Graphical abstract
Keywords: Siderophore, GNATs, SidF, X-ray crystallography, SAXS
Highlights
-
•
Siderophore iron acquisition is essential for the virulence of Aspergillus fumigatus.
-
•
SidF and SidL transacetylases generate siderophores intermediates in A. fumigatus.
-
•
SidF X-ray structure reveals a two-domain architecture with tetrameric assembly.
-
•
AlphaFold modelling demonstrates structural similarity between SidF and SidL.
-
•
SidF can utilize acetyl-CoA complementing transacetylase activity in SidL knockouts.
Abstract
Siderophore-mediated iron acquisition is essential for the virulence of Aspergillus fumigatus, a fungus causing life-threatening aspergillosis. Drugs targeting the siderophore biosynthetic pathway could help improve disease management. The transacetylases SidF and SidL generate intermediates for different siderophores in A. fumigatus. A. fumigatus has a yet unidentified transacetylase that complements SidL during iron deficiency in SidL-lacking mutants.
We present the first X-ray structure of SidF, revealing a two-domain architecture with tetrameric assembly. The N-terminal domain contributes to protein solubility and oligomerization, while the C-terminal domain containing the GCN5-related N-acetyltransferase (GNAT) motif is crucial for the enzymatic activity and mediates oligomer formation. Notably, AlphaFold modelling demonstrates structural similarity between SidF and SidL. Enzymatic assays showed that SidF can utilize acetyl-CoA as a donor, previously thought to be a substrate of SidL but not SidF, and selectively uses N5-hydroxy-L-ornithine as an acceptor.
This study elucidates the structure of SidF and reveals its role in siderophore biosynthesis. We propose SidF as the unknown transacetylase complementing SidL activity, highlighting its central role in A. fumigatus siderophore biosynthesis. Investigation of this uncharacterized GNAT protein enhances our understanding of fungal virulence and holds promise for its potential application in developing antifungal therapies.
Introduction
Aspergillus fumigatus is a ubiquitous saprophytic fungus living on organic debris. Nevertheless, it is also an opportunistic pathogen that can cause life-threatening invasive pulmonary aspergillosis (Latgé, 1999). While the incidence is higher among immunocompromised patients and those with underlying pulmonary diseases, the population at risk is steadily expanding, also due to SARS–CoV2 infections (Fisher et al., 2022, Janssen et al., 2021, Koehler et al., 2021). Additionally, evidence is emerging that resistance to triazole antifungals, the predominant class of drugs used to treat fungal diseases (Denning et al., 2016), is increasing worldwide (Fisher et al., 2022). This trend underscores the importance of monitoring Aspergillus as a potential drug-resistant threat (Cdc, 2019).
Iron, with its redox properties, is a vital cofactor for essential biochemical reactions in diverse life forms (Gutteridge and Halliwell, 2018). However, strict homeostasis is necessary to balance its potential toxicity (Anderson and Frazer, 2017, Gutteridge and Halliwell, 2018, Katsarou and Pantopoulos, 2020). Moreover, the low environmental bioavailability of iron in the environment, driven by its propensity to form insoluble ferric (Fe3+) hydroxides, necessitates specific acquisition mechanisms, particularly for pathogens exploiting host iron stores (Gerwien et al., 2018, Gutteridge and Halliwell, 2018). A. fumigatus employs two main iron uptake strategies: low-affinity ferrous (Fe2+) iron and high-affinity ferric iron uptake systems including siderophore-mediated iron acquisition (SIA) and reductive iron assimilation (RIA) (Arputhanantham et al., 2021, Misslinger et al., 2021). Siderophores are low molecular mass, ferric iron-specific chelators (Misslinger et al., 2021) and this research focuses on SIA, a key virulence factor during host infection (Aguiar et al., 2022, Hissen et al., 2005, Schrettl et al., 2004). A. fumigatus produces two fusarinine-type siderophores, termed fusarinine C (FsC) and triacetylfusarinine C (TAFC), and two ferrichrome-type siderophores, termed ferricrocin (FC) and hydroxyferricrocin (HFC) (Happacher et al., 2023, Misslinger et al., 2021). FsC, TAFC, and FC are secreted to capture environmental iron (Aguiar et al., 2022, Happacher et al., 2023, Misslinger et al., 2021, Schrettl et al., 2007); FC is also employed for intracellular iron handling within hyphae, and HFC is employed for conidial iron storage (Misslinger et al., 2021, Schrettl et al., 2007, Wallner et al., 2009).
Both fusarinine- and ferrichrome-type siderophores share the same starting point, the conversion of ornithine to N5-hydroxyornithine by the monooxygenase SidA (Schrettl et al., 2004), then the pathways diverge (Fig. 1). Ferrichrome biosynthesis utilizes the transacetylase SidL and an additional uncharacterized enzyme for N5-acetylation (Blatzer et al., 2011), followed by assembling FC from serine, glycine, and N5-acetyl-N5-hydroxyornithine, catalyzed by the nonribosomal peptide synthetase (NRPS) SidC (Schrettl et al., 2007). An unknown enzyme hydroxylates FC to hydroxyferricrocin. Fusarinine-type siderophore synthesis involves mevalonate conversion to anhydromevalonyl-CoA by the mevalonyl-CoA ligase SidI and the mevalonyl-CoA hydratase SidH (Yasmin et al., 2012). Anhydromevalonyl-CoA is transferred to N5-hydroxyornithine by the transacylase SidF to yield N5-anhydromevalonyl-N5-hydroxyornithine (Supplementary Fig. S1) (Schrettl et al., 2007). The NRPS SidD then links three N5-anhydromevalonyl-N5-hydroxyornithine units to form FsC, which SidG subsequently acetylates to yield TAFC (Gutteridge and Halliwell, 2018, Misslinger et al., 2021, Wang and Pantopoulos, 2011, Weinberg, 1975).
