Abstract
The myoglobin of the polychaete annelid Ophelia bicornis was isolated, purified to homogeneity and characterized. The primary structure, obtained from cDNA and protein sequencing, consists of 139 amino acid residues. The alignment with other globin sequences showed that O. bicornis myoglobin misses the pre-A helix and the first six residues of the A helix. The presence of a PheB10-GlnE7 haem distal residue pair is in agreement with the measured oxygen affinity (P50=0.85 mmHg; 1 mmHg=0.133 kPa) and the only slightly higher autoxidation rate constant (0.28 h−1) with respect to that of the sperm whale myoglobin mutant E7 His→Gln (0.21 h−1) and to elephant myoglobin (0.1 h−1). Oxygen-binding co-operativity was found to be absent under all the examined experimental conditions. The resistance of O. bicornis myoglobin towards autoxidation seems to confirm the important role of part of the A helix in the stability of the globin. The higher pKa of the acid–alkaline ferric transition of O. bicornis with respect to Asian elephant myoglobin, as well as the higher absorbance ratio of its ferric form to the oxy form measured in the Soret region (γmet/γoxy) with respect to that of the African elephant myoglobin, suggested a stronger interaction between the distal glutamine and the water molecule at the sixth co-ordinate position.
Keywords: autoxidation rate, body wall myoglobin, haem distal residue pair, Ophelia bicornis, oxygen binding, polychaete annelid
Abbreviations: amu, atomic mass units; Caps, 3-(cyclohexylamino)propane-1-sulphonic acid; ESI, electrospray ionization; h50, oxygen binding co-operativity at 50% saturation; IEF isoelectricfocusing; IT, ion-trap; Mb, myoglobin; P50, partial pressure of oxygen required to saturate 50% of the haems; RP-HPLC, reverse-phase HPLC; RT, reverse transcriptase; TFA, trifluoroacetic acid
INTRODUCTION
In polychaete annelids, respiratory proteins may occur: (i) intracellularly, in circulating erythrocytes, as single-chain single-domain Hbs, either monomeric and multimeric (Glycera dibranchiata [1]) or dimeric (Travisia foetida [2]); (ii) intracellularly, in the cytoplasm of specific tissues, such as the body wall (Arenicola marina [3]), the proboscis (Glycera robusta [4]) and the ventral nerve (Aphrodite aculeata [5]), in general as monomeric Mbs (myoglobins), with few exceptions, for example, the dimeric Mb from the body wall of Travisia foetida [2]); (iii) intracellularly, as myo-haemerythrin (Hediste diversicolor [6]) or as haemerythrin (Magelona papillicornis [7]); (iv) extracellularly, as multi-subunit giant Hbs freely dissolved in the haemolymph (erythrocruorin in Perinereis cultrifera [8], chlorocruorin, with the protohaem replaced by a chlorocruorohaem in Sabella spallanzanii [9], and in Serpula vermicularis, with both a protohaem and a chlorocruorohaem [10]).
Mbs may be found as the sole respiratory pigment, as in Aph. aculeata [11], or in addition to coelomic cell Hb (Glycera robusta [4]), erythrocruorin (Abarenicola pacifica [12]) and chlorocruorin (Potamilla sp. [13]).
While the presence of an extracellular Hb in the polychaete Ophelia bicornis has been described previously [14], in the present paper, we demonstrate, for the first time, the presence of a Mb in the body wall of this species. The structure, the physicochemical characteristics and the functional properties of the Mb are described and discussed in comparison with Mbs from other invertebrates.
EXPERIMENTAL
Mb purification
Live specimens of O. bicornis were collected along the shore in the island of Sardinia near Pula (Cagliari). Fresh collected worms, rinsed with cold seawater, were homogenized on ice in a small volume of cold 50 mM Tris/HCl buffer, pH 7.5, and 10 mM CaCl2, and the supernatant obtained was brought to 40% ammonium sulphate saturation. After centrifugation at 15000 g for 15 min, the precipitate was discarded, and the supernatant was precipitated with 90% ammonium sulphate saturation. The red precipitate obtained was dissolved in a small volume of the same buffer and loaded on to a Sephadex G-100 column (6 cm×50 cm) equilibrated with the same buffer. The haem-containing fractions, retarded by the column, were concentrated, dialysed against 20 mM Tris/HCl and 1 mM EDTA, pH 8.4, loaded on to a DEAE-cellulose column (3 cm×20 cm) equilibrated with the same buffer, and eluted under isocratic conditions.
Body walls from 30 worms were carefully dissected free from gills and viscera, rinsed with filtered seawater and homogenized as described above, to determine the localization of the purified haem protein.
The homogeneity of the purified protein was verified by IEF (isoelectric focusing)/PAGE on a thin-layer 5% polyacrylamide slab gel (pH 5–8) [15].
Approximate molecular mass was estimated by SDS/PAGE (18% polyacrylamide) [16] using low-molecular-mass standards (Sigma), and molecular mass was determined exactly by ESI (electrospray ionization)-IT (ion-trap)-MS (as described below).
Tryptic and CNBr digestion
Globin (3 mg) obtained by acid/acetone extraction [17] was dissolved in 1 ml of 6 M guanidinium chloride, 0.1 M Tris/HCl and 1 mM EDTA, pH 8.3. Dithiothreitol was added in a 2 mM excess over the disulphide bonds of the protein. Nitrogen was passed through the solution, which was then sealed under an inert atmosphere and incubated at 37 °C for 1 h. At the end of the incubation, 4-vinylpyridene was added in a 1.1-fold molar excess over total thiol groups. The mixture was incubated overnight under nitrogen, quenched with 2-mercaptoethanol, submitted to dialysis against water and freeze-dried.
For CNBr digestion, modified globin (1 mg) was dissolved in 70% methanoic (formic) acid (200 μl), and 1 mg of CNBr (in 70% methanoic acid) was added under nitrogen. The reaction mixture was stirred for 24 h at 25 °C, then 500 μl of water was added, and the solution was freeze-dried.
MS experiments
The instrument used was an Xcalibur LCQ Deca XP Plus mass spectrometer (ThermoFinnigan, San Jose, CA, U.S.A.), equipped with an ESI source and an IT analyser. The molecular mass of intact Mb was determined by direct infusion of a solution prepared by mixing 200 μl of 0.175 mM Mb with 800 μl of 0.2% ethanoic (acetic) acid in methanol/water (50:50, v/v). Electronebulization was performed at a flow rate of 10 μl/min with a sheath gas flow rate of 34 arbitrary units. The electrospray capillary temperature was 280 °C, the source voltage was 4 kV and the capillary voltage was 30 V. The IT analyser operated in positive mode in the 300–2000 m/z range, and spectra were acquired every 3 ms.
