Abstract
Genomic DNA of Ostreococcus tauri, a fully sequenced marine unicellular alga from the phytoplankton, was used to amplify a gene coding for a typical front-end desaturase involved in polyunsaturated fatty acid biosynthesis. Heterologous expression in Saccharomyces cerevisiae revealed very high desaturation activity with Δ6-regioselectivity. Short-time kinetic experiments showed that the desaturase product was detected in the acyl-CoA pool 5 min after addition of the exogenous substrate to the yeast medium and long before its appearance in the total fatty acids. When this desaturase was co-expressed with the acyl-CoA Δ6-elongase from Physcomitrella patens and the lipid-linked Δ5-desaturase from Phaeodactylum tricornutum, high proportions of arachidonic or eicosapentaenoic acid were obtained, because nearly all of the Δ6-desaturated products were elongated. Furthermore, the product/educt ratios calculated in each glycerolipid for the Δ6-desaturase or for the acyl-CoA Δ6-elongase were in about the same range, whereas this ratio showed a very uneven profile in the case of the lipid-linked Δ5-desaturase. Finally, a sequence-based comparison of all the functionally characterized Δ6-desaturases showed that this enzyme was not related to any previously described sequence. Altogether, our data suggest that this desaturase from O. tauri is an acyl-CoA Δ6-desaturase, the first one cloned from a photosynthetically active organism.
Keywords: acyl-carrier, acyl-CoA desaturase, desaturation, marine unicellular alga, Ostreococcus tauri, polyunsaturated fatty acid
Abbreviations: FADS2, fatty acid desaturase 2; FAME, fatty acid methyl ester; ORF, open reading frame; OtD6, Ostreococcus tauri Δ6-desaturase; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PSE1, polyunsaturated-fatty-acid-specific elongase 1; PtD5, Phaeodactylum tricornutum Δ5-desaturase
INTRODUCTION
A major role for unsaturated fatty acids is to keep the membranes of all eukaryotic cells in an appropriate state of fluidity. In these organisms, fatty acid desaturases are responsible for the insertion of cis-double bonds into pre-formed fatty acid chains in reactions that require oxygen and reducing equivalents. Many genes coding for desaturases with different regioselectivities and from various organisms have already been cloned [1]. With the exception of the soluble acyl-ACP (acyl carrier protein)-desaturases found in plant plastids, all of these enzymes contain several hydrophobic regions corresponding to four putative transmembrane domains and three characteristic histidine clusters that probably co-ordinate the di-iron centre of the active site [2,3]. ‘Front-end desaturases’ are referred to as desaturases that introduce a new double bond between the carboxy group of a fatty acid and a pre-existing double bond [4]. They contribute to the biosynthesis of polyunsaturated fatty acids and insert double bonds at position Δ4, Δ5, Δ6 or Δ8. At the amino acid level, front-end desaturases are characterized by the presence at the N-terminus of a fused cytochrome b5-domain that most likely serves as immediate electron donor in the desaturation reaction [5].
Δ6-Desaturases are front-end desaturases that introduce Δ6-double bonds, usually converting linoleic acid (C18:2,Δ9,12) and α-linolenic acid (C18:3,Δ9,12,15) into γ-linolenic acid (C18:3,Δ6,9,12) and stearidonic acid (C18:4,Δ6,9,12,15) respectively. It was shown that human diets deficient in γ-linolenic acid could result in several health disorders, which could be reverted by the administration of this particular fatty acid [6]. These positive effects on human health led to an increasing demand for γ-linolenic acid, which is only found in some micro-organisms and a few higher plants with poor agronomic performance. The production of γ-linolenic acid in conventional oilseed crops by plant biotechnology was therefore considered to be a rewarding goal. This possibility led to the cloning of genes coding for Δ6-desaturase from several organisms. The first one was obtained from the cyanobacterium Synechocystis [7], but, in the meantime, sequences from many different species became available [1].
Because the purification of membrane-bound desaturases by conventional methods is difficult, the functional characterization of most of these Δ6-desaturases was based on their heterologous expression in various host organisms followed by appropriate fatty acid analysis. Using such systems, it was shown that all Δ6-desaturases had the same fatty acid specificity, but that enzymes from different organisms may differ with respect to the acyl-carrier they use as substrate. In cyanobacteria, the Δ6-desaturases use exclusively the acyl group at the sn-1 position of the membrane lipid monogalactosyldiacylglycerol as substrate [8]. Okayasu et al. [9] purified the Δ6-desaturase from rat liver microsomes to homogeneity and showed by in vitro experiments that linoleoyl-CoA was the reaction product. This study represents the only report on the in vitro biochemical characterization of a purified Δ6-desaturase and suggests that mammalian Δ6-desaturases are acyl-CoA desaturases. Regarding the corresponding enzymes from fungi and higher plants, in vitro assays with microsomes from Mucor circinelloides and from maturing cotyledons of borage (Borago officinalis) demonstrated that Δ6-desaturation mainly occurred at the sn-2 position of PC (phosphatidylcholine) [10,11], suggesting the presence of lipid-linked desaturases in these organisms.
In a previous study [12], we have expressed front-end desaturases from various organisms in Saccharomyces cerevisiae and conducted detailed analyses of lipids and acyl-CoAs to identify the acyl-carriers used as substrate. These experiments indicated that the Δ5- and Δ6-desaturases from fungi (Mortierella alpina and Phytophthora megasperma), algae (Phaeodactylum tricornutum), lower plants (the mosses Physcomitrella patens and Ceratodon purpureus) and lower animals (Caenorhabditis elegans) accepted predominantly as substrate the acyl groups esterified at the sn-2 position of PC, and thus have to be classified as lipid-linked desaturases. Since yeast cells have only a limited capacity to re-equilibrate the sn-2-bound Δ5- or Δ6-desaturated acyl groups of PC with the acyl-CoA pool, this mode of action resulted in an enrichment of the desaturase product at this particular position [12]. Moreover, when such a lipid-linked Δ6-desaturase was coexpressed with the acyl-CoA Δ6-elongase PSE1 (polyunsaturated-fatty-acid-specific elongase 1) from Phys. patens [13], the elongation was rather inefficient. In contrast, the expression of the human Δ6-desaturase FADS2 (fatty acid desaturase 2) led to a more equal distribution of the desaturation product among the various lipids, while its co-expression with PSE1 resulted in the elongation of more than 90% of the Δ6-desaturated product [12]. These results represented independent in vivo evidence for the above-mentioned in vitro experiments which had identified the human Δ6-desaturase as an acyl-CoA desaturase.