Fig. 1.
A. fumigatus siderophore biosynthetic pathway. Involved enzymes are boxed in blue and described in the text. Siderphores are boxed in brown. Red arrows denote SidF function identified in this study. Peroxisome and cytoplasm are shaded in purple and yellow, respectively. Adapted from “Plant cell with organelles”, by BioRender.com (2024). Retrieved from https://app.biorender.com/biorender-templates. Created with BioRender.com. / Mahidol University.
GCN5-related N-acetyltransferases (GNATs), an ubiquitous superfamily, encompasses enzymes across all life domains, crucial for diverse processes from bacterial antibiotic resistance to circadian rhythms (Vetting et al., 2005). Typically, the enzyme acylates the primary amine of an acceptor molecule using a donor molecule such as acetyl-CoA (AcCoA). Their conserved fold consists of β-sheets and α-helices (β1-H1-H2-β2-β3-β4-H3-β5-H4-β6) with a characteristic conserved V-shaped cleft for acyl-CoA binding (β4-β5) and a variable acceptor substrate binding site reflecting GNAT functional diversity (Ud-Din et al., 2016). The catalytic site is placed between the donor and acceptor sites (Baumgartner et al., 2021).
Although many GNATs have been extensively studied, most are still unexplored. Given the established role of siderophore biosynthesis as a crucial virulence factor in A. fumigatus, targeting enzymes within this pathway holds promise for novel antifungal drug development. SidF has unique characteristics and warrants further investigation, despite sharing functional similarities with SidL. This study integrates protein structural analysis with enzymatic assays to comprehensively characterize SidF.
Our study revealing the high-resolution structure and biochemical characterization of SidF enabled us to propose that SidF might complement SidL during iron starvation, opening new avenues for therapeutic intervention against A. fumigatus infections.
Materials and Methods
Plasmid Construction
A synthetic SidF DNA construct (UniProt Q4WF55), optimized for Escherichia coli expression, was obtained from Genscript and cloned into the NdeI/BamHI restriction sites of the pET-28a(+)-TEV vector. The recombinant protein was expressed with an N-terminal His-tag. The theoretical molecular weight and isoelectric point of SidF were calculated to be 55.7 kDa and 6.54, respectively. To generate N- and C-terminal deletion mutants, the “around the horn” PCR method (Moore and Prevelige, 2002) was employed using primers listed in Supplementary Table S1.
Protein expression and purification
SidF expression was induced by either autoinduction or 1 mM IPTG at 30 °C overnight in E. coli BL21(DE3). Lysis buffer was 50 mM Tris pH 8.0, 500 mM NaCl, 0.5 mM ethylenediaminetetraacetic acid (EDTA), 10 mM β-mercaptoethanol (BME), and 20 mM imidazole. Cell lysis was performed by sonication. The sample was centrifuged at 12000xg for 30 min at 4 °C and then purified by using Ni-NTA in 50 mM Tris pH 8, 200 mM NaCl, and 20 mM imidazole binding buffer. Then, washed and eluted with the same buffer containing 20 mM imidazole and 250 mM imidazole, respectively. Elution fraction was loaded onto a Superdex™ 200 Increase 10/300 GL Tricorn™ size-exclusion chromatography (SEC) column (GE HealthCare) equilibrated with 50 mM Tris pH 8.0, 200 mM NaCl at a flow rate of 0.5 mL·min−1. The peak at a retention volume of 12.18 mL was concentrated and used for crystallization.
SidF N-terminal domain and SidF C-trim were expressed and purified with the same procedure. Except that the SEC column for the N-terminal domain was Superdex™ 75 Increase 10/300 GL Tricorn™.
Crystallization, diffraction Experiment, data processing and model Building
SidF crystallization conditions were searched using commercial screens from Molecular Dimension and Hampton Research at a protein concentration of 10 mg/ml by the micro batch-under the volatile oil method at 20 °C. Crystals obtained from 0.2 M potassium thiocyanate, and 20 % polyethylene glycol (PEG) 3350 were used for diffraction experiments. The crystals were flash-frozen in liquid N2. Cryoprotection was achieved by supplementing the reservoir solution with 12 % glycerol. X-ray data were collected at a cryogenic temperature (100 K) at the ELETTRA 11.2C beamline, Elettra Synchrotron Trieste, Italy (Lausi et al., 2015).
Data were reduced by XDS (Kabsch, 2010) and scaled by Aimless (Evans and Murshudov, 2013). The predicted three-dimensional structure, generated by the ColabFold AlphaFold2 (Jumper et al., 2021, Mirdita et al., 2022), was used as a template for molecular replacement via MOLREP (Vagin and Teplyakov, 2010). The process succeeded with R/Rfree = 0.25/0.30. The structure was refined by REFMAC5 (Murshudov et al., 2011) and built in COOT (Emsley et al., 2010) in CCP4Cloud (Krissinel et al., 2022) or CCP4i2 (Potterton et al., 2018). Data collection and refinement statistics are shown in Table 1. Models were visualized using Chimera (Pettersen et al., 2004) and ChimeraX (Pettersen et al., 2021).