The peptide mixtures obtained by trypsin and CNBr digestion were freeze-dried and submitted to RP-HPLC (reverse-phase HPLC)–ESI-IT-MS analysis using a Vydac C8 middle-bore column (100 mm×2.0 mm; 5 μm particle diameter). The eluents were: (eluent A) 0.056% aqueous TFA (trifluoroacetic acid) and (eluent B) 0.050% TFA in acetonitrile/water (80:20, v/v). The gradient applied was linear from 0 to 55% of B in 40 min at a flow-rate of 0.30 ml/min. The T splitter addressed a flow rate of approx. 0.20 ml/min towards the diode-array detector and a flow-rate of approx. 0.10 ml/min towards the ESI source. The diode array detector was set in the range 214–276 nm. Mass spectra were collected every 3 ms in the positive-ion mode. MS spray voltage was 4.50 kV, and the capillary temperature was 220 °C. MS/MS experiments were performed by detecting the parent ions with a peak width of 2–4 m/z values and by using 40% of the maximum activation amplitude.
Deconvolution of average ESI mass spectra was automatically performed either by the software provided with the Deca-XP instrument (Bioworks Browser) or by MagTran 1.0 software [18]. Mass values determined on digested samples were compared with the expected average and monoisotopic values calculated using the PeptideMass program. Theoretical MS/MS spectra were generated utilizing the MS-Product program, available at the Protein Prospector site (http://prospector.ucsf.edu/).
Protein sequencing
N-terminal amino acid residues were determined by using an automated Procise 49XHT Protein Sequencer (Applied Biosystems, Foster City CA, U.S.A.). Further confirmation of the sequence was obtained by HPLC–ESI-MS/MS experiments performed on the tryptic mixture.
cDNA sequencing
Total RNA was isolated from O. bicornis using the EZNA Mollusc RNA kit (Omega Bio-Tek). Two degenerate primers were designed based on the amino acid sequence: OpF1 (5′-GGNGTNGCNGAYACNTGGGC-3′), a 20-mer with 512 redundancies, corresponding to the sense strand predicted by the peptide fragment GVADTWA, and OpR1 (5′-GCRTCNGCYTGNGCYTGCAT-3′), a 20-mer with 128 redundancies, corresponding to the antisense strand predicted by the peptide fragment MQAQADA. RT (reverse transcriptase)-PCR was carried out using the Enhanced Avian HS RT-PCR kit (Sigma) with an anchored oligo-(dT)23 primer. A PCR was then performed with the OpF1 and the OpR1 primers using Platinum Taq DNA Polymerase High Fidelity (Invitrogen), with a denaturation step at 94 °C for 5 min, followed by 35 cycles at 94 °C for 1.5 min, 45 °C for 2 min and 72 °C for 1 min. The PCR program was concluded by an extension at 72 °C for 7 min. With this approach, a 170 bp product was amplified, cleaned, cloned in the pCRII vector using the TA cloning kit (Invitrogen), and used to transform Escherichia coli INVαF' cells. Eight recombinant clones were isolated and sequenced. A specific primer OpF2 (5′-CTGAAGGGTGACCCAAACATG-3′), derived from the obtained DNA sequence, was used, together with the anchored oligo(dT)23 primer in a PCR to obtain the 3′ end of the cDNA. The PCR amplification was performed for 35 cycles as follows: 94 °C for 30 s, 55 °C for 30 s and 72 °C for 1 min.
Oxygen equilibria
Oxygen dissociation curves were obtained by the tonometric method [19] using a Varian Cary 50 spectrophotometer at low protein concentrations (≈20 μM) and by the thin layer dilution technique [20] using a HP8453 diode array spectrophotometer at high protein concentrations (0.6 mM). Oxygen-binding experiments were performed in 50 mM Tris/HCl, 0.1 M NaCl in the 6.5–8.5 pH range at various temperatures, in the presence or absence of 5 mM sodium lactate.
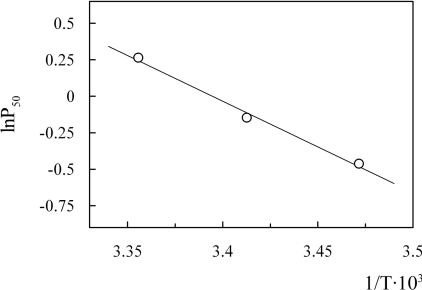
The overall oxygenation enthalpy (ΔH), corrected for the heat of solubilization (−12.5 kJ·mol−1) was calculated from the integrated van't Hoff equation [21]:
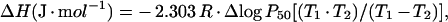 |
where T is the absolute temperature, R=8.31 J·K−1·mol−1 and P50 is the partial pressure of the ligand required to saturate 50% of the haems. Over the temperature range examined (15–25 °C), van't Hoff plots were linear within the experimental error. Confidence limits of the data are ±8% for P50 and ±15% for ΔH values.
Autoxidation rate
Autoxidation rate was measured at a Mb concentration of 10 μM (on haem basis) at 37 °C in air-equilibrated 0.1 M potassium phosphate buffer, pH 7.0, containing 1 mM EDTA and 3 mmol of catalase and superoxide dismutase per mole of haem [22]. Changes in absorption spectrum of the oxy form were recorded between 370 and 700 nm at measured intervals of time with a HP8453 diode array spectrophotometer. The reaction was also followed by the absorbance change at 581 nm (α-peak of MbO2). For the final state of the runs, the Mb was converted completely into its ferric form by the addition of potassium ferricyanide.
Dependence of the autoxidation rate from pH was measured in 0.1 M buffer at 25 °C under the same experimental conditions reported above, except that the buffers used were: sodium acetate/ethanoic acid (pH 5.0), Mes/NaOH (pH 6.0), potassium phosphate (pH 7.0), Taps/NaOH (pH 8.0), Caps [3-(cyclohexylamino)propane-1-sulphonic acid]/NaOH (pH 9.0 and 10.0), and phosphate (pH 11.0, 11.5 and 12.0). The pH of the reaction mixture was checked before and after the run.
The acid–alkaline equilibrium of ferric O. bicornis Mb was determined as a function of pH with a HP8453 diode array spectrophotometer at 10 °C using a protein concentration of 10 μM (on haem basis). Buffers used were 0.1 M Mes, Taps and Caps.
Structural and evolutional analysis of globin sequence
The primary structure of O. bicornis Mb was aligned with relevant vertebrate and invertebrate globins by means of the non-vertebrate [23] and the vertebrate [24] template II. Penalty scores were calculated by adding the scores of all the positions presenting a disallowed residue.
RESULTS AND DISCUSSION
Isolation and structural characterization of O. bicornis Mb
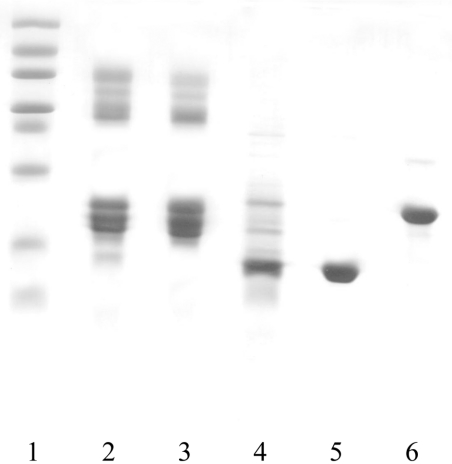
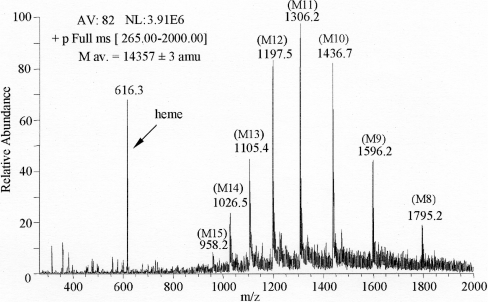
Purified O. bicornis Mb appeared on SDS/PAGE as a single polypeptide chain with a molecular mass lower than that of pig Mb (Figure 1), as confirmed by ESI-IT-MS analysis, which provided a mass value for the globin of 14357±3 amu (atomic mass units) and a mass value of 616.3 amu for the haem (Figure 2). The experimental average mass of 14462±3 amu obtained by direct ESI-MS infusion after protection with 4-vinylpyridene (Δmass=105 amu with respect to the unreacted protein) allowed us to establish the presence of a single cysteine residue for the globin molecule.