Ostreococcus tauri is a marine photosynthetic picoeukaryote belonging to the Prasinophyceae and presenting a minimal cellular organization, as well as the smallest genome described for a free-living autotrophic eukaryote [14]. O. tauri contains polyunsaturated fatty acids, such as arachidonic acid (C20:4,Δ5,8,11,14), eicosapentaenoic acid (C20:5,Δ5,8,11,14,17) and docosahexaenoic acid (C22:6,Δ4,7,10,13,16,19). Using the data issued from a sequencing program covering approx. 98% of the O. tauri genome, we have already cloned the two unique ORFs (open reading frames) coding for ELO-type elongase present in this alga. Heterologous expression in yeast showed that these genes encoded the Δ5- and Δ6-elongase required for the biosynthesis of docosahexaenoic acid [15], indicating that O. tauri apparently contains a minimal number of elongases. In the case of fatty acid desaturases, we could annotate as many as 13 sequences. Although this apparently high number may indicate a possible redundancy, this will only be clarified once all the different ORFs have been functionally characterized.
In the present study, we report on the in vivo characterization of one of these desaturases in S. cerevisiae as a Δ6-desaturase. Using the methodology mentioned above [12], we provide strong evidence that this enzyme from O. tauri represents the first front-end acyl-CoA desaturase to be reported from the plant kingdom.
MATERIALS AND METHODS
Materials
Restriction enzymes, polymerases and DNA-modifying enzymes were obtained from New England Biolabs (Frankfurt, Germany). All other chemicals were from Sigma (St. Louis, MO, U.S.A.). Addition of fatty acids to culture media was made using 100 mM stock solutions prepared in absolute ethanol.
PCR amplification and cloning of the O. tauri desaturase sequence
The gene coding for the putative desaturase was amplified by PCR using 5′-GGTACCACATAATGTGCGTGGAGACGGAAAATAACG-3′ and 5′-CTCGAGTTACGCCGTCTTTCCGGAGTGTTGGCC-3′ as forward and reverse primers respectively, Vent DNA polymerase, an O. tauri cell pellet as DNA template and a standard PCR program. The primers used introduced KpnI and XhoI cloning sites (underlined) upstream and downstream of the start and stop codons (in boldface) respectively. A 1400 bp band was obtained, phosphorylated with T4 polynucleotide kinase and cloned blunt-ended in pUC18. The ORF was released by KpnI/XhoI digestion and was cloned in pYES2 using the same sites, yielding pYES2-OtD6 (where OtD6 is O. tauri Δ6-desaturase). The construction of the other yeast expression vectors used in the present study was reported previously [12].
Expression in S. cerevisiae
S. cerevisiae strain INVSc1 from Invitrogen (Karlsruhe, Germany) was transformed with plasmid DNA using the method of Dohmen et al. [16]. After selection on minimal medium agar plates without uracil, pre-cultures were grown at 30 °C in minimal medium lacking uracil, but containing 2% (w/v) raffinose and 1% (v/v) Tergitol Nonidet P40. When these pre-cultures had reached a D600 of 0.1, they were transferred to 20 °C, and the expression was induced by supplementing galactose to 2% (w/v) and adding the appropriate fatty acids to a final concentration of 250 μM (unless otherwise indicated). After 48 h at 20 °C, cells were harvested by centrifugation at 1200 g for 5 min and the pellet was washed once with 10 mM perchloric acid before being used for fatty acid and/or acyl-CoA analysis. This resuspension in perchloric acid removed all non-esterified (‘free’) fatty acids, which, in our previous study [12], had been separated from esterified fatty acids by transmethylation with sodium methoxide. The host strain transformed with the empty vector(s) was used as negative control in all experiments.
For kinetic experiments, 50 ml pre-cultures were grown for 24 h at 30 °C in the presence of galactose to a D600 of approx. 1.5 in order to obtain cells expressing the heterologous enzyme(s) and sufficient material for analysis. The kinetic experiments were then started by transferring the yeast cells to 20 °C and adding the exogenous fatty acid. After the indicated times, 5 ml aliquots were harvested, added to 1/10 volume of 6.6 M perchloric acid and subjected to a short centrifugation at 1200 g (1 min). The supernatant was removed, and the cell pellet was resuspended in 2.5 ml of 10 mM perchloric acid. A 1/5 volume was used for fatty acid analysis, while the rest was used for acyl-CoA extraction. In both cases, cells were sedimented by short centrifugation at 1200 g (1 min), the supernatant was removed, and the resulting pellets were used immediately or were frozen in liquid nitrogen and stored at −80 °C until needed.
Lipid and fatty acid analysis
For the analysis of lipids, expressions were conducted for 24 h at 20 °C with 120 ml cultures. Cells were harvested by centrifugation at 1200 g for 5 min, washed with 30 ml of 0.1 M NaHCO3, and lipid extracts were prepared and used for various analytical procedures (isolation of individual compounds, total and positional analysis of fatty acids) as described previously [12]. Quantification of the different lipid components was based on GLC analysis of FAMEs (fatty acid methyl esters) using C17:1,Δ10 as internal standard.
Acyl-CoA extraction and analysis
Acyl-CoAs from approx. 4 ml of yeast culture (D600 of 1.5) were extracted using a modified version of the method of Schjerling et al. [17]. Frozen cells were resuspended in 800 μl of ice-cold water in glass tubes, and 3 ml of chloroform/methanol (2:1, v/v) and glass beads were added. Cells were broken by vortex-mixing 5 times for 1 min with intermittent cooling on ice. Volumes of 1 ml each of chloroform and water were successively added, and the tubes were vortex-mixed further for 1 min. The samples were centrifuged at 1200 g for 10 min at 4 °C, and the upper and lower phases were gently removed. The interphase containing the acyl-CoAs was dried together with the glass beads under argon. Subsequently, 400 μl of freshly made extraction buffer, 10 μl of saturated ammonium sulphate and 1.2 ml of chloroform/methanol (1:2, v/v) were added. The extraction buffer was made by mixing 2 ml of 50 mM potassium phosphate buffer, pH 7.2, 2 ml of isopropanol, 50 μl of ethanoic (acetic) acid and 80 μl of 50 mg/ml BSA. After vortex-mixing for 3 min, the samples were incubated for 20 min at room temperature (20 °C) before being centrifuged at 1200 g for 5 min. The supernatant fraction was removed and dried under argon at 50 °C. The acyl-CoAs in the residue were converted into etheno-derivatives and were analysed by HPLC as described previously [18,19].