Table 1.
Diffraction data collection and refinement statistics.
| Data collection | SidF (7qf6) | SidF Nter (8kd8) |
|---|---|---|
| Space group | P 1 | C 2221 |
| Cell dimensions | ||
| a, b, c (Å) | 80.27 80.84 179.84 | 62.2 110.67 69.47 |
| α, β, γ(°) | 101.168 91.668 117.252 | 90 90 90 |
| Resolution (Å) | 174.82–1.87(1.90–1.87) | 29.42 – 2.58(2.70–2.58) |
| Rmerge Rpim |
0.061(0.459) 0.038(0.290) |
0.095(0.6) 0.04(0.255) |
| I/σI CC half |
17(3.6) 0.999(0.929) |
18.1(4.4) 0.998(0.942) |
| Completeness (%) Redundancy |
97.1 (96.2) 6.8(6.8) |
99.8(99.2) 12.1(12.3) |
| Refinement | ||
| Resolution (Å) | 174.82–1.87 | 29.42 – 2.58 |
| Total No. reflections | 4,925,967 | 95,327 |
| Rwork / Rfree | 0.17/0.21 | 0.17/ 0.24 |
| No. atoms Protein Ions ligands Waters B-factors Protein Ions ligands Waters |
30,901 28,812 2 114 1973 33.37 60.21 43.07 34.79 |
2712 2681 − − 31 53.88 − − 48.41 |
| R.M.S. deviations | ||
| Bond lengths (Å) | 0.015 | 0.013 |
| Bond angles (°) | 1.972 | 2.192 |
SidF N-terminal domain and SidF C-trim were purified as the full-length protein. SidF N-terminal crystals were obtained from 0.2 M potassium sodium tartrate tetrahydrate, 20 % w/v polyethylene glycol 3,350 and subsequently cryoprotected with 40 % Morpheus® precipitant mix 3. The structure was solved by molecular replacement using MrBUMP (Keegan and Winn, 2008). SidF C-trim crystals were obtained from 0.1 M Sodium cacodylate pH 6.5, 40 % v/v MPD, 5 % w/v PEG 8000 but no X-ray diffraction was observed. For SidF C-trim, X-ray diffraction experiment was performed at a cryogenic temperature at beamline P13 operated by EMBL Hamburg at the PETRA III storage ring (DESY, Hamburg, Germany) (Cianci et al., 2017).
Computational tools
A conserved domain search was performed using the National Center for Biotechnology Information (NCBI) tool (Lu et al., 2020) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The Basic Local Alignment Search Tool (BLAST) was used to compare protein sequences to sequence databases (Altschul et al., 1990). Protein structures database search was retrieved from the Protein Data Bank (PDB) (https://www.rcsb.org). Protein structure comparison was conducted using the PDBeFold service at European Bioinformatics Institute (Krissinel, 2007) (https://www.ebi.ac.uk/msd-srv/ssm), RCSB PDB tool (pairwise structure alignment) (https://www.rcsb.org/alignment), or US-align (multiple structure alignment) (Zhang et al., 2022) (https://zhanggroup.org/US-align/). Analysis of macromolecular interfaces was done using 'Protein Interfaces, Surfaces and Assemblies' service PISA at the European Bioinformatics Institute (Krissinel and Henrick, 2007) (https://www.ebi.ac.uk/pdbe/prot_int/pistart.html). The computed models of SidF and SidL were generated by the web-based service AlphaFold2 (40) (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb).
Small angle X-ray scattering (SAXS)
SAXS was measured using a laboratory SAXS instrument (SAXSpoint 2.0, Anton Paar) at Institute of Biotechnology of the Czech Academy of Sciences (IBT CAS) (Vestec, Czech Republic); the instrument was equipped with X-ray source MetalJet C2 (Excillum) and detector Eiger R 1 M. The X-ray wavelength was 1.34 Å.
SidF N-terminal domain and SidF C-terminal domain were measured using a batch mode: sample (5 mg/ml in 50 mM Tris-HCl, 200 mM NaCl pH 8) was loaded to thin wall quartz capillary (diameter of 1.5 mm) and exposed for 15 min with sample to detector distance of 571 mm. The corresponding buffer was measured under the same condition. Scattering empty capillaries and capillaries filled with water were measured for 5 min to perform absolute scale calibration.
SidF full-length was analyzed with SEC-SAXS mode: the SAXS system was connected to a fast protein liquid chromatography (FPLC) system AKTAGo (GE Healthcare. Samples were loaded onto a Superdex™ 200 Increase 10/300 GL (GE HealthCare) at 20 °C; the separation was performed with flow a rate of 0.7 ml/min which was lowered to 0.05 ml/min upon peak detection with absorbance at 280 nm in AktaGO. Data were collected at a distance of 791 mm. The measurement cell allows in-situ measurement of UV–Vis absorption spectra (CaryUV 60, Agilent technologies) which were continuously measured in a range from 200 nm to 400 nm with a repetition rate of 30 s.
Data processing was conducted in PRIMUS [41] and custom software. SEC-SAXS chromatographs were analyzed using RAW (Hopkins et al., 2017). The molecular mass was calculated from Bayesian Inference. Comparison of scattering profiles was done in CRYSOL [42] and FoXS [43]. EOM (Tria et al., 2015) was further used for validation of the flexible loops: 10,000 models were of the flexible N-terminus was generated using RANCH (Tria et al., 2015), and the most probable ensemble selected using GAJOE (Tria et al., 2015).Rigid body modelling model of C-trim was calculated using CORAL (Petoukhov et al., 2012). Essential SAXS data acquisition, sample details, data analysis, and modelling fitting are given in Supplementary Table S2.