Figure 1. Purification of O. bicornis Mb.
SDS/PAGE of selected fractions: lane 1, low-molecular-mass standard; lane 2, total homogenate after 40–90% ammonium sulphate precipitation; lane 3, unretarded Sephadex G100 fractions; lane 4, retarded Sephadex G100 fractions; lane 5, purified O. bicornis Mb after DEAE-cellulose; lane 6, purified pig Mb.
Figure 2. ESI mass spectrum of O. bicornis holo-Mb.
Analysis was performed by direct infusion of 35 μM Mb in 0.16% ethanoic acid in methanol/water (50:50, v/v). The IT analyser operated in positive-ion mode in the 300–2000 m/z range, and spectra were acquired every 3 ms. AV, number of averaged spectra; NL, normalization level; M av., mass average.
IEF analysis performed under native conditions showed the homogeneity of the purified Mb from O. bicornis (Figure 3) and indicated a pI value in good agreement with that calculated on the basis of the established amino acid sequence (theoretical pI equal to 6.19). To determine the localization of the haem protein, the body walls of 30 worms, stripped of all internal organs and blood vessels, were carefully washed with sea water, homogenized and processed as described for total homogenate in the experimental session. The resulting SDS/PAGE electrophoretic pattern was identical with that obtained from total homogenate (results not shown), suggesting that the characterized haemoprotein can be considered to be a Mb.
Figure 3. Thin layer 5% polyacrylamide IEF of purified O. bicornis Mb (pH range 5–8).
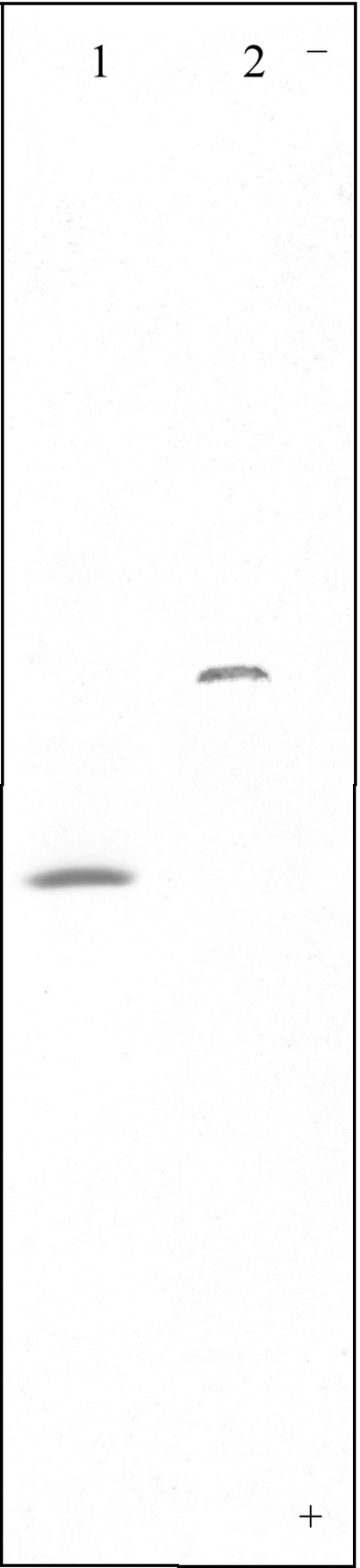
Lane 1: O. bicornis Mb; lane 2: human HbA.
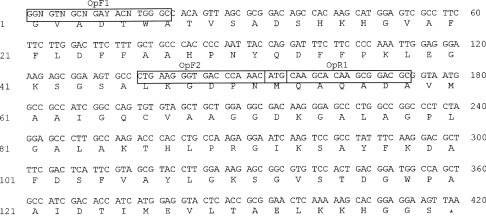
The nucleotide sequence of the cDNA is shown in Figure 4, together with the deduced sequence of 139 residues. The theoreti- cal average mass of 14359 amu, computed from the globin sequence, is in a good agreement with the experimental average mass value determined by ESI-MS.
Figure 4. Sequence of the 420 bp cDNA encoding the O. bicornis globin.
The single-letter amino acid designations of the globin chain residues are listed below the corresponding codons. Primers are boxed and named. The first 20 nucleotides correspond to the degenerate OpF1 primer.
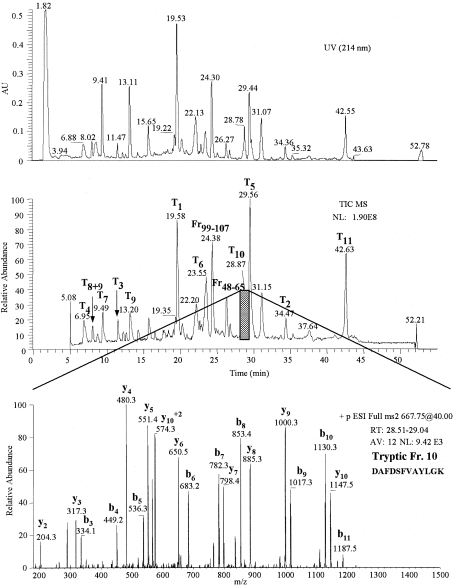
The primary structure of O. bicornis Mb was also reconstructed on the basis of sequences of relevant peptides generated by trypsin and CNBr digestion of the modified apo-Mb. Sequences were determined by both Edman degradation and HPLC–ESI-MS/MS analysis. The results of the HPLC–ESI-MS separation of the tryptic mixture, together with the MS/MS fragmentation pattern of T10 are reported in Figure 5, as an example. The reconstructed primary structure covered approx. 90% of the cDNA sequence (Figure 6).
Figure 5. RP-HPLC–ESI-MS analysis of the tryptic digest of O. bicornis globin (after 4-vinylpyridene treatment).
Upper panel: UV profile obtained at 214 nm. Middle panel: total ion current (TIC MS) profile. Lower panel: MS/MS spectrum of T10 tryptic peptide eluting between 28.51–29.04 min, obtained by the fragmentation of the bi-charged ion at 667.7 m/z. The analysis of the mass spectrum allowed us to recognize the y and b fragmentation series. AV, number of averaged spectra; NL, normalization level.
Figure 6. Comparison between the primary structure of O. bicornis Mb deduced from cDNA and that deduced from Edman degradation and HPLC–ESI-MS data.