RESULTS
Isolation of a front-end desaturase clone from genomic DNA
An intron-free gene coding for a putative fatty acid desaturase was recognized in the draft genome of the microalga O. tauri and was amplified by PCR using appropriate primers and PCR program. The deduced ORF (1371 bp; GenBank® accession number AY746357) encoded a polypeptide of 456 amino acids which contained all the signatures of microsomal membrane-bound front-end desaturases [2–4]: several hydrophobic stretches representing potential transmembrane helices, three conserved histidine clusters with a His→Gln substitution in the third box and a cytochrome b5 domain fused at the N-terminus (results not shown). At the protein level, the putative desaturase from O. tauri was most similar to the Δ5-desaturase of Thraustochytrium sp., but it also presented high sequence similarities to Δ6-desaturases from various organisms (see Supplementary Table 1 at http://www.BiochemJ.org/bj/389/bj3890483add.htm).
Functional characterization in S. cerevisiae
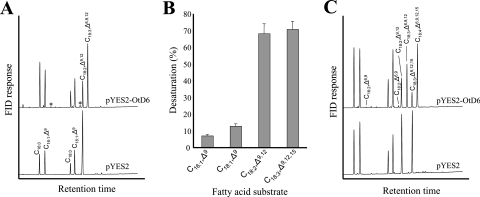
In order to confirm the activity of the putative desaturase and to determine its regiospecificity, the yeast strain INVSc1 was transformed with the expression vector pYES2 containing the O. tauri ORF or the empty vector as control and was grown in the presence of linoleic acid (C18:2,Δ9,12) or eicosatrienoic acid (C20:3,Δ8,11,14) as potential substrates for Δ6- or Δ5-desaturation respectively. In the presence of C20:3,Δ8,11,14, the fatty acid profiles of both the control and the desaturase-expressing strain were identical (results not shown), whereas, in the presence of linoleic acid, the yeast expressing the O. tauri ORF produced a new predominant fatty acid (Figure 1A). GLC–MS analyses identified this new fatty acid as γ-linolenic acid (C18:3,Δ6,9,12), indicating that the gene we had cloned encoded a Δ6-fatty acid desaturase.
Figure 1. Activity, substrate specificity and substrate selectivity of OtD6 expressed in S. cerevisiae.
Cultures of the yeast strain INVSc1 transformed with pYES2 or pYES2-OtD6 were grown for 48 h at 20 °C and used for fatty acid analysis as indicated in the Materials and methods section. (A) OtD6 activity in the presence of 250 μM C18:2,Δ9,12. These analyses reflect results obtained in five independent expression experiments. (B) OtD6 substrate specificity in the presence (C18:2,Δ9,12 and C18:3,Δ9,12,15) or absence (C16:1,Δ9 and C18:1,Δ9) of 250 μM exogenously supplied fatty acids. Each value is the mean±S.D. for five independent experiments. (C) OtD6 substrate selectivity in the presence of 110 μM C18:2,Δ9,12 and 140 μM C18:3,Δ9,12,15. These analyses reflect results obtained in three independent expression experiments. The minor components C16:2,Δ6,9 and C18:2,Δ6,9 are marked with an asterisk (*). FID, flame ionization detector.
As shown in Figure 1(A), approx. 73% of the available substrate was desaturated. Since all the other Δ6-desaturases so far expressed in our laboratory under similar conditions converted only 20–40% of C18:2,Δ9,12 into C18:3,Δ6,9,12, OtD6 displayed a much higher activity. On the other hand, the substrate specificity of this desaturase was similar to that of other Δ6-desaturases (Figure 1B). Apart from linoleic acid, OtD6 desaturated α-linolenic acid (C18:3,Δ9,12,15) at similarly high proportions (71% conversion), whereas the mono-unsaturated fatty acids C16:1,Δ9 and C18:1,Δ9 were less well accepted (7% and 12% conversion respectively). All the other substrates tested (C14:1,Δ9, C18:1,Δ11, C20:2,Δ11,14, C20:3,Δ8,11,14, C20:3,Δ11,14,17 and C20:4,Δ8,11,14,17) were not accepted (results not shown). When the substrate selectivity of OtD6 was assayed by providing simultaneously the four potential substrates described in Figure 1(B), C18:2,Δ9,12 was slightly less efficiently desaturated (59% conversion), whereas the conversion of C18:3,Δ9,12,15 was higher (82%; Figure 1C). Altogether, these heterologous expressions in yeast showed that we had cloned from O. tauri a gene coding for a Δ6-desaturase that had a very high catalytic activity and a slight preference for α-linolenic acid.
Lipid analysis of a yeast culture expressing OtD6
As a first step towards the identification of the actual acyl-carrier used as substrate by OtD6, we analysed the lipids from a yeast culture that had expressed this desaturase in the presence of 250 μM C18:2,Δ9,12 (Table 1). After 24 h of expression, approx. 62% of the substrate had been desaturated, and the newly formed C18:3,Δ6,9,12 represented as much as 32.8% of the total fatty acids in the lipid extract. Similar proportions of γ-linolenic acid were detected in the two major components of the lipid extract, the NL (neutral lipid) fraction (32.2%) and PC (34.4%). In addition, C18:3,Δ6,9,12 was also present in all the other phospholipids, with proportions varying from 14.6% in phosphatidylinositol to 22.2% in PE (phosphatidylethanolamine). The fatty acid positional distribution in PC and PE showed that the sn-1 positions of both phospholipids were enriched in C16:0, C16:1,Δ9 and C18:0, whereas C18:1,Δ9, C18:2,Δ9,12 and C18:3,Δ6,9,12 were preferentially localized at the sn-2 position (Table 1). In PC and PE, the proportion of C18:2,Δ9,12 at the sn-2 position was about twice as high as that found at the sn-1 position, while γ-linolenic was 18 times and 6 times more abundant at the sn-2 position than at the sn-1 position of PC and PE respectively. Such uneven distributions may be explained by previous labelling studies that have shown that exogenously supplied fatty acids are not evenly distributed in all the different lipids and sn-positions, because yeast acyltransferases preferentially fill certain lipids and positions and discriminate against others [20]. The sn-2 position of PC is indeed a major entry point for unsaturated fatty acids from the acyl-CoA pool, whereas the sn-1 position is maintained with high proportions of saturated fatty acids (Table 1) [12,20].
Table 1. Fatty acid composition of different lipid fractions from a yeast expressing the Δ6-desaturase from O. tauri.