For all three proteins (SidF full-length, N-terminal domain, and C-terminal domain), Guinier analysis revealed linear behavior in the low-q region (Fig. 3A inset) indicating the presence of monodisperse particles of similar size. We noted that SidF N-terminal domain and C-trim show some sign of protein aggregation (Supplementary Fig. S2). Dimensionless Kratky plot of all three protein show bell-shaped profile indicating folded protein. Pair distribution functions (P(r)) for all three proteins approached zero at the maximum intra-particle distance (dmax), confirming their folded and monodisperse nature. Moreover, the radius of gyration (Rg) and scattering intensity at zero scattering angle (I(0)) values obtained from Guinier and P(r) analyses were in good agreement for all three proteins (Supplementary Table S2).
Fig. 3.
SAXS results for SidF full-length (black), N-terminal domain (cyan) and C-trim (yellow). (A) I(q) versus q as log-linear plots with the inset showing the Guinier fits for estimation of Rg and I(0), with the thick lines defining the data range used for parameter estimation. (B) Dimensionless Kratky plots for the data in (A). (C) P(r) versus r profiles normalized to equal areas. (D-F) The comparative analysis of the calculated SAXS profiles of the SidF variant crystal structures and the experimental SAXS data. Experimental SAXS scattering curve is shown in scattering with error bars, while the calculated scattering profile from crystal structures are shown in different colors. Chi-square value is indicated. (D) CRYSOL-derived models for SidF Full-length monomer, dimers, and tetramer (PDB ID 7qf6) fitted to log(I) versus q. The scattering profile of SidF tetramer calculated with EOM shown in red. (E) FoXS-derived models for SidF N-terminal domain (PDB ID 8k8d) fitted to log(I) versus q. The scattering profile of SidF tetramer calculated with EOM shown in blue. (F) FoXS-derived models for SidF Full-length monomer, dimers, and tetramer (PDB ID 7qf6) fitted to log(I) versus q of SidF C-trim. The scattering profile of the ab-intio model SidF tetramer calculated with CORAL shown in red.
Broad-Substrate screen for Gcn5-Related N-acetyltransferases
The broad-substrate screen was adapted from M. Kuhn et al, 2013 (Kuhn et al., 2013). The experiments were conducted in conventional 96-well PCR plates or tubes in a reaction volume of 50 µL. The reaction consisted of 50 mM Tris pH 8.0, 0.5 mM AcCoA, and 5 mM substrates. A wide variety of acceptors were used including cadaverine, L-lysine, L-glutamic acid, glycine, kanamycin A, ampicillin, chloramphenicol, nicotinamide adenine dinucleotide (NAD), urea, guanidine HCl, L-ornithine, oxidized glutathione (GSSH), hydroxylamine, and N5-hydroxy-L-ornithine. Cadaverine was freshly prepared before use. The reaction was initiated by adding 0.06 µM freshly prepared enzyme diluted in 50 mM Tris pH 8.0 and 200 mM NaCl. The reaction was carried out for 8 min at 25 °C before being stopped with 50 µL of 100 mM Tris pH 8.0 and 6 M guanidine HCl. For free thiols detection, 200 µL of developer (0.2 mM 5,5′-dithiobis (2-nitrobenzoic acid) (DTNB), 100 mM Tris pH 8.0, and 1 mM EDTA) was immediately added and incubated for 10 min at room temperature. 290 µL of reaction was transferred to 96-well clear polystyrene flat-bottom plate and the absorbance was measured at 405 nm with 595 nm background subtraction in a Tecan Infinite F200 microplate reader. Each reaction had its control which was the identical condition without enzyme. All assays were performed in duplicate for each protein purification batch. A standard curve was created by preparing 2-fold dilutions of N-acetyl-L-cysteine ranging from 500 to 3.9 µM and measuring in duplicate for each independent assay.
Results and Discussion
SidF structural characterization
The SidF monomer comprises two domains, with the N-terminal domain resembling the C-terminal domain.