Edman degradation was performed on the intact globin (N-term) and on the underlined peptides originated by trypsin (tryptic) and CNBr digestion. HPLC–ESI-MS/MS experiments were performed on the underlined tryptic peptides (MS/MS); experimental mass values ([M+H]+) are underlined, theoretical values are in brackets.
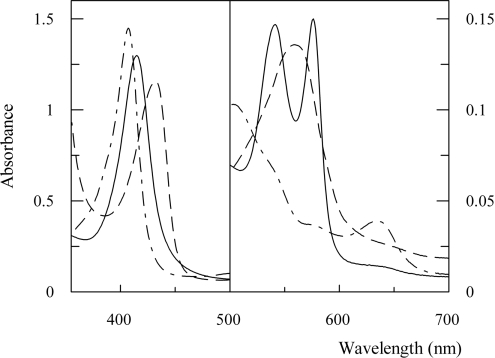
The absorption spectrum of O. bicornis Mb in the oxy, dithionite-reduced and acidic met forms is reported in Figure 7. The oxy form showed a typical spectrum, with an absorbance ratio of α/β corresponding to 1.02. In other invertebrate Mbs (Aplysia limacina [25], Nassa mutabilis [26]), reversed peak heights have been found. The visible spectrum of the dithionite-reduced form showed a broad peak. This characteristic, up to now never observed in a Mb spectrum, has been already observed in deoxyerythrocruorin from different species, and it has been related to the heterogeneity shown by that multimeric protein [27]. Since O. bicornis Mb appeared homogeneous both in IEF and ESI-IT-MS, the broadening of the spectrum could not be ascribed to the heterogeneity of the sample.
Figure 7. Absorption spectra of O. bicornis Mb.
OxyMb (——), acidic metMb (–·–·–) and dithionite-reduced Mb (– – –) in 10 mM Tris/HCl buffer, pH 8.0, for the oxy and deoxy forms, and pH 7.0 for the met form; Protein concentration was 10 μM.
It has been proposed to divide Mbs into two groups considering the value of the absorbance ratio of the acidic met form Soret peak to that of the oxy form (γmet/γoxy). In fact, several Mbs lacking the distal histidine residue showed a blue-shift and a marked intensity decrease of the Soret peak that resulted in an absorbance ratio of the Soret peak of the acidic met form (408 nm) to that of the oxy form (414 nm) (γmet/γoxy) lower than 1.0, while the Mbs presenting the usual histidine residue at the distal site showed γmet/γoxy values higher than 1.0 [28].
The Soret peak of the acidic met form of O. bicornis Mb showed its maximum at 408 nm, with an absorbance ratio (γmet/γoxy) equal to 1.11 (Figure 7). These features, typical of the hexa-co-ordinate high-spin complex, suggested that, even if the distal histidine is substituted by glutamine, a water molecule should be co-ordinated to the haem iron in O. bicornis acid met Mb. Other residues that may be involved in this phenomenon are E10, B10, E11 and CD3. However, Mbs from African elephant and O. bicornis, which have phenylalanine and valine in B10 and E11 respectively, differ in their γmet/γoxy values, at lower than 1.0 and equal to 1.11 respectively [29]. Moreover, the acid ferric derivative of Apl. limacina Mb showed the blue-shift of the absorption maximum wavelength in the Soret region (at 400 nm), but the spectrum of the single mutant E10 Arg→Thr of Apl. limacina Mb did not show it. It has been hypothesized that the shorter and polar threonine chain at position E10, in place of the long and flexible arginine, may increase solvent accessibility to the distal pocket, thus leading to water-co-ordination [25]. All this evidence suggested that the absence of the distal histidine residue is not a molecular feature sufficient to determine the blue-shift and the decrease in intensity observed in the Soret region of the acid met form of different Mbs.
Previous papers on Mbs lacking the distal histidine residue have shown that the pKa of the ferric acid–alkaline transition may increase or decrease significantly with respect to sperm whale and horse Mbs. For instance, it was shown that the pKa of Apl. limacina Mb, equal to 7.5, increased to 9.4 in the mutant E7 Val→His, to 10.2 in the mutant E10 Arg→Thr and to 10.9 in the double mutant E10 Arg→Thr, E7 Val→His [25].
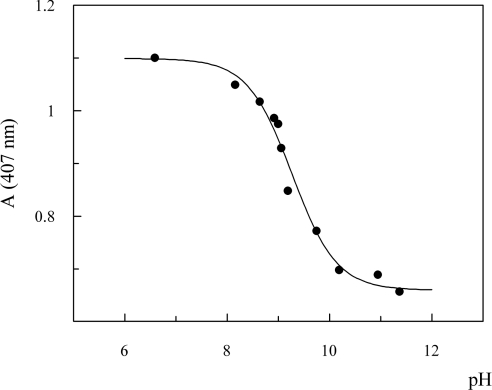
The measurements of the acid–alkaline transition of ferric O. bicornis Mb in the Soret region allowed us to calculate the pKa of the transition (Figure 8), which resulted equal to 9.27. The pKa value obtained, together with the γmet/γoxy value (>1.0), strongly suggested that, in O. bicornis, the glutamine E7 is able to stabilize the water molecule at the sixth co-ordinate position. This interaction should be stronger than that existing in Asian elephant Mb, that showed a pKa value of 8.5 [30], in sperm whale and horse Mb (pKa equal to 8.99 and 8.93 respectively [31]).
Figure 8. Titration curve for the acid–alkaline equilibrium of ferric O. bicornis Mb.
Absorbance of 10 μM ferric Mb was measured at 407 nm, and 10 °C in the 6.6–11.4 pH range, using 0.1 M Mes, Taps and Caps buffers.
Functional characteristics
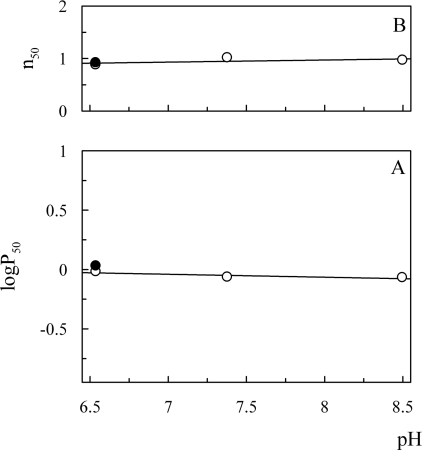
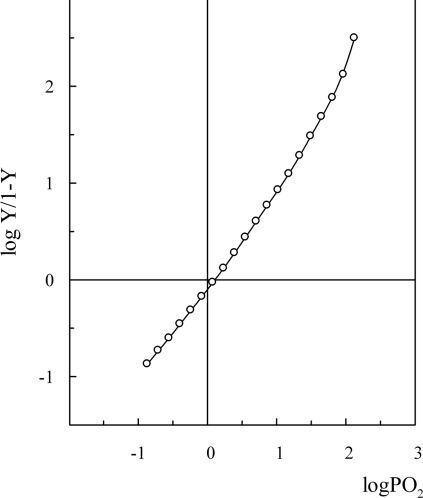
Oxygen dissociation curves of O. bicornis Mb at low (≈20 μM) and high (≈0.6 mM) protein concentration showed absence of co-operativity (h50=1) at all examined pHs and high oxygen affinity [P50=0.85 and 1.3 mmHg (1 mmHg=0.133 kPa) respectively], not pH-dependent (Figures 9 and 10). The oxygen affinity of O. bicornis was found similar to that of Ar. marina MbI and MbII (P50=0.39 and 0.72 mmHg respectively [32]), Aph. aculeata nerve Mb (P50=1.1 mmHg [5]), G. dibranchiata Mb (P50=0.5 mmHg [33]), human Mb (P50=0.65 mmHg [34]). Since the adults live buried in intertidal habitats consisting of relatively fine sand, emerging from sea water for long periods [35], it is not surprising that the oxygen-binding properties of this globin are compatible with an oxygen-storage function, also considering the presence of a giant Hb freely dissolved in the haemolymph [14].