The INVSc1 yeast strain transformed with pYES2-OtD6 was grown 24 h at 20 °C in the presence of 250 μM C18:2,Δ9,12 and was used to isolate different lipid fractions. Positional analyses were carried out for PC and PE as described in the Materials and methods section. Educt/product ratio was calculated using values corresponding to the percentage of total fatty acids. Quantification of the different lipid components was based on GLC analysis of FAMEs using C17:1,Δ10 as internal standard. Each value is the mean±S.D. for three independent experiments. PI, phosphatidylinositol; PS, phosphatidylserine; NL, neutral lipid.
| Lipid extract | PC | sn-1-PC | sn-2-PC | PI | PS | PE | sn-1-PE | sn-2-PE | NL | |
|---|---|---|---|---|---|---|---|---|---|---|
| Fraction | ||||||||||
| C16:0 | 23.5±1.6 | 27.2±4.2 | 53.2±4.1 | 1.4±0.2 | 33.9±3.5 | 30.6±1.2 | 30.9±4.0 | 41.3±2.7 | 1.7±1.3 | 22.4±0.4 |
| C16:1,Δ9 | 8.8±0.3 | 7.8±1.4 | 10.3±0.5 | 5.9±1.0 | 3.7±0.7 | 12.7±3.9 | 15.2±0.8 | 19.7±2.4 | 3.8±0.8 | 7.7±0.3 |
| C18:0 | 8.0±2.0 | 12.4±0.5 | 20.1±1.6 | 0.6±0.1 | 20.8±6.0 | 4.8±1.9 | 4.3±2.2 | 5.5±2.5 | 0.9±0.7 | 9.5±1.4 |
| C18:1,Δ9 | 6.7±0.9 | 4.6±0.2 | 3.7±0.7 | 5.2±1.1 | 10.4±2.7 | 17.9±2.0 | 8.3±1.0 | 6.7±2.5 | 11.1±3.9 | 7.0±0.3 |
| C18:2,Δ9,12 | 19.6±0.9 | 13.4±0.8 | 8.8±2.5 | 18.1±0.8 | 13.6±2.2 | 17.5±3.5 | 18.8±1.6 | 13.7±1.7 | 25.0±1.0 | 21.0±0.9 |
| C18:3,Δ6,9,12 | 32.8±2.5 | 34.4±3.8 | 3.8±0.6 | 68.9±1.9 | 14.6±4.1 | 16.2±2.6 | 22.2±2.1 | 7.8±2.0 | 47.4±6.9 | 32.2±0.9 |
| Educt/product ratio | 1.67 | 2.57 | 0.43 | 3.81 | 1.07 | 0.93 | 1.18 | 0.57 | 1.90 | 1.53 |
| Total acyl chains (%) | 100 | 22.0±0.2 | – | – | 7.1±0.6 | 7.4±0.2 | 16.8±0.2 | – | – | 46.7±1.5 |
Most importantly, the data presented in Table 1 show that C18:3,Δ6,9,12 was not particularly enriched in PC when compared with the lipid extract, which is in marked contrast with the situation resulting from the expression of Δ6-desaturases specific for the sn-2 position of PC [12]. In addition, these data show relatively high proportions of C18:3,Δ6,9,12 in all of the different lipid species. Altogether, these analyses suggest that the Δ6-desaturase from O. tauri may accept as substrate acyl-CoA thioesters rather than lipid-linked acyl groups.
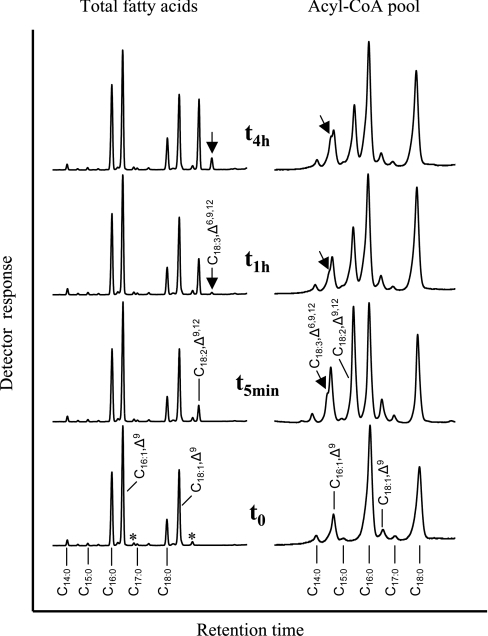
Kinetics of desaturation with parallel analysis of acyl-CoAs and total acyl groups
To evaluate further whether the desaturase from O. tauri is an acyl-CoA desaturase, the changes occurring in the acyl-CoA pool within a very short time after the addition of the substrate to the growth medium were analysed. To carry out such an analysis, a transgenic yeast culture was grown for 24 h in the presence of galactose for permanent expression of OtD6 before the kinetic analysis was started by adding C18:2,Δ9,12. Before substrate addition (t0 in Figure 2), the total fatty acid profile was dominated by mono-unsaturated (C16:1,Δ9 and C18:1,Δ9) and saturated (C16:0 and C18:0) fatty acids. As indicated by the asterisks, it also contained small proportions of C16:2,Δ6,9 and C18:2,Δ6,9, which resulted from the expression of OtD6 during the 24 h preceding the addition of C18:2,Δ9,12. In the acyl-CoA pool, the major fatty acids were C16:0 and C18:0 (45 and 30% of the total respectively), whereas C16:1,Δ9 and C18:1,Δ9 represented only 11 and 5% respectively. After 5 min in the presence of C18:2,Δ9,12 (t5min in Figure 2), this latter represented only 5% of the total fatty acids, whereas it was the major component of the acyl-CoA thioesters, representing 28% of this pool. At this time point, C18:3,Δ6,9,12 was not detected in the total fatty acids, but it was clearly present in the acyl-CoA pool. Only after 1 h (t1h in Figure 2) did C18:3,Δ6,9,12 appear as a trace component in the total fatty acids, while its proportion in the acyl-CoA pool was as high as after 5 min. After 4 h (t4h in Figure 2), C18:3,Δ6,9,12 accounted for 3% of the total fatty acids, whereas it was as abundant as C16:1,Δ9 in the acyl-CoA pool (each representing approx. 7% of the total). The detection of the desaturase product in the acyl-CoA pool before its appearance in the total fatty acids strongly suggested that OtD6 may be an acyl-CoA desaturase rather than a lipid-linked desaturase.
Figure 2. Kinetic analysis of fatty acid changes in acyl-CoAs and lipids of a yeast culture expressing OtD6.
A culture of the yeast strain INVSc1 transformed with pYES2-OtD6 was grown for 24 h at 30 °C in the presence of galactose, before being supplemented with 250 μM C18:2,Δ9,12. Just before adding C18:2,Δ9,12 (t0) as well as 5 min, and 1 and 4 h (t5min, t1h and t4h) after substrate addition, the fatty acid composition of the transgenic yeast pellets (left-hand part; GLC) and of their acyl-CoAs (right-hand part; HPLC) were determined as indicated in the Materials and methods section. These analyses reflect results obtained in four independent kinetic experiments. The minor components C16:2,Δ6,9 and C18:2,Δ6,9 are marked with an asterisk (*).