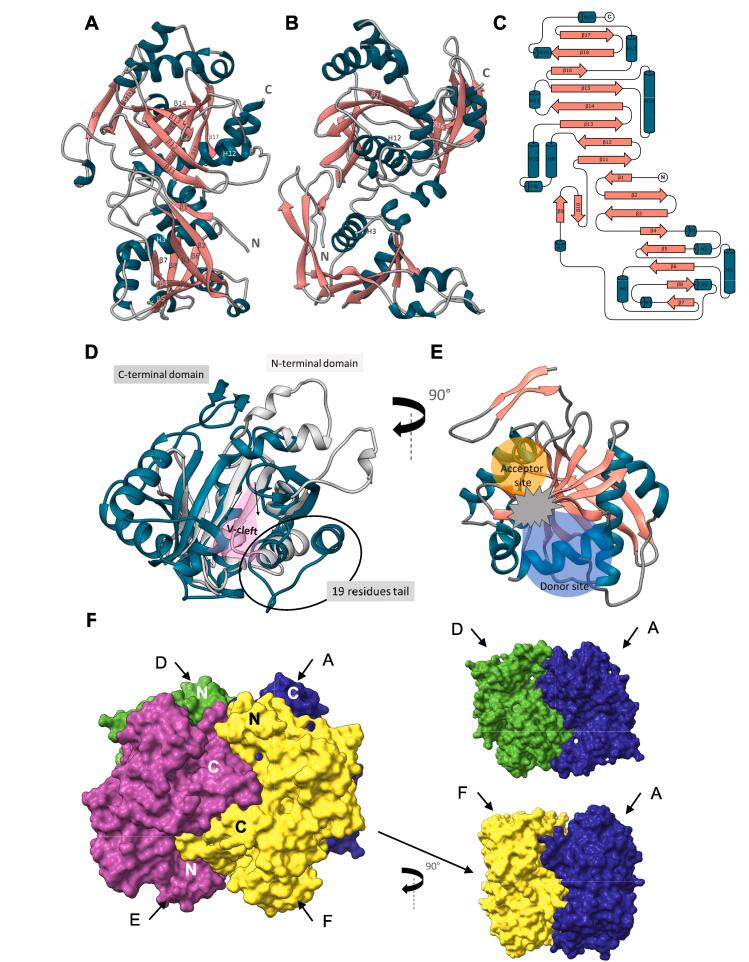
The SidF monomer contains 18 β-strands and 14 helices (both 3–10 and α-helix) divided into an N-terminal (residues 1–191) and a C-terminal domain (residues 227–462) (Fig. 2). There is no electron density for the His-tag and the residues 1–24. The SidF N-terminal domain is composed of 8 β-strands (1–8) that form a barrel-like structure, wrapping around helix 3 (H3) and fringed with loops and helixes between β-strands (Fig. 2). Similarly, the C-terminal domain shows a barrel-like structure containing 10 β-strands (11–18) wrapping around helix 11 (H11). The gaps of the half-barrel structure in each domain are facing each other. The two domains are connected by a long loop.
Fig. 2.
Crystal structures of SidF. (A) Ribbon representation of a SidF monomer (PDB entry 7qf6). Helices are colored green and labeled with “H”, β-strands are colored salmon and labeled with “β”, and loop regions are colored grey. (B) The same SidF monomer rotated 90° counter-clockwise along the vertical axis. (C) Topology diagram of SidF monomer generated with TopDraw [56]. β represent the β-strand and H represent helix. (D) Superposition of the N-terminal and C-terminal domains of SidF. The C-terminal domain (residues 200–462) is colored identically to Fig. 1A, while the N-terminal domain is shown in dark grey. The superposition highlights the absence of the V-cleft donor binding site in the N-terminal domain. The 19-residue C-terminal tail is circled. (E) The C-terminal domain of SidF rotated 90° counter-clockwise along the vertical axis. This view reveals the characteristic GNAT fold with the putative acceptor, donor, and catalytic sites clearly visible. (F) Surface representation of the SidF tetramer colored uniquely by chain. The right panel depicts the arrangement of A:D and A:F dimers within a tetramer.
Searching the PDB with the search term “Gcn5-related N-acetyltransferases,” gives 105 protein structures representative of each UniProt accession with an Enzyme Commission number (EC number) 2.3.1.- (accessed on 23 October 2023). Of these, 73.3 % are small (calculated mass 12.8–31.8 kDa) and contain only the GNAT motif. SidF has a mass of about 53.3 kDa, larger than typical GNAT proteins, and consists of two structural similar N- and C-terminal domains. It was previously reported that the GNAT N-myristoyl transferase monomer harbors two GNAT domains with an internal two-fold symmetry. Despite the poor sequence identity of the two domains, this arrangement was proposed to be the outcome of gene duplication (Weston et al., 1998). This could possibly be the case for SidF.
However, the amino acid sequence search using NCBI conserved domain tools identified the conserved GNAT motif only in the SidF C-terminal domain and not in the N-terminal domain. The protein BLAST (BLASTP) identified many homologs of SidF with coverage of 93–100 % encoded by several other Aspergillus spp. and various ascomycetes. A Position-Specific Iterated BLAST (PSI-BLAST) search identified a wider range of distantly related homologs from ascomycetes and γ-proteobacteria, albeit with reduced coverage (40–87 %). These lower-coverage homologs aligned primarily to the C-terminal region of SidF, encompassing the conserved GNAT domain. Moreover, the N-terminus of SidF is not conserved outside the phylum Ascomycota.
By using PDBeFold (accessed on 4 January 2023), the structure similarity search agreed with the sequence search, and the conserved acetyltransferase at the C-terminal domain of SidF matched with several acetyltransferase proteins. Superimposition of SidF C-terminal domain onto DfoC (Salomone-Stagni et al., 2018) from Erwinia amylovora (PDB ID: 5O7O) yielded a low sequence identity (30 %) but a high structural conservation (RMSD 1.274 Å). Similar results were obtained when comparing SidF C-terminal domain to human N-alpha-acetyltransferase 80 (Rebowski et al., 2020) (PDB ID: 6NBE, 12 % identity, 2.66 Å RMSD). In another search, the N-terminal domain alone gave the similar results, suggesting a similarity between the two domains of SidF.
To further explore the similarity between the N- and C-terminal domains, the SidF model was bisected at residue 200 and the two domains were superimposed (Fig. 2D). Pairwise structure alignment showed an RMSD of 4.28 and a Template Modeling (TM)-score of 0.44, indicating that the two protein structures compared are different both locally and globally. Though the comparison gave a TM-score below the cut-off of 0.5, we noted that the N-terminal domain is not completely dissimilar to the C-terminal domain. The main difference was the absence of the substrate-binding V-cleft in the SidF N-terminal domain (Fig. 2D), which explains the absence of the GNAT motif in the N-terminal domain and the poor structure alignment score.
