against the suggestion [3,4] that the two hydrophobic side chains close to the crystallographically observed Ins(1,4,5)P3-binding site are required. Rather, it seems more reasonable to suggest that the major contribution to membrane activity comes from the C-terminal half of PH-PLCδ, based on the studies of PH-PLCδ/p130 chimaeras by Balla and co-workers [11]. Surface-lying hydrophobic residues in this region may be primarily responsible for the membrane activity of PH-PLCδ, and identifying which of these make key contributions will require analysis of multiple mutants and careful controls for protein misfolding. We consider it highly unlikely that the observed membrane insertion and irreversible binding represent an artifactual surface denaturation of the PH domain in our monolayer and SPR studies. Indeed, a similar Ins(1,4,5)P3-resistant fraction of membrane-associated PH-PLCδ1 is observed in studies of vesicle binding using a centrifugation approach [7]. Moreover, the fact that the K32A/W36N/R38K mutant of PH-PLCδ1 showed decreased membrane activity despite expressing at lower levels argues against a non-specific denaturation effect.
It is not yet clear precisely what is the nature of this irreversible mode of interaction that involves membrane insertion and is seen only for approx. 20–30% of the protein in our studies. Indeed, at first thought, it seems surprising that binding due to membrane insertion accounts for only approx. 20–30% of the total PH-PLCδ1 binding to a membrane containing 20% PtdIns(4,5)P2. However, if each ‘binding site’ required for PH-PLCδ1 to associate with the membrane in this way involves 20–25 lipid molecules (and is phosphoinositide-independent), these numbers would be expected. In other words, it is very difficult to compare the stoichiometry of a precise headgroup recognition event [binding to PtdIns(4,5)P2] with the stoichiometry of a binding/insertion event that appears to be defined by the overall nature of the membrane (or monolayer). As shown in Figure 3(C), PH-PLCδ1 inserts much more efficiently into PtdIns(4,5)P2-containing monolayers with a PC/PE/cholesterol mixture as background than when a pure PC background was used. In fact, PH-PLCδ1 inserted no more efficiently into a PC/PtdIns(4,5)P2 (4:1) mixture than it did into a neutral PC/PE/cholesterol (1:1:1) monolayer lacking anionic phospholipids altogether (compare Figures 3A and 3C). This finding suggests that the PE and cholesterol in the background lipid mixture play an important role in determining the extent of insertion (i.e. in creating the ‘binding sites’ for PH-PLCδ1 mentioned above). PE and cholesterol are type II (cone-shaped) lipids [29,30]. They decrease the lateral pressure among the lipid headgroups at the membrane/water interface, creating interfacial insertion sites that may well facilitate protein binding [31,32,33]. Indeed, PE has been reported to stimulate membrane insertion of the catalytic domain of leader peptidase (△2–75) from Escherichia coli. The size of the insertion sites, the amount of free space present between the phospholipid headgroups, increases with a decrease in lipid headgroup size and was estimated to be 15±7 Å/lipid molecule (1 Å=10−10 m) for 1,2-dioleoyl-PE, relative to 1,2-dioleoyl-PC [34]. These studies underscore the fact that, although model membranes consisting of only PC with small amounts of a lipid of interest [e.g. PtdIns(4,5)P2] can give information about specific surface interactions, it is important to study lipid mixtures that more closely reflect biological membranes to obtain information on overall protein–membrane interactions, particularly membrane insertion.
Finally, it is worth considering the implications of our results for the use of PH-PLCδ1 as a probe for the localization of cellular PtdIns(4,5)P2. Although most of the reversible binding observed in our SPR studies did reflect binding of PH-PLCδ1 to PtdIns-(4,5)P2 at the membrane surface, it is clear from both our SPR and monolayer analyses that a significant portion of the binding is not dependent on this phospholipid. Long-lived interactions that cannot be reversed by adding excess Ins(1,4,5)P3 may represent PtdIns(4,5)P2-independent membrane association that results from membrane insertion. Since membrane insertion of PH-PLCδ1 is probably highly sensitive to lipid packing, it will also be significantly influenced by both local lipid composition and membrane curvature (see [35]). As such, these factors may play an important role in defining the subcellular localization of a fraction of the PH domain probe. Caution should therefore be exercised before assuming that all of a GFP–PH-PLCδ1 fusion protein (where GFP stands for green fluorescent protein) is located in regions of high PtdIns(4,5)P2 concentration in vivo, and comparative studies with more than one PtdIns(4,5)P2 probe should ideally be used to control for such additional modes of binding [9].
Acknowledgments
This work was supported by the Human Frontier Science Program (to K. N. J. B.) and by grant no. GM056846 from the National Institutes of Health (to M. A. L.). We thank S. Schmid, S. McLaughlin, K. Ferguson, E. Kooijman and B. de Kruijff for stimulating discussions.