Co-expression of OtD6 with a acyl-CoA Δ6-elongase
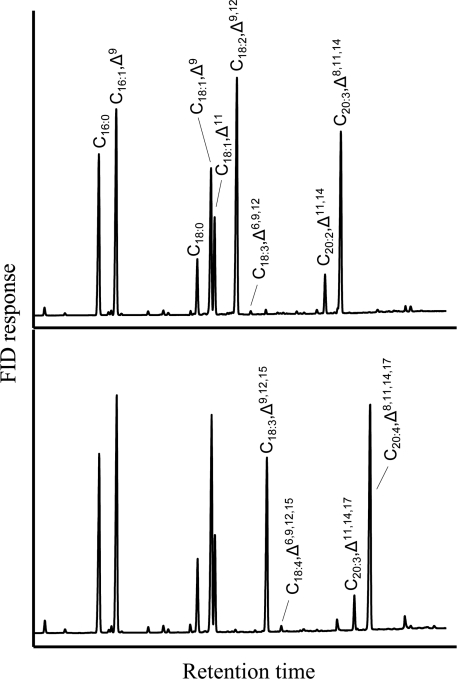
For additional confirmation that OtD6 may be an acyl-CoA desaturase, it was co-expressed with the acyl-CoA Δ6-elongase from the moss Phys. patens PSE1 [13] in the presence of exogenous linoleic (Figure 3A) or α-linolenic acid (Figure 3B). In such co-expression experiments, the final elongated products (C20:3,Δ8,11,14 and C20:4,Δ8,11,14,17 respectively) were present in high proportions, whereas the Δ6-desaturated fatty acid intermediates (C18:3,Δ6,9,12 and C18:4,Δ6,9,12,15 respectively) were barely detectable (Figure 3). Indeed, more than 98% of the fatty acids produced by the Δ6-desaturase were elongated further to the corresponding C20-polyunsaturated fatty acids. In Figure 3, the presence of C18:1,Δ11, C20:2,Δ11,14 and C20:3,Δ11,14,17 correspond to elongated products of C16:1,Δ9, C18:2,Δ9,12 and C18:3,Δ9,12,15, confirming that PSE1 displays minor activity upon Δ9-desaturated fatty acid substrates [13].
Figure 3. Fatty acid composition of a yeast culture co-expressing OtD6 and PSE1.
A culture of the yeast strain INVSc1 co-transformed with pYES2-OtD6 and pESC-LEU-PSE1 was grown for 48 h at 20 °C in the presence of 250 μM C18:2,Δ9,12 (A) or C18:3,Δ9,12,15 (B). FAMEs from cell pellets were analysed by GLC as indicated in the Materials and methods section. These analyses reflect results obtained in five independent expression experiments. FID, flame ionization detector.
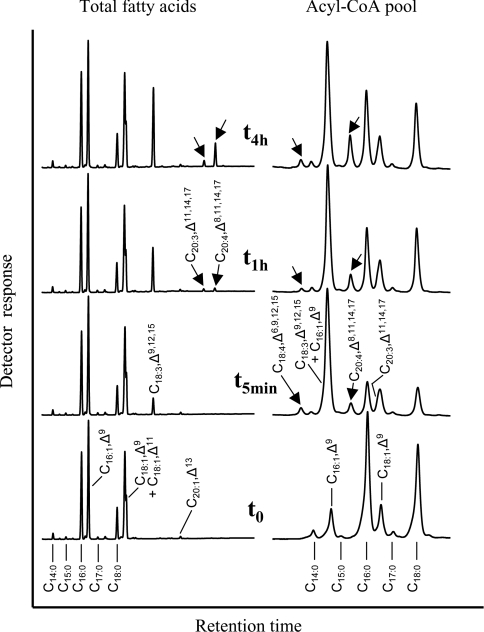
We then carried out a kinetic analysis with the yeast strain co-expressing OtD6 and PSE1 using α-linolenic acid (C18:3,Δ9,12,15) as exogenous substrate (Figure 4). The profiles at t0 (after 24 h of growth in the presence of galactose for permanent expression of both heterologous enzymes, but before addition of C18:3,Δ9,12,15) differed from those shown in Figure 2 by the presence in total fatty acids of C18:1,Δ11 and C20:1,Δ13, which were due to the elongation of endogenous C16:1,Δ9 by PSE1. After 5 min in the presence of C18:3,Δ9,12,15, this latter represented the only new component of the total fatty acids, whereas the Δ6-desaturated C18:4,Δ6,9,12,15 and the subsequently elongated product C20:4,Δ8,11,14,17 were, in addition, detected in the acyl-CoA pool. Figure 4 also shows that C20:3,Δ11,14,17, resulting from the elongation of C18:3,Δ9,12,15 by PSE1 and co-elution with C18:1,Δ9, could also be detected in the acyl-CoA pool after this short time. After 1 and 4 h, the C20-polyunsaturated fatty acids C20:3,Δ11,14,17 and, most of all, C20:4,Δ8,11,14,17 (resulting from the elongation of C18:3,Δ9,12,15 and C18:4,Δ6,9,12,15 respectively) accumulated in the total fatty acids, although the Δ6-desaturated fatty acid C18:4,Δ6,9,12,15 was not detected in this fraction. In contrast, this intermediate was detected in the acyl-CoA pool in nearly unchanged proportion after 1 and 4 h (Figure 4). This result is the diagnostically most relevant difference compared with the expression of the Δ6-desaturase on its own (Figure 2), where the Δ6-desaturated product (C18:3,Δ6,9,12) appeared in both the acyl-CoA pool and, with an appreciable time lag, in the total lipids. In the additional presence of the elongase, the Δ6-desaturated product (C18:4,Δ6,9,12,15) was detected in the acyl-CoA pool, but not in the total fatty acids (Figure 4). Under these conditions, it seems most likely that the yeast acyltransferases cannot compete with the elongase, so that nearly all C18:4,Δ6,9,12,15 is elongated instead of being esterified into the glycerolipids. Altogether, these co-expression experiments strongly suggested that both OtD6 and PSE1 used substrates from the acyl-CoA pool.
Figure 4. Kinetic analysis of fatty acid changes in acyl-CoAs and lipids of a yeast culture co-expressing OtD6 and PSE1.
A culture of the yeast strain INVSc1 co-transformed with pYES2-OtD6 and pESC-LEU-PSE1 was grown for 24 h at 30 °C in the presence of galactose, before being supplemented with 250 μM C18:3,Δ9,12,15. Just before adding C18:3,Δ9,12,15 (t0) as well as 5 min, and 1 and 4 h (t5min, t1h and t4h) after substrate addition, the fatty acid composition of the transgenic yeast cells (left-hand part; GLC) and of their acyl-CoAs (right-hand part; HPLC) were determined as indicated in the Materials and methods section. These analyses reflect results obtained in three independent kinetic experiments.