SidF is observed as a unique tetramer in the crystal structure and in solution
Numerous enzymes within the GNAT superfamily are known to exist as oligomers. While the majority of GNATs are typically observed in dimeric states, it's worth mentioning that examples of monomeric, trimeric, tetrameric, hexameric, and even dodecameric configurations have also been documented (Ud-Din et al., 2016). Moreover, the GNAT domain can also be part of a large protein with multiple domains (Salomone-Stagni et al., 2018). The structure of SidF differs from that of other GNAT proteins, which form dimers from each small monomer with a continuous β-sheet (Vetting et al., 2005), an inserted β-sheet (Magalhaes et al., 2008), or a helical bundle at the dimer interface (Shi et al., 2014). The SidF monomer has two similar (TM score 0.44) independent folding units in the N- and C-terminal halves which are connected by a long loop (Fig. 2). Eight chains (two tetramers) of SidF are observed in the asymmetric unit of the P1 symmetry crystal at a maximum resolution of 1.87 Å. The SidF tetramer exhibits an antiparallel arrangement, wherein the N-terminus of one chain interacts with the C-terminus of its adjacent chain (Fig. 2F). The observed inter-chain interactions of A: D and E: F encompass both N- to C'-terminal and C- to C'-terminal domain pairings. According to PISA, SidF chains A and D engage extensively, forming a dimer with 43 hydrogen bonds and 31 salt bridges, yielding a high Complex Formation Significance Score (CSS) of 0.946 (Krissinel and Henrick, 2007). Similarly, SidF chains E and F create another dimer, featuring 43 hydrogen bonds and 29 salt bridges, with a CSS of 0.946. The interaction extends further as the adjacent dimers link chains A to F and D to E to form a tetramer. SidF chains A and F are stabilized by 31 hydrogen bonds, 15 salt bridges, with a CSS of 0.727. Additionally, SidF chains D to E form an interaction characterized by 33 hydrogen bonds, 16 salt bridges, with a CSS of 0.727. The predominant inter-chain interactions of A: F and D: E observed involve the N-to C’-terminus of an adjacent chain, with a minor contribution from C- to C’-terminus interactions. These inter-chain interfaces collectively contribute to the robustness and stability of the tetramer assembly (no interaction between chain A: E and D: F) (Fig. 2F). However, the tetramer-tetramer interaction is not significant with a CSS value of 0. The SidF molecular mass of 157 kDa was estimated by SEC corresponds to a lighter mass than the tetramer, as compared to standard proteins.
SAXS analysis was used to confirm the tetrameric nature of SidF in solution (Fig. 3A and Supplementary Table S2). The Rg determined through Guinier analysis was 40.49 ± 0.26 Å. Dimensionless Kratky plot displays the expected globular protein bell-shaped curve, with a slight shift to the right and a maximum of 1.17 at qRg = 1.80 (Fig. 3B). The P(r) versus r profile shows a bell shape and a long tail suggesting a dominant compact globular domain with the presence of an extended or flexible region (Fig. 3C). The P(r) function also revealing a dmax of 190 Å. The molecular mass estimation for SidF, derived from Bayesian Inference, yielded a value of 208 kDa corresponding to SidF tetrameric oligomerization. Nevertheless, the maximum dimension does not align with the measurement observed in the X-ray structure, which is only 97.9 Å (Supplementary Fig. S3). The 1–24 N-terminal residues are disordered as suggested by the low Alphafold model confidence score and confirmed by the X-ray structure observation. Together with evidence from SAXS, variations in the measured dmax values may influenced by these flexible terminal residues.
The comparative analysis involved aligning the SAXS profiles derived from the SidF crystal structure with the experimental SAXS data for SidF (Fig. 3D). The tetrameric SidF structure exhibited a satisfactory fit to the experimental data, yielding a chi-square value of 2.7 and ruling out the possibility of monomeric or dimeric arrangements. EOM was used to take the flexible loop into account, which resulted in improving the fit to the chi-square of 1.093, where the compact core is identical with the crystal structure.
Structure analysis of SidF N-terminal domain
The full length of the SidF X-ray structure shows discrete N- and C-terminal domains. The two domains are connected by a loop and 12 hydrogen bonds, with no part of the N-terminal domain intervening in the C-terminal domain. To further explore the function of SidF N- and C-terminal domains, the SidF full-length was divided into two domains at residue 200 (Supplementary Fig. S4), and each one was expressed and purified under the conditions used for the full-length protein. SidF N-terminus (residue 1–199) was successfully expressed and purified, while the expressed SidF C-terminal domain (residue 200–462) was insoluble. Both protein constructs were expressed with an N-terminal His-tag.