Figure 9. Effect of pH on oxygen affinity (A) and co-operativity (B) of O. bicornis Mb.
Experimental conditions: 20 μM Mb in 50 mM Tris/HCl, and 0.1 M NaCl at 20 °C in the absence (○) and presence of 5 mM sodium lactate (●). n50, h50.
Figure 10. Oxygen-binding curve of O. bicornis Mb.
The experiment was performed at 20 °C in 50 mM Tris/HCl, pH 7.4, and 0.1 M NaCl at a protein concentration of 0.6 mM, by using the thin layer dilution technique. Y is the fraction of oxygen-binding sites occupied by oxygen; PO2, partial pressure of oxygen.
Lactate, described as an heterotrophic modulator of oxygen affinity in sperm whale and horse heart Mb [36], did not affect oxygen affinity and co-operativity of O. bicornis Mb under the experimental conditions used (Figure 9).
The enthalpy of oxygenation at pH 7.4 was measured to be −38.9 kJ/mol (Figure 11). This value is lower than that of Ar. marina MbI and MbII (−62 and −64 kJ/mol respectively [32]), human Mb (−56 kJ/mol [34]), Aplysia Mb (−57 kJ/mol [37]) and Biomphalaria glabrata Mb (−69.5 kJ/mol [38]).
Figure 11. van't Hoff plot of O. bicornis Mb.
Experimental conditions: 20 μM Mb in 50 mM Tris/HCl, pH 7.4, and 0.1M NaCl.
The distal E7 histidine residue, known to play a key role in stabilizing the bound oxygen [39], is replaced by glutamine in virtually all the prokaryotic and protist globins, as well as in some invertebrate globins (i.e. Cerebratulus lacteus Hb [40], B. glabrata Mb [38], Lucina pectinata HbI [41], Calyptogena kaikoi Mb [42] and Urechis caupo Hb [43]). This finding suggested that glutamine might be the ancestral residue at this position [40].
It has been shown that the E7 His→Gln mutation introduced by site-directed mutagenesis in wild-type sperm whale Mb lowered oxygen affinity ∼5-fold and increased the rate of autoxidation 3-fold [44]. In O. bicornis Mb, a glutamine residue is present at E7, but the oxygen affinity is similar to that of native sperm whale Mb and many other non-vertebrate Mbs [32].
The position B10 in non-vertebrate globins is usually occupied by a large hydrophobic residue (leucine, phenylalanine, tyrosine, methionine or tryptophan) whose side chain can be turned into the haem pocket and is thus involved in the control of oxygen affinity through stabilization of the ligand binding, as described in nematode [45] and trematode Hbs [46]. In O. bicornis, the residue at B10 is phenylalanine. The single mutation B10 Leu→Phe, introduced by site-directed mutagenesis in wild-type sperm whale Mb, produced a 10-fold increase in oxygen affinity [47], while the double mutation E7 His→Gln, B10 Leu→Phe abolished the effect of decreasing/increasing oxygen affinity caused by the single mutations [48]. Phenylalanine is rarely present at the B10 position, and, to our knowledge, it has been described in association with glutamine at E7 in L. pectinata HbI [41], U. caupo Hb [43], Caenorhabditis elegans Hb [49], Asian elephant Mb (Elephas maximus [50]) and African elephant Mb (Loxodonta africana [51]) only. The oxygen affinity of Asian elephant Mb does not differ from the typical vertebrate Mbs [52], and O. bicornis Mb falls at about mid range of the oxygen affinities reported for non-vertebrate Mbs [53].
The autoxidation rate constant of O. bicornis Mb measured at pH 7 and 37 °C was equal to 0.28 h−1, higher with respect to native sperm whale Mb (0.054 h−1) and its engineered mutant with glutamine at E7 and phenylalanine at B10 [53], but very similar to the single mutant with glutamine at E7 (0.24 h−1) [22]. It is worthwhile to outline that the autoxidation rate constants at neutral pH of other engineered Mb mutants, where distal histidine is replaced with glycine, valine, phenylalanine, cysteine, methionine, lysine, arginine, aspartate, threonine or tyrosine, were found to be increased 40–350 times [49].
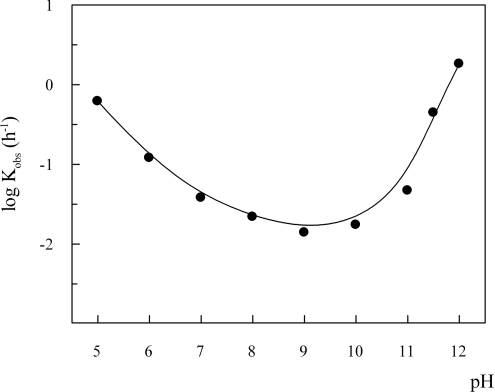
The stability profile of O. bicornis oxyMb (Figure 12) showed a minimum of the rate constants around pH 9 and increased values both at higher and lower pH. It was reported that the autoxidation rate of sperm whale Mb has a minimum at pH 9.2 [54], while different Mbs lacking the distal histidine residue (Aplysia kurodai [55] and Dolabella auricularia [56]) did not show any pH-dependence of the autoxidation rate above pH 8. On the contrary, the African elephant Mb was found to be as resistant to autoxidation as sperm whale MbO2 in the pH range 7–12 [50]. Engineered sperm whale Mb mutants with replaced distal histidine showed a reduced or null pH-dependence of autoxidation rate above pH 8–9 [57], being the E7 His→Gln mutant the only exception [51].
Figure 12. Dependence on pH of O. bicornis Mb autoxidation rate constant.
Measurements were performed at 10 μM Mb, in 0.1 M buffer under air-saturated conditions and in the presence of 1 mM EDTA and 3 mmol of catalase and superoxide dismutase per mole of haem, at 25 °C. Buffers used were: acetate (pH 5.0), Mes (pH 6.0), potassium phosphate (pH 7.0), Taps (pH 8.0), Caps (pH 9.0 and 10.0) and phosphate (pH 11.0, 11.5 and 12.0).
Structural aspects of the deduced amino acid sequence
The globin sequence of O. bicornis could be unambiguously aligned with sperm whale Mb, Ar. marina MbI and MbII [58], Aph. aculeata Mb [59], Ce. lacteus body wall and neural Hb [40] (Figure 13). The alignment was confirmed by: (i) the exclusion of polar residues from 28 out of 31 hydrophobic internal sites [23] (the residues at positions A4, A6 and A8 are lacking in O. bicornis); (ii) the alignment of proline C2 which determines the folding of the BC corner; (iii) the presence of glycine B6 essential for the near crossing of the B and E helices; and (iv) the presence of tryptophan H8, which is typical of invertebrate globins.