Reconstitution of arachidonic and eicosapentaenoic acid biosynthesis
Finally, we compared in a single co-expression experiment the effects of acyl-CoA- and lipid-linked desaturases on the fatty acid patterns of the yeast lipids by co-expressing the lipid-linked Δ5-desaturase from Phae. tricornutum (PtD5), together with the two enzymes used above. As shown in Table 2, using C18:2,Δ9,12 or C18:3,Δ9,12,15 as exogenous substrate, the biosynthesis of arachidonic (C20:4,Δ5,8,11,14) and eicosapentaenoic acid (C20:5,Δ5,8,11,14,17) were efficiently reconstituted. The end products C20:4,Δ5,8,11,14 or C20:5,Δ5,8,11,14,17 accounted for as much as 4.5% of the total fatty acids, which is approx. 20 times higher than the proportion obtained in similar reconstitution experiments using lipid-linked Δ6-desaturases [12,21]. The comparatively high yields presented in Table 2 were due to the very high activity of OtD6, as well as to the very efficient elongation, since more than 95% of OtD6 products were elongated further. In contrast, only approx. 25% of the elongated products were Δ5-desaturated further, most probably because not all of the elongated fatty acids were targeted to the sn-2-position of PC, which represents the favourite substrate of the lipid-linked desaturase.
Table 2. Reconstitution of polyunsaturated fatty acid biosynthesis in yeast.
The yeast strains INVSc1, transformed with pYES2-OtD6 and pESC-LEU-PSE1-PtD5 or the corresponding empty vectors, were grown for 48 h at 20 °C in the presence of 250 μM C18:2,Δ9,12 or C18:3,Δ9,12,15. FAMEs from whole cells were prepared and analysed by GLC as indicated in the Materials and methods section. Each value corresponds to the percentage of total fatty acids and is the mean±S.D. for five independent experiments.
| C18:2,Δ9,12 exogenously supplied | C18:3,Δ9,12,15 exogenously supplied | |||
|---|---|---|---|---|
| Fatty acid | Empty vectors | OtD6+PSE1+PtD5 | Empty vectors | OtD6+PSE1+PtD5 |
| C16:0 | 19.7±0.8 | 17.6±1.5 | 16.4±0.3 | 14.7±0.7 |
| C16:1,Δ9 | 22.2±1.1 | 19.9±1.6 | 24.6±0.4 | 22.4±1.6 |
| C18:0 | 6.8±0.5 | 6.0±0.8 | 6.7±0.2 | 6.0±0.2 |
| C18:1,Δ9 and C18:1,Δ11 | 15.8±0.5 | 23.2±2.9 | 21.1±0.7 | 27.4±2.9 |
| C18:2,Δ9,12 | 35.4±1.5 | 13.8±3.5 | – | – |
| C18:3,Δ6,9,12 | – | 0.5±0.1 | – | – |
| C18:3,Δ9,12,15 | – | – | 31.0±1.5 | 9.5±3.9 |
| C18:4,Δ6,9,12,15 | – | – | – | 0.5±0.1 |
| C20:2,Δ11,14 | – | 0.8±0.4 | – | – |
| C20:3,Δ8,11,14 | – | 13.6±2.2 | – | – |
| C20:4,Δ5,8,11,14 | – | 4.5±0.9 | – | – |
| C20:3,Δ11,14,17 | – | – | – | 0.6±0.2 |
| C20:4,Δ8,11,14,17 | – | – | – | 134±3.6 |
| C20:5,Δ5,8,11,14,17 | – | – | – | 4.7±0.4 |
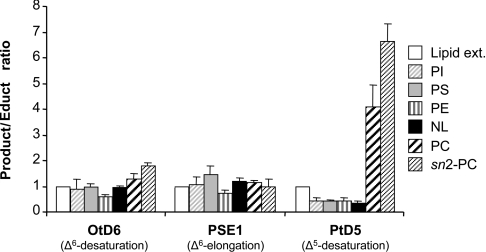
In order to see whether the presumed differences in acyl-carriers would be reflected in the fatty acid compositions of the different lipid components, we made a detailed lipid analysis of a transgenic yeast culture producing C20:5,Δ5,8,11,14,17. As shown in Figure 5, the product/educt ratios obtained for both the Δ6-desaturase OtD6 and the Δ6-elongase PSE1 were in about the same range in all the different lipids, including the sn-2 position of PC. In contrast, and most importantly, the ratio calculated for the Δ5-desaturase PtD5 showed large variations. It was lower in all lipid components than in the lipid extract, with the exception of PC, where it was 4–5 times higher. This ratio even reached a value of approx. 7 at the sn-2 position of PC (Figure 5), confirming our previous characterization of the Δ5-desaturase from Phae. tricornutum as a lipid-linked desaturase preferentially acting at this particular position [12]. These co-expression results again suggested that, in contrast with the Phae. tricornutum Δ5-desaturase, but similar to the Δ6-elongase PSE1, the Δ6-desaturase of O. tauri uses acyl-CoAs as substrates.
Figure 5. Product to educt ratios in different lipid fractions for Δ6-desaturation by OtD6, Δ6-elongation by PSE1 and Δ5-desaturation by PtD5 in a yeast culture co-expressing the three enzymes.
A culture of the yeast strain INVSc1 co-transformed with pYES2-OtD6 and pESC-LEU-PSE1-PtD5, was grown for 24 h at 20 °C in the presence of 250 μM C18:3,Δ9,12,15 and was used for lipid analysis. The fatty acid compositions of the different lipid fractions were determined as indicated in the Materials and methods section. The ratios were calculated as (product/educt) using values corresponding to the percentage of total fatty acids. All ratios were normalized to the ratio of the total lipid extract, set as 1. Each value is the mean±S.D. from three independent experiments. Lipid ext., lipid extract; PI, phosphatidylinositol; PS, phosphatidylserine; NL, neutral lipid.