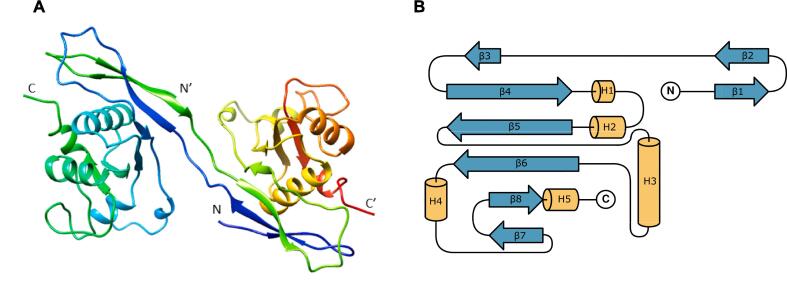
The crystal structure of the SidF N-terminal domain was solved as a monomer in C2221 space group at a maximum resolution of 2.58 Å. Its monomer displayed the same topology observed in the full-length protein, except that the residues 25–49 extended outward instead of being tucked in (Fig. 4). The N-terminal His-tag and the residues 1–24 were not visible in the electron density maps. Upon generating symmetry mates, the extended N-terminal region interacted with the corresponding region from another monomer, resulting in a stable dimer with a CSS score of 1 (according to PISA).
Fig. 4.
(A) Ribbon representation of the SidF N-terminal domain dimer generated from crystal symmetry (PDB ID 8kd8). (B) Topology diagram of SidF N-terminal domain monomer generated with TopDraw (Bond, 2003). β represent the β-strand and H represent helix.
While several nearby symmetry mates were produced, no interactions were predicted (CSS score = 0), indicating a lack of stability and suggesting the possibility of crystal contact artifacts rather than biological contacts. Besides, the protein was eluted from size exclusion chromatography column with approximate mass of 53.84 kDa supporting the formation of a stable dimer.
To confirm the dimeric nature of the SidF N-terminal domain, SAXS investigations were conducted (Supplementary Table S2). Unexpectedly, SAXS analysis did not support dimer conformation. The Guinier analysis determined the Rg of 23.23 ± 0.36 Å. Dimensionless Kratky plot exhibited a slight rightward shift from the ideal globular protein profile, suggesting a slightly elongated shape with a maximum value of 1.28 at qRg = 1.98 (Fig. 3B). Moreover, the plot was noisier compared to the other two proteins and showed early sign of flexibility (high value at high q region) at qRg > 3 potentially due to the low sample concentration. Even though, it was run at 5 mg/ml same as SidF C-trim, the SidF N-terminal domain is much smaller, thus giving less signal. Some degree of observed flexibility of the N-terminal domain might suggest the need for interaction may be from the C-terminal domain to stabilize the structure.The P(r) curve revealed characteristics resembling a prolate ellipsoid particle, with a dmax of 89 Å. Bayesian Inference estimated the molecular mass as 21.2 kDa, corresponding to the monomeric state, which contradicts the dimeric arrangement observed in the crystal structure.
Comparing the calculated SAXS profiles from the dimeric crystal structure of the SidF N-terminal domain to the SAXS experimental data (Fig. 3E) resulted in a poor fit with a chi-square value of 5.67. However, using the monomer structure significantly improved the fit, yielding a chi-square value of 2.02. Further refinement using the EOM led to a further improvement in the fit, with a chi-square value of 1.168.
PISA analysis of the SidF structure indicates that the N-terminal domain possesses a propensity to exist as a monomer in solution. The formation of the X-ray quaternary structure is driven by hydrogen bonding interactions of the C-terminal domain, with the N-terminal domain potentially contributing to inter-chain interactions with the C-terminal domain of other chains. These findings suggest that the N-terminal domain alone lacks the essential structural elements required for stable dimer or tetramer formation in solution.
In the crystal structure, the phenomenon characterized by the partial exchange of N-terminal residues between monomers, resulting in the formation of a continuous β-sheet, bears resemblance to the established dimerization interface observed in typical GNAT family proteins (Ud-Din et al., 2016). However, the biological significance of this dimeric form remains uncertain, as it lacks evidence from the SAXS analysis. The observed N-terminal crystallographic dimerization of SidF might be attributed to crystal packing. The flexibility of the N-terminal residues could also influence the protein's conformation in solution. Further studies are needed to experimentally validate these findings.
Structure analysis of SidF C-trim
Attempts to isolate the C-terminal domain in a soluble form were unsuccessful. To improve C-terminal solubility, we employed a domain truncation strategy targeting predicted disordered regions at both the N- and C-termini of the C-terminal domain, using structural data from full length SidF crystal structure and AlphaFold modelling (Supplementary Fig. S4). Despite attempts to generate soluble C-terminal truncates for further characterization, none were successful. Notably, a construct encompassing the full-length protein with a C-terminal truncation of 19 amino acids (residues 1–443) (Fig. 2D and Supplementary Fig. S4), designated SidF C-trim, remained soluble.
The SidF C-trim domain displayed comparable protein expression levels to the full-length protein. SEC analysis indicated a molecular mass of approximately 206.25 kDa, consistent with the full-length protein, suggesting a potential tetrameric quaternary structure. However, subsequent SAXS data challenged this hypothesis (Supplementary Table S2). Please note that, although the SidF C-trim was successfully crystallized, it unfortunately failed to diffract X-rays.
SAXS analysis revealed that the SidF C-trim exists as a dimer in solution. Guinier analysis yielded an Rg of 36.81 ± 0.31 Å. Dimensionless Kratky plot showed a right shift with a maximum value of 1.24 at qRg = 1.95 (Fig. 3B) exhibiting an extended characteristic. The P(r) function showed a similar profile to the full-length protein with a slight early peak and a long tail indicating a prolate ellipsoidal particle with the presence of an extended region with a dmax of 150 Å (Fig. 3C). Bayesian Inference estimated the molecular mass of the C-trim construct to be 109.1 kDa, consistent with dimeric oligomerization.