Figure 13. Alignment of O. bicornis Mb with selected globin sequences.
The top row of the alignment shows the helical notation (A–H) of sperm whale Mb (sw fold). Phys, Physeter catadon Mb; AremI, Ar. marina MbI; AremII, Ar. marina MbII; Aphr, Aph. aculeata Mb; Ophe, O. bicornis Mb; Cerb, Ce. lacteus body wall Hb; Cern, Ce. lacteus neural Hb; Equus, Equus caballus Mb; mini, miniMb from horse Mb.
The O. bicornis Mb alignment matched both non-vertebrate [23] and vertebrate [24] template II quite well, as shown by the low penalty scores reported in Table 1, indicating that all the major determinants of the globin fold are preserved. Comparison with other globin sequences revealed 25.2% identity with Ar. marina MbII, 23.9% with Ar. marina MbI and Aph. aculeata nerve Mb, and 20.4% with Ce. lacteus body wall mini-Hb.
Table 1. Penalty scores of O. bicornis Mb and some globin sequences against the non-vertebrate (NV) [23] and vertebrate (V) [24] globin templates.
Ophe, O. bicornis Mb; AremI, Ar. marina MbI; AremII, Ar. marina MbII; Aphr, Aph. aculeata Mb; Cerb, Ce. lacteus body wall Hb; Cern, Ce. lacteus neural Hb; Phys, Physeter catadon Mb.
| A-motif | BC-motif | E-motif | FG-motif | H-motif | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NV | V | NV | V | NV | V | NV | V | NV | V | NV | V | |
| Ophe | 0.0 | 0.0 | 0.0 | 0.7 | 0.2 | 0.7 | 0.0 | 1.2 | 0.5 | 0.5 | 0.7 | 3.1 |
| AremI | 0.0 | 1.5 | 0.0 | 1.2 | 0.7 | 0.5 | 0.0 | 2.4 | 0.0 | 0.0 | 0.7 | 5.6 |
| AremII | 0.0 | 1.0 | 0.0 | 1.2 | 0.7 | 0.5 | 0.0 | 2.4 | 0.0 | 0.0 | 0.7 | 5.1 |
| Aphr | 0.0 | 0.0 | 0.0 | 0.0 | 1.5 | 2.9 | 0.0 | 0.4 | 0.0 | 0.0 | 1.5 | 3.3 |
| Cerb | 0.5 | 1.5 | 0.0 | 0.7 | 1.4 | 2.3 | 0.7 | 0.9 | 0.2 | 0.0 | 2.8 | 5.4 |
| Cern | 0.5 | 1.5 | 0.0 | 0.7 | 1.4 | 2.3 | 0.7 | 0.9 | 0.0 | 0.0 | 2.6 | 5.4 |
| Phys | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.5 |
From the alignment of the globin sequences of Figure 13, it is also evident that O. bicornis Mb misses the pre-A helix and the first six residues of the A helix. This feature is in common with C. lacteus neural and body wall mini-Hbs, but, in these globins, three more residues of the A helix are missing, the B helix is seven residues shorter and the C-terminal of the H helix is 14 residues shorter. The mini-Mb obtained from horse heart by proteolytic digestion [60] lacks the pre-A and the A helix, almost the entire B helix (12 residues) and the C-terminal part of the H helix. Since Ce. lacteus Hbs are stable enough to allow the measurement of oxygen equilibria over several hours [40], while the mini-Mb from horse oxidizes rapidly in air [60], it has been hypothesized that the presence of part of the A helix plays an important role in stabilizing the globin towards autoxidation [40]. The results obtained on O. bicornis Mb, with regard to autoxidation rate, are in agreement with this hypothesis.
Acknowledgments
Dr Maria Cristina Gambi is gratefully acknowledged for O. bicornis classification. This work was supported by MIUR (Ministero dell'Istruzione, dell'Università e della Ricerca), CNR (Consiglio Nazionale delle Ricerche) and local universities funds.
References
- 1.Hoffmann R. J., Mangum C. P. The function of coelomic cell hemoglobin in the polychaete Glycera dibranchiata. Comp. Biochem. Physiol. 1970;36:211–228. doi: 10.1016/0010-406x(70)90001-0. [DOI] [PubMed] [Google Scholar]
- 2.Terwilliger R. C., Garlick R. L., Terwilliger N. B. Characterization of the hemoglobins and myoglobin of Travisia foetida. Comp. Biochem. Physiol. 1980;66B:261–266. [Google Scholar]
- 3.Weber R. E., Pauptit E. Molecular and functional heterogeneity in myoglobin from the polychaete Arenicola marina L. Arch. Biochem. Biophys. 1972;148:322–324. doi: 10.1016/0003-9861(72)90149-x. [DOI] [PubMed] [Google Scholar]
- 4.Terwilliger R. C., Garlick R. L., Terwilliger N. B. Hemoglobins of Glycera robusta: structures of coelomic cell hemoglobin and body wall myoglobin. Comp. Biochem. Physiol. 1976;54B:149–153. doi: 10.1016/0305-0491(76)90073-0. [DOI] [PubMed] [Google Scholar]
- 5.Wittenberg B. A., Briehl R. W., Wittenberg J. B. Haemoglobins of invertebrate tissues. Biochem. J. 1965;96:363–371. doi: 10.1042/bj0960363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takagi T., Cox J. A. Primary structure of myohemerythrin from the annelid Nereis diversicolor. FEBS Lett. 1991;285:25–27. doi: 10.1016/0014-5793(91)80716-g. [DOI] [PubMed] [Google Scholar]
- 7.Wells R. M., Dales R. P. Oxygenational properties of haemerythrin in the blood of Magelona papillicornis Muller (Polychaeta: Magelonidae) Comp. Biochem Physiol. 1974;49A:57–64. doi: 10.1016/0300-9629(74)90541-6. [DOI] [PubMed] [Google Scholar]
- 8.Chiancone E., Ascoli F., Giardina B., Vecchini P., Antonini E., Musmeci M. T., Cinà R., Zagra M., D'Amelio V., De Leo G. Physicochemical and functional properties of Perinereis cultrifera (Grube) erythrocruorin. Biochim. Biophys. Acta. 1977;494:1–8. doi: 10.1016/0005-2795(77)90129-5. [DOI] [PubMed] [Google Scholar]
- 9.Antonini E., Rossi-Fanelli A., Caputo A. Studies on chlorocruorin. I. The oxygen equilibrium of Spirographis chlorocruorin. Arch. Biochem. Biophys. 1962;97:336–342. doi: 10.1016/0003-9861(62)90086-3. [DOI] [PubMed] [Google Scholar]
- 10.Terwilliger R. C. The respiratory pigment of the serpulid polychaete, Serpula vermicularis L: structure of its chlorocruorin and hemoglobin (erythrocruorin) Comp. Biochem. Physiol. 1978;61B:463–469. [Google Scholar]
- 11.Lankester E. R. A contribution to the knowledge of haemoglobin. Proc. R. Soc. London Ser. B. 1872;21:70–80. [Google Scholar]
- 12.Garlick R. L., Terwilliger R. C. Structure and oxygen equilibrium of hemoglobin and myoglobin from the pacific lugworm Abarenicola pacifica. Comp. Biochem. Physiol. 1977;57B:177–184. [Google Scholar]