DISCUSSION
Whereas front-end acyl-CoA desaturases have been described in animals, our data are the first to suggest that Δ6-acyl-CoA-desaturases also exist in the plant kingdom. So far, only the presence of acyl-CoA desaturases introducing the first double bond into saturated acyl chains had been suggested in higher plants and algae [1]. Nevertheless, a detailed biochemical characterization and a convincing demonstration that such desaturases actually act on acyl-CoAs are still lacking. Examples studied in more detail are the Δ5-desaturase from meadowfoam (Limnanthes alba) involved in the synthesis of C20:1,Δ5 [22,23] and the Δ9-desaturase from white spruce responsible for the conversion of stearic into oleic acid [24]. The sequences of these desaturases showed high similarities to cyanobacterial lipid-linked Δ9-desaturases and animal acyl-CoA Δ9-desaturases [23,24]. Based on in vitro assays with cell-free seed extracts from meadowfoam or lysates from yeast cells expressing the spruce enzyme, it had been suggested that acyl-CoAs were the actual substrates [22,24]. But, in both cases, the product was not isolated in the thioester form. This detail is particularly important in view of the ubiquitous presence of highly active acyltransferases that catalyse a rapid incorporation of acyl groups from the acyl-CoA pool into various lipids in cells of yeast [20] and higher plants [25]. These activities may complicate the identification of the actual substrate of plant desaturases tentatively assigned as acyl-CoA desaturases in in vitro assays, and the isolation of the product as an acyl-CoA may therefore represent more convincing evidence. Putative Δ9-acyl-CoA desaturases were also identified in the genome of Arabidopsis thaliana by sequence analogy [26], but Heilmann et al. [27] demonstrated recently that three of these putative acyl-CoA desaturases are in fact responsible for the introduction of a Δ7-double bond into lipid-linked palmitoyl residues of monogalactosyldiacylglycerol. These results show clearly that the acyl-carrier specificity of any desaturase cannot reliably be deduced from sequence similarities alone, and that each enzyme has to be characterized individually, either in vitro with identification of the actual product status or by in vivo studies as exemplified by our present investigation.
The only front-end Δ6-desaturase characterized so far in vitro is the human FADS2 enzyme [9]. This enzyme was purified to homogeneity and was used in detergent-containing assays in the presence of additional cytochrome b5 and cytochrome b5 reductase to show that the desaturated product was γ-linolenoyl-CoA [9]. In retrospect, a surprising detail of these reconstituted systems is the fact that the activity was absolutely dependent on the addition of exogenous cytochrome b5, although the cloning of the gene showed that FADS2 contains an N-terminal cytochrome b5 domain expected to function as immediate electron donor [28,29]. Since OtD6 represents the first Δ6-acyl-CoA-desaturase characterized from the plant kingdom, we also tried to provide in vitro evidence to support our conclusions. For this purpose, we tried to demonstrate in vitro desaturation activity using microsomes from yeast cultures expressing OtD6 in the presence of linoleoyl-CoA and appropriate co-factors. Despite repeated attempts with different microsomal preparations, HPLC analysis of the acyl-CoA fraction could never detect any desaturation activity. Nevertheless, using the same set of independent in vivo experiments that had been worked out previously [12], we provide strong evidence that OtD6 is an acyl-CoA desaturase. The interpretation of these results may not be as straightforward as the cell-free demonstration of a desaturated acyl-CoA product, but diagnostically relevant differences allow a clear distinction between acyl-CoA- and lipid-linked desaturases. In addition, this in vivo technology avoids possible artifacts which may be caused by the presence of detergents as reported for human FADS2 [9] or for the spinach lipid-linked Δ12-desaturase (FAD6) [30].
In conclusion, OtD6 represents the first plant acyl-CoA Δ6-desaturase to be characterized biochemically in vivo. The fact that its sequence is not related to any other functionally characterized Δ6-desaturases (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/389/bj3890483add.htm) suggests that it may represent a previously undescribed type of desaturase. Whether this reflects a biochemically unusual type of desaturase or the particular phylogenetic history of O. tauri cannot be answered at present, because the structural requirements for accepting different forms of acyl substrates cannot be recognized in the amino acid sequences of desaturases. Fortunately, the continuously increasing number and variety of cloned desaturases may provide the chance to identify the structural motifs responsible not only for the regioselectivity, but also for acyl-carrier specificity of these enzymes. Finally, because of the efficient elongation obtained in our reconstitution experiments in yeast, front-end acyl-CoA desaturases of plant origin may represent new and promising tools for the biotechnological production of very-long-chain polyunsaturated fatty acids in transgenic oilseed crops [31,32].
Online data
Acknowledgments
This research has been supported by a Marie Curie Fellowship of the European Community programme Human Potential under contract number HPMF-CT-1999-00148 (for F.D.) and by the Bundesministerium für Bildung und Forschung project NAPUS 2000 (part FK 0312252 F; for A.A.). Financial support by the BASF Plant Science (Ludwigshafen, Germany) and Fonds der Chemischen Industrie is also gratefully acknowledged. H.M. thanks the Génopole Languedoc-Roussillon for its participation in the genome sequencing program of O. tauri.
References
- 1.Sperling P., Ternes P., Zank T. K., Heinz E. The evolution of desaturases. Prostaglandins Leukotrienes Essent. Fatty Acids. 2003;68:73–95. doi: 10.1016/s0952-3278(02)00258-2. [DOI] [PubMed] [Google Scholar]
- 2.Shanklin J., Cahoon E. B. Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:611–641. doi: 10.1146/annurev.arplant.49.1.611. [DOI] [PubMed] [Google Scholar]
- 3.Shanklin J., Whittle E., Fox B. G. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- 4.Aitzetmüller K., Tsevegsüren N. Seed fatty acids, ‘front-end’-desaturases and chemotaxonomy – a case study in the Ranunculaceae. J. Plant. Physiol. 1994;143:538–543. [Google Scholar]
- 5.Sperling P., Heinz E. Desaturases fused to their electron donor. Eur. J. Lipid Sci. Technol. 2001;103:158–180. [Google Scholar]
- 6.Fan Y. Y., Chapkin R. S. Importance of dietary γ-linolenic acid in human health and nutrition. J. Nutr. 1998;128:1411–1414. doi: 10.1093/jn/128.9.1411. [DOI] [PubMed] [Google Scholar]
- 7.Reddy A. S., Nuccio M. L., Gross L. M., Thomas T. L. Isolation of a Δ6-desaturase gene from the cyanobacterium Synechocystis sp. strain PCC 6803 by gain-of-function expression in Anabaena sp. strain PCC 7120. Plant Mol. Biol. 1993;22:293–300. doi: 10.1007/BF00014936. [DOI] [PubMed] [Google Scholar]