References
- 1.Garcia P., Gupta R., Shah S., Morris A. J., Rudge S. A., Scarlata S., Petrova V., McLaughlin S., Rebecchi M. J. The pleckstrin homology domain of phospholipase C-delta 1 binds with high affinity to phosphatidylinositol 4,5-bisphosphate in bilayer membranes. Biochemistry. 1995;34:16228–16234. doi: 10.1021/bi00049a039. [DOI] [PubMed] [Google Scholar]
- 2.Lemmon M. A., Ferguson K. M., O'Brien R., Sigler P. B., Schlessinger J. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc. Natl. Acad. Sci. U.S.A. 1995;92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferguson K. M., Lemmon M. A., Schlessinger J., Sigler P. B. Structure of a high affinity complex between inositol-1,4,5-trisphosphate and a phospholipase C pleckstrin homology domain. Cell (Cambridge, Mass.) 1995;83:1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- 4.Singh S. M., Murray D. Molecular modeling of the membrane targeting of phospholipase C pleckstrin homology domains. Protein Sci. 2003;12:1934–1953. doi: 10.1110/ps.0358803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemmon M. A. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 6.Lemmon M. A., Ferguson K. M. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- 7.Kavran J. M., Klein D. E., Lee A., Falasca M., Isakoff S. J., Skolnik E. Y., Lemmon M. A. Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J. Biol. Chem. 1998;273:30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- 8.Yu J. W., Mendrola J. M., Audhya A., Singh S., Keleti D., DeWald D. B., Murray D., Emr S. D., Lemmon M. A. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 9.Balla T., Bondeva T., Varnai P. How accurately can we image inositol lipids in living cells? Trends Pharmacol. Sci. 2000;21:238–241. doi: 10.1016/s0165-6147(00)01500-5. [DOI] [PubMed] [Google Scholar]
- 10.Levine T. P., Munro S. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr. Biol. 1998;8:729–739. doi: 10.1016/s0960-9822(98)70296-9. [DOI] [PubMed] [Google Scholar]
- 11.Várnai P., Lin X., Lee S. B., Tuymetova G., Bondeva T., Spät A., Rhee S. G., Hajnóczky G., Balla T. Inositol lipid binding and membrane localization of isolated pleckstrin homology (PH) domains. Studies on the PH domains of phospholipase C delta 1 and p130. J. Biol. Chem. 2002;277:27412–27422. doi: 10.1074/jbc.M109672200. [DOI] [PubMed] [Google Scholar]
- 12.DiNitto J. P., Cronin T. C., Lambright D. G. Membrane recognition and targeting by lipid-binding domains. Science STKE. 2003;213:re16. doi: 10.1126/stke.2132003re16. [DOI] [PubMed] [Google Scholar]
- 13.Gillooly D. J., Simonsen A., Stenmark H. Cellular functions of phosphatidylinositol 3-phosphate and FYVE domain proteins. Biochem. J. 2001;355:249–258. doi: 10.1042/0264-6021:3550249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stahelin R. V., Long F., Diraviyam K., Bruzik K. S., Murray D., Cho W. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J. Biol. Chem. 2002;277:26379–26388. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- 15.Kutateladze T. G., Capelluto D. G., Ferguson C. G., Cheever M. L., Kutateladze A. G., Prestwich G. D., Overduin M. Multivalent mechanism of membrane insertion by the FYVE domain. J. Biol. Chem. 2004;279:3050–3057. doi: 10.1074/jbc.M309007200. [DOI] [PubMed] [Google Scholar]
- 16.Kutateladze T., Overduin M. Structural mechanism of endosome docking by the FYVE domain. Science. 2001;291:1793–1796. doi: 10.1126/science.291.5509.1793. [DOI] [PubMed] [Google Scholar]
- 17.Cheever M. L., Sato T. K., de Beer T., Kutateladze T. G., Emr S. D., Overduin M. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 2001;3:613–618. doi: 10.1038/35083000. [DOI] [PubMed] [Google Scholar]
- 18.Stahelin R. V., Burian A., Bruzik K. S., Murray D., Cho W. Membrane binding mechanisms of the PX domains of NADPH oxidase p40phox and p47phox. J. Biol. Chem. 2003;278:14469–14479. doi: 10.1074/jbc.M212579200. [DOI] [PubMed] [Google Scholar]