- 13.Fox H. M. Chlorocruorin and haemoglobin. Nature (London) 1947;160:825. doi: 10.1038/160825a0. [DOI] [PubMed] [Google Scholar]
- 14.Mezzasalma V., di Stefano L., Piazzese S., Zagra M., Salvato B., Tognon G., Ghiretti-Magaldi A. Physicochemical and structural properties of the extracellular hemoglobin of Ophelia bicornis. Biochim. Biophys. Acta. 1985;829:135–143. [Google Scholar]
- 15.Masala B., Manca L. Detection of the common Hb F Sardinia [Aγ(E19)Ile→Tyr] Clin. Chim. Acta. 1991;198:195–202. doi: 10.1016/0009-8981(91)90353-e. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli L. K. Cleavage of structural proteins during the assembly of the head of bacteriophage. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Rossi-Fanelli A., Antonini E., Caputo A. Pure native globin from human hemoglobin: preparation and some physico-chemical properties. Biochim. Biophys. Acta. 1958;28:221. doi: 10.1016/0006-3002(58)90462-1. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z., Marshall A. G. A universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. J. Am. Soc. Mass Spectrom. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 19.Giardina B., Amiconi G. Measurement of binding of gaseous and nongaseous ligands to hemoglobin by conventional spectrophotometric procedures. Methods Enzymol. 1981;76:417–427. doi: 10.1016/0076-6879(81)76133-0. [DOI] [PubMed] [Google Scholar]
- 20.Dolman D., Gill S. J. Membrane-covered thin layer optical cell for gasreaction studies of hemoglobin. Anal. Biochem. 1978;87:127–134. doi: 10.1016/0003-2697(78)90576-6. [DOI] [PubMed] [Google Scholar]
- 21.Wyman J. Linked functions and reciprocal effects in hemoglobin: a second look. Adv. Protein Chem. 1964;19:223–286. doi: 10.1016/s0065-3233(08)60190-4. [DOI] [PubMed] [Google Scholar]
- 22.Brantley R. E., Jr, Smerdon S. J., Wilkinson A. J., Singleton E. W., Olson J. S. The mechanism of autoxidation of myoglobin. J. Biol. Chem. 1993;268:6995–7010. [PubMed] [Google Scholar]
- 23.Moens L., Vanfleteren J., Van de Peer Y., Kapp O., Czeluzniak J., Goodman M., Blaxter M., Vinogradov S. Globins in nonvertebrate species: dispersal by horizontal gene transfer and evolution of the structure–function relationships. Mol. Biol. Evol. 1996;13:324–333. doi: 10.1093/oxfordjournals.molbev.a025592. [DOI] [PubMed] [Google Scholar]
- 24.Bashford D., Chlothia C., Lesk A. A. Determinants of a protein fold: unique features of the globin amino acid sequences. J. Mol. Biol. 1987;196:199–216. doi: 10.1016/0022-2836(87)90521-3. [DOI] [PubMed] [Google Scholar]
- 25.Cutruzzolà F., Travaglini-Allocatelli C., Brancaccio A., Brunori M. Aplysia limacina myoglobin cDNA cloning: an alternative mechanism of oxygen stabilization as studied by active-site mutagenesis. Biochem. J. 1996;314:83–90. doi: 10.1042/bj3140083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geraci G., Sada A., Cirotto C. Cooperative, low-molecular-weight dimeric myoglobins from the radular muscle of the gastropod mollusc Nassa mutabilis L. Eur. J. Biochem. 1977;77:555–560. doi: 10.1111/j.1432-1033.1977.tb11698.x. [DOI] [PubMed] [Google Scholar]
- 27.Rossi Fanelli M. R., Chiancone E., Vecchini P., Antonini E. Studies on erythrocruorin. I. Physicochemical properties of earthworm erythrocruorin. Arch. Biochem. Biophys. 1970;141:278–283. doi: 10.1016/0003-9861(70)90133-5. [DOI] [PubMed] [Google Scholar]
- 28.Shikama K., Matsuoka A. Spectral properties unique to the myoglobins lacking the usual distal histidine residue. J. Mol. Biol. 1989;209:489–491. doi: 10.1016/0022-2836(89)90012-0. [DOI] [PubMed] [Google Scholar]
- 29.Korenaga S., Igarashi J., Matsuoka A., Shikama K. A primitive myoglobin from Tetrahymena pyriformis: its heme environment, autoxidizability, and genomic DNA structure. Biochim. Biophys. Acta. 2000;1543:131–145. doi: 10.1016/s0167-4838(00)00187-4. [DOI] [PubMed] [Google Scholar]
- 30.Bartnicki D. E., Mizukami H., Romero-Herrera A. E. Interaction of ligands with the distal glutamine in elephant myoglobin. J. Biol. Chem. 1983;258:1599–1602. [PubMed] [Google Scholar]
- 31.Antonini E., Brunori M. The derivatives of ferric hemoglobin and myoglobin. In: Neuberger A., Tatum E. L., editors. Hemoglobin and Myoglobin in their Reactions with Ligands. Amsterdam: North-Holland Publishing; 1971. pp. 40–54. [Google Scholar]
- 32.Weber R. E., Vinogradov S. N. Nonvertebrate hemoglobins: functions and molecular adaptations. Physiol. Rev. 2001;81:569–628. doi: 10.1152/physrev.2001.81.2.569. [DOI] [PubMed] [Google Scholar]
- 33.Terwilliger R. C., Terwilliger N. Proboscis myoglobin of Glycera dibranchiata. Comp. Biochem. Physiol. 1981;70B:169–171. [Google Scholar]
- 34.Rossi-Fanelli A., Antonini E. Studies on the oxygen and carbon monoxide equilibria of human myoglobin. Arch. Biochem. Biophys. 1958;77:478–492. doi: 10.1016/0003-9861(58)90094-8. [DOI] [PubMed] [Google Scholar]
- 35.Maltagliati F., Casu M., Castelli A. Morphological and genetic evidence supports the existence of two species in the genus Ophelia (Annelida, Polychaeta) from the Western Mediterranean. Biol. J. Linn. Soc. 2004;83:101–113. [Google Scholar]
- 36.Giardina B., Ascenzi P., Clementi M. E., De Sanctis G., Rizzi M., Coletta M. Functional modulation by lactate of myoglobin. J. Biol. Chem. 1996;271:16999–17001. doi: 10.1074/jbc.271.29.16999. [DOI] [PubMed] [Google Scholar]
- 37.Rossi-Fanelli A., Antonini E., Povoledo D. Symposium on Protein Structure. In: Neuberger A., editor. London: Methuen; 1958. p. 140. [Google Scholar]