- 8.Wada H., Murata N. Membrane lipids in cyanobacteria. In: Siegenthaler P.-A., Murata N., editors. Lipids in Photosynthesis: Structure, Function and Genetics. Dordrecht: Kluwer Academic Publishers; 1998. pp. 65–81. [Google Scholar]
- 9.Okayasu T., Nagao M., Ishibashi T., Imai Y. Purification and partial characterization of linoleoyl-CoA desaturase from rat liver microsomes. Arch. Biochem. Biophys. 1981;206:21–28. doi: 10.1016/0003-9861(81)90061-8. [DOI] [PubMed] [Google Scholar]
- 10.Stymne S., Stobart A. K. Biosynthesis of γ-linolenic acid in cotyledons and microsomal preparations of the developing seeds of common borage (Borago officinalis) Biochem. J. 1986;240:385–393. doi: 10.1042/bj2400385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson F. M., Fraser T. C., Smith M. A., Lazarus C., Stobart A. K., Griffiths G. Biosynthesis of C18 polyunsaturated fatty acids in microsomal membrane preparations from the filamentous fungus Mucor circinelloides. Eur. J. Biochem. 1998;252:513–519. doi: 10.1046/j.1432-1327.1998.2520513.x. [DOI] [PubMed] [Google Scholar]
- 12.Domergue F., Abbadi A., Ott C., Zank T. K., Zahringer U., Heinz E. Acyl-carriers used as substrates by the desaturases and elongases involved in very long-chain polyunsaturated fatty acids biosynthesis reconstituted in yeast. J. Biol. Chem. 2003;278:35115–35126. doi: 10.1074/jbc.M305990200. [DOI] [PubMed] [Google Scholar]
- 13.Zank T. K., Zähringer U., Beckmann C., Pohnert G., Boland W., Holtorf H., Reski R., Lerchl J., Heinz E. Cloning and functional characterisation of an enzyme involved in the elongation of Δ6-polyunsaturated fatty acids from the moss Physcomitrella patens. Plant J. 2002;31:255–268. doi: 10.1046/j.1365-313x.2002.01354.x. [DOI] [PubMed] [Google Scholar]
- 14.Derelle E., Ferraz C., Lagoda P., Eychenié S., Cooke R., Regad F., Sabau X., Courties C., Delseny M., Demaille J., et al. DNA libraries for sequencing the genome of Ostreococcus tauri (Chlorophyta, Prasinophyceae): the smallest free-living eukaryotic cell. J. Phycol. 2002;38:1150–1156. [Google Scholar]
- 15.Meyer A., Kirsch H., Domergue F., Abbadi A., Sperling P., Bauer J., Cirpus P., Zank T. K., Moreau H., Roscoe T. J., et al. Novel fatty acid elongases and their use for the reconstitution of docosahexaenoic acid biosynthesis. J. Lipid Res. 2004;45:1899–1909. doi: 10.1194/jlr.M400181-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Dohmen R. J., Strasser A. W., Honer C. B., Hollenberg C. P. An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast. 1991;7:691–692. doi: 10.1002/yea.320070704. [DOI] [PubMed] [Google Scholar]
- 17.Schjerling C. K., Hummel R., Hansen J. K., Borsting C., Mikkelsen J. M., Kristiansen K., Knudsen J. Disruption of the gene encoding the acyl-CoA-binding protein (ACB1) perturbs acyl-CoA metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:22514–22521. doi: 10.1074/jbc.271.37.22514. [DOI] [PubMed] [Google Scholar]
- 18.Larson T. R., Graham I. A. Technical Advance: a novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant J. 2001;25:115–125. doi: 10.1046/j.1365-313x.2001.00929.x. [DOI] [PubMed] [Google Scholar]
- 19.Larson T. R., Edgell T., Byrne J., Dehesh K., Graham I. A. Acyl CoA profiles of transgenic plants that accumulate medium-chain fatty acids indicate inefficient storage lipid synthesis in developing oilseeds. Plant J. 2002;32:519–527. doi: 10.1046/j.1365-313x.2002.01440.x. [DOI] [PubMed] [Google Scholar]
- 20.Wagner S., Paltauf F. Generation of glycerophospholipid molecular species in the yeast Saccharomyces cerevisiae: fatty acid pattern of phospholipid classes and selective acyl turnover at sn-1 and sn-2 positions. Yeast. 1994;10:1429–1437. doi: 10.1002/yea.320101106. [DOI] [PubMed] [Google Scholar]
- 21.Beaudoin F., Michaelson L. V., Hey S. J., Lewis M. J., Shewry P. R., Sayanova O., Napier J. A. Heterologous reconstitution in yeast of the polyunsaturated fatty acid biosynthetic pathway. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6421–6426. doi: 10.1073/pnas.110140197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreau R. A., Pollard M. R., Stumpf P. K. Properties of a Δ5-fatty acyl-CoA desaturase in the cotyledons of developing Limnanthes alba. Arch. Biochem. Biophys. 1981;209:376–384. doi: 10.1016/0003-9861(81)90295-2. [DOI] [PubMed] [Google Scholar]
- 23.Cahoon E. B., Marillia E. F., Stecca K. L., Hall S. E., Taylor D. C., Kinney A. J. Production of fatty acid components of meadowfoam oil in somatic soybean embryos. Plant Physiol. 2000;124:243–251. doi: 10.1104/pp.124.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marillia E. F., Giblin E. M., Covello P. S., Taylor D. C. A desaturase-like protein from white spruce is a Δ9 desaturase. FEBS Lett. 2002;526:49–52. doi: 10.1016/s0014-5793(02)03112-5. [DOI] [PubMed] [Google Scholar]
- 25.Stymne S., Stobart A. K., Glad G. The role of the acyl-CoA pool in the synthesis of polyunsaturated 18-carbon fatty acids and triacylglycerol production in the microsomes of developing safflower seeds. Biochim. Biophys. Acta. 1983;752:198–208. doi: 10.1016/0005-2760(83)90113-3. [DOI] [PubMed] [Google Scholar]
- 26.Fukuchi-Mizutani M., Tasaka Y., Tanaka Y., Ashikari T., Kusumi T., Murata N. Characterization of Δ9 acyl-lipid desaturase homologues from Arabidopsis thaliana. Plant Cell Physiol. 1998;39:247–253. doi: 10.1093/oxfordjournals.pcp.a029364. [DOI] [PubMed] [Google Scholar]
- 27.Heilmann I., Pidkowich M. S., Girke T., Shanklin J. Switching desaturase enzyme specificity by alternate subcellular targeting. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10266–10271. doi: 10.1073/pnas.0402200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho H. P., Nakamura M. T., Clarke S. D. Cloning, expression, and nutritional regulation of the mammalian Δ6 desaturase. J. Biol. Chem. 1999;274:471–477. doi: 10.1074/jbc.274.1.471. [DOI] [PubMed] [Google Scholar]
- 29.Marquardt A., Stohr H., White K., Weber B. H. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66:175–183. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt H., Heinz E. Direct desaturation of intact galactolipids by a desaturase solubilized from spinach (Spinacia oleracea) chloroplast envelopes. Biochem. J. 1993;289:777–782. doi: 10.1042/bj2890777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abbadi A., Domergue F., Bauer J., Napier J. A., Welti R., Zahringer U., Cirpus P., Heinz E. Biosynthesis of very-long-chain polyunsaturated fatty acids in transgenic oilseeds: constraints on their accumulation. Plant Cell. 2004;16:2734–2748. doi: 10.1105/tpc.104.026070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Domergue F., Abbadi A., Heinz E. Relief for fish stocks: oceanic fatty acids in transgenic oilseeds. Trends Plant Sci. 2005;10:112–116. doi: 10.1016/j.tplants.2005.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.