- 19.Tuzi S., Uekama N., Okada M., Yamaguchi S., Saitô H., Yagisawa H. Structure and dynamics of the phospholipase C-delta1 pleckstrin homology domain located at the lipid bilayer surface. J. Biol. Chem. 2003;278:28019–28025. doi: 10.1074/jbc.M300101200. [DOI] [PubMed] [Google Scholar]
- 20.Rebecchi M., Boguslavsky V., Boguslavsky L., McLaughlin S. Phosphoinositide-specific phospholipase C-δ1: effects of monolayer surface pressure and electrostatic surface potentials on activity. Biochemistry. 1992;31:12748–12753. doi: 10.1021/bi00166a006. [DOI] [PubMed] [Google Scholar]
- 21.Jost M., Simpson F., Kavran J. M., Lemmon M. A., Schmid S. L. Phosphatidylinositol-4,5-bisphosphate is required for endocytic coated vesicle formation. Curr. Biol. 1998;8:1399–1402. doi: 10.1016/s0960-9822(98)00022-0. [DOI] [PubMed] [Google Scholar]
- 22.Burger K. N. J., Demel R. A., Schmid S. L., de Kruijff B. Dynamin is membrane-active: lipid insertion is induced by phosphoinositides and phosphatidic acid. Biochemistry. 2000;39:12485–12493. doi: 10.1021/bi000971r. [DOI] [PubMed] [Google Scholar]
- 23.Erb E. M., Chen X., Allen S., Roberts C. J., Tendler S. J., Davies M. C., Forsen S. Characterization of the surfaces generated by liposome binding to the modified dextran matrix of a surface plasmon resonance sensor chip. Anal. Biochem. 2000;280:29–35. doi: 10.1006/abio.1999.4469. [DOI] [PubMed] [Google Scholar]
- 24.Yu J. W., Lemmon M. A. All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol-3-phosphate. J. Biol. Chem. 2001;276:44179–44184. doi: 10.1074/jbc.M108811200. [DOI] [PubMed] [Google Scholar]
- 25.Demel R. A. Monomolecular layers in the study of biomembranes. Subcell. Biochem. 1994;23:83–120. doi: 10.1007/978-1-4615-1863-1_3. [DOI] [PubMed] [Google Scholar]
- 26.Marsh D. Lateral pressure in membranes. Biochim. Biophys. Acta. 1996;1286:183–223. doi: 10.1016/s0304-4157(96)00009-3. [DOI] [PubMed] [Google Scholar]
- 27.Hirose K., Kadowaki S., Tanabe M., Takeshima H., Iino M. Spatiotemporal dynamics of inositol 1,4,5-trisphosphate that underlies complex Ca2+ mobilization patterns. Science. 1999;284:1527–1530. doi: 10.1126/science.284.5419.1527. [DOI] [PubMed] [Google Scholar]
- 28.Stahelin R. V., Long F., Peter B. J., Murray D., De Camilli P., McMahon H. T., Cho W. Contrasting membrane interaction mechanisms of AP180 N-terminal homology (ANTH) and epsin N-terminal homology (ENTH) domains. J. Biol. Chem. 2003;278:28993–28999. doi: 10.1074/jbc.M302865200. [DOI] [PubMed] [Google Scholar]
- 29.Burger K. N. J. Greasing membrane fusion and fission machineries. Traffic. 2000;1:605–613. doi: 10.1034/j.1600-0854.2000.010804.x. [DOI] [PubMed] [Google Scholar]
- 30.Epand R. M. Lipid polymorphism and protein-lipid interactions. Biochim. Biophys. Acta. 1998;1376:353–368. doi: 10.1016/s0304-4157(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 31.Cantor R. S. Lipid composition and the lateral pressure profile in bilayers. Biophys. J. 1999;76:2625–2639. doi: 10.1016/S0006-3495(99)77415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies S. M. A., Epand R. M., Kraayenhof R., Cornell R. B. Regulation of CTP: phosphocholine cytidylyltransferase activity by the physical properties of lipid membranes: an important role for stored curvature strain energy. Biochemistry. 2001;40:10522–10531. doi: 10.1021/bi010904c. [DOI] [PubMed] [Google Scholar]
- 33.van den Brink-van der Laan E., Killian J. A., de Kruijff B. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim. Biophys. Acta. 2004;1666:275–288. doi: 10.1016/j.bbamem.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 34.van den Brink-van der Laan E., Dalbey R. E., Demel R. A., Killian J. A., de Kruijff B. Effect of nonbilayer lipids on membrane binding and insertion of the catalytic domain of leader peptidase. Biochemistry. 2001;40:9677–9684. doi: 10.1021/bi002903a. [DOI] [PubMed] [Google Scholar]
- 35.Holthuis J. C., Burger K. N. J. Sensing membrane curvature. Dev. Cell. 2003;5:821–822. doi: 10.1016/s1534-5807(03)00366-6. [DOI] [PubMed] [Google Scholar]