- 38.Dewilde S., Winnepenninckx B., Arndt M. H. L., Nascimento D. G., Santoro M. M., Knight M., Miller A. N., Kerlavage A. R., Geoghagen N., Van Marck E., et al. Characterization of the myoglobin and its coding gene of the mollusc Biomphalaria glabrata. J. Biol. Chem. 1998;273:13583–13592. doi: 10.1074/jbc.273.22.13583. [DOI] [PubMed] [Google Scholar]
- 39.Phillips S. E., Schoenborn B. P. Neutron diffraction reveals oxygen-histidine hydrogen bond in oxymyoglobin. Nature (London) 1981;292:81–82. doi: 10.1038/292081a0. [DOI] [PubMed] [Google Scholar]
- 40.Vandergon T. L., Riggs C. K., Gorr T. A., Colacino J. M., Riggs A. F. The mini-hemoglobins in neural and body wall tissue of the nemertean worm Cerebratulus lacteus. J. Biol. Chem. 1998;273:16998–17011. doi: 10.1074/jbc.273.27.16998. [DOI] [PubMed] [Google Scholar]
- 41.Antommattei-Perez F. M., Rosado-Ruiz T., Cavilla C. L., Lopez-Garriga J. The cDNA-derived amino acid sequence of hemoglobin I from Lucina pectinata. J. Protein Chem. 1999;18:831–836. doi: 10.1023/a:1020623011363. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki T., Kawamichi H., Ohtsuki R., Iwai M., Fujikura K. Isolation and cDNA-derived amino acid sequences of hemoglobin and myoglobin from the deep-sea clam Calyptogena kaikoi. Biochem. Biophys. Acta. 2000;1478:152–158. doi: 10.1016/s0167-4838(99)00210-1. [DOI] [PubMed] [Google Scholar]
- 43.Garey J. R., Riggs A. F. The hemoglobin of Urechis caupo: the cDNA-derived amino acid sequence. J. Biol. Chem. 1986;261:16446–16450. [PubMed] [Google Scholar]
- 44.Springer B. A., Egerberg K. D., Sligar S. G., Rohlfs R. J., Mathews A. J., Olson J. S. Discrimination between oxygen and carbon monoxide and inhibition of autoxidation by myoglobin. J. Biol. Chem. 1989;264:3057–3060. [PubMed] [Google Scholar]
- 45.De Baere I., Perutz M., Kiger L., Marden M. C., Poyart C. Formation of the two hydrogen bonds from the globin to the heme-linked oxygen molecule in Ascaris hemoglobin. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1594–1597. doi: 10.1073/pnas.91.4.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rashid A. K., Van Hauwaert M., Haque M., Siddiqi A. H., Lasters I., De Maeyer M., Griffon N., Marden M. C., Dewilde S., Clauwaert J., et al. Trematode myoglobins, functional molecules with distal tyrosine. J. Biol. Chem. 1997;272:2992–2999. doi: 10.1074/jbc.272.5.2992. [DOI] [PubMed] [Google Scholar]
- 47.Carver T. E., Brantley R. E., Singleton E. W., Arduini R. M., Quillin M. L., Phillips G. N., Jr, Olson J. S. A novel site-directed mutant of myoglobin with an unusual high O2 affinity and low autoxidation rate. J. Biol. Chem. 1992;267:14443–14450. [PubMed] [Google Scholar]
- 48.Zhao X., Vyas K., Bao D., Rajarathnam K., La Mar G. N., Li T., Phillips G. N., Jr, Eich R. F., Olson J. S., Ling J., Bocian D. A double mutant of sperm whale myoglobin mimics the structure and function of elephant myoglobin. J. Biol. Chem. 1995;270:20763–20774. doi: 10.1074/jbc.270.35.20763. [DOI] [PubMed] [Google Scholar]
- 49.Mansell J. B., Timms K., Tate W. P., Moens L., Trotman C. N. A. Expression of a globin gene in Caenorhabditis elegans. Biochem. Mol. Biol. Int. 1993;30:643–647. [PubMed] [Google Scholar]
- 50.Bisig D. A., Di Iorio E. E., Diederichs K., Winterhalter K. H., Piontek K. Crystal structure of Asian elephant (Elephas maximus) cyano-metmyoglobin at 1.78-Å resolution. Phe29(B10) accounts for its unusual ligand binding properties. J. Biol. Chem. 1995;270:20754–20762. doi: 10.1074/jbc.270.35.20754. [DOI] [PubMed] [Google Scholar]
- 51.Tada T., Watanabe Y., Matsuoka A., Ikeda-Saito M., Imai K., Ni-hei Y., Shikama K. African elephant myoglobin with an unusual autoxidation behaviour: comparison with the H64Q mutant of sperm whale myoglobin. Biochim. Biophys. Acta. 1998;1387:165–176. doi: 10.1016/s0167-4838(98)00118-6. [DOI] [PubMed] [Google Scholar]
- 52.Romero-Herrera A. E., Goodman M., Dene H., Bartnicki D. E., Mizukami K. An exceptional amino acid replacement on the distal side of the iron atom in proboscidean myoglobin. J. Mol. Evol. 1981;17:140–147. doi: 10.1007/BF01733907. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki T., Imai K. Evolution of myoglobin. Cell. Mol. Life Sci. 1998;54:979–1004. doi: 10.1007/s000180050227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brooks J. The oxidation of haemoglobin to methaemoglobin by oxygen. II. The relation between the rate of oxidation and the partial pressure of oxygen. Proc. R. Soc. London Ser. B. 1935;118:560–577. [Google Scholar]
- 55.Shikama K., Matsuoka A. Aplysia oxymyoglobin with an unusual stability property: kinetic analysis of the pH dependence. Biochemistry. 1986;25:3898–3903. [Google Scholar]
- 56.Suzuki T. Amino acid sequence of myoglobin from the mollusc Dolabella auricularia. J. Biol. Chem. 1986;261:3692–3699. [PubMed] [Google Scholar]
- 57.Suzuki T., Watanabe Y., Nagasawa M., Matsuoka A., Shikama K. Dual nature of the distal histidine residue in the autoxidation reaction of myoglobin and hemoglobin. Eur. J. Biochem. 2000;267:6166–6174. doi: 10.1046/j.1432-1327.2000.01685.x. [DOI] [PubMed] [Google Scholar]
- 58.Kleinschmidt T., Weber R. E. Primary structures of Arenicola marina isomyoglobins: molecular basis for functional heterogeneity. Biochim. Biophys. Acta. 1998;1383:55–62. doi: 10.1016/s0167-4838(97)00181-7. [DOI] [PubMed] [Google Scholar]
- 59.Dewilde S., Blaxter M., Van Hauwaert M., Vanfleteren J., Esmans E. L., Marden M., Griffon N., Moens L. Globin and globin gene structure of the nerve myoglobin of Aphrodite aculeata. J. Biol. Chem. 1996;271:19865–19870. doi: 10.1074/jbc.271.33.19865. [DOI] [PubMed] [Google Scholar]
- 60.De Sanctis G., Falcioni G., Giardina B., Ascoli F., Brunori M. Mini-myoglobin: the structural significance of haem–ligand interactions. J. Mol. Biol. 1988;200:725–733. doi: 10.1016/0022-2836(88)90483-4. [DOI] [PubMed] [Google Scholar]