Comparative analysis of the experimental SAXS data with the calculated dimeric forms extracted from the SidF full-length X-ray structure AD and AF dimers yielded chi-square values of 13.04 and 13.54, respectively (Fig. 3F). These relatively high chi-square values suggest a potential discrepancy between the solution-state conformation of the C-trim dimer and the dimeric interfaces observed in the crystal structure. Employing CORAL, which combines rigid body modelling with ab-intio modelling, resulted in a new arrangement of the SidF dimer with chi-square values of 1.87 (Fig. 3F and Supplementary Fig. S5).
Analysis of the full-length SidF crystal structure revealed the presence of 14 hydrogen bonds within the 19C-terminal residues of the AD dimer interface, whereas no such hydrogen bonds were observed in the equivalent region of the AF dimer. This suggests that the C-terminal 19 residues may be crucial for the formation of the AD dimer. However, when allow the flexibility of N- and C-terminal domains using CORAL, a new arrangement of the SidF C-trim dimer generate more complex interaction different from the crystal structure (Supplementary Fig. S5).
While the exact function of the N-terminal domain remains unidentified, its role in maintaining the solubility of the whole protein is plausible. The SidF N-terminal domain may be essential for the stability of the C-terminal domain, which is crucial for tetramer formation. The 19-residue tail likely contributes to tetramer formation through interactions between dimers.
SidF has most likely N5-acetyl-N5-hydroxy-L-ornithine transacetylase activity
In vitro analysis of SidF’s donor substrate specificity
According to the Uniprot database, A. fumigatus (ATCC MYA-4609) has 92 N-acetyltransferases (NAT) (EC number 2.3.1.-) proteins that belong to different protein families (pfam) (Table S3). Notably, SidF and SidL are the only two Aspergillus GNATs classified within the pfam13523, which exhibits a broad taxonomic distribution, predominantly bacterial (86.9 %) with a smaller fungal representation (12.2 %). Despite sharing the same functional class, SidF and SidL display significant differences (Blatzer et al., 2011, Misslinger et al., 2021). The SidF encoding gene is on chromosome 3 clustered with other siderophore biosynthesis genes, while the SidL encoding gene is on chromosome 1. SidF, is localized in the peroxisome and coregulated with other siderophore genes by transcription factor SreA in response to iron levels, distinct from cytosolic SidL, whose regulation appears to be largely independent of iron availability or SreA (Blatzer et al., 2011, Gründlinger et al., 2013). Interestingly, A. fumigatus possesses an unidentified N5-acetyl-N5-hydroxy-L-ornithine transacetylase activity that complements SidL function during iron limitation, although its molecular identity remains unknown (Blatzer et al., 2011).
Although SidF and SidL are different in many aspects such as gene location, gene regulation, and protein subcellular location, a BLASTP sequences search demonstrated significant similarity between SidF and SidL, with an alignment score of 155, 56 % coverage, 31.63 % percent identity, and an E-value of 5 x 10-46 (Supplementary Fig. S6). The distribution of the Blast hits was on the C-terminal half of the protein. Furthermore, SidF and SidL belong to the same Pfam family, suggesting that they share similar protein folding and functional characteristics. SidF catalyzes the transfer of an anhydromevalonyl group from anhydromevalonyl-CoA to the N5-hydroxyornithine primary amine. Similarly, SidL transfer an acetyl group from acetyl-CoA to the N5-hydroxyornithine primary amine. Structural modeling using AlphaFold revealed a high degree of similarity between SidL and SidF, with an RMSD of 3.14 Å, a reference coverage of 74 %, and a TM-score of 0.76 despite only 29 % sequence identity (Fig. 5A).
Fig. 5.
(A) Superposition of the SidF crystal structure and SidL AlphaFold model.
To evaluate functional similarity, in vitro assays were performed to assess the acetyltransferase activity of SidF. The assays utilized acetyl-CoA, the preferred donor substrate for SidL, in conjunction with N5-hydroxyornithine as the acceptor. Acetyl transfer was monitored using the DTNB assay, which detects exposed thiol groups generated during the reaction. The findings confirmed that SidF can utilize acetyl-CoA as a donor substrate (Fig. 5B and Table 2), leading to the hypothesis that SidF may compensate for SidL (Fig. 1) in previous knockout experiments (Blatzer et al., 2011). Nevertheless, further studies are required to elucidate the mechanism underlying this compensatory activity in vivo.
Table 2.
Broad-Substrate Screen for acetyltransferase function using AcCoA as a donor.
| Acceptor substrate | SidF variant | 1Activity (µmol/min*mg) | p-values | |
|---|---|---|---|---|
| 2Own background | 3SidF WT | |||
| Cadaverine | SidF WT | 0.650 (0.745) | 0.123 | 1.644E-07 |
| L-Lysine | −0.008 (0.010) | 0.148 | 5.447E-07 | |
| L-Glutamic acid | −0.017 (0.024) | 0.186 | 5.360E-07 | |
| L-Ornithine | −0.018 (0.015) | 0.050 | 5.381E-07 | |
| Glycine | 0.013 (0.107) | 0.797 | 4.821E-07 | |
| Kanamycin A | 0.003 (0.140) | 0.970 | 4.303E-07 | |
| Ampicilin | 0.025 (0.039) | 0.221 | 5.549E-07 | |
| Chloramphenicol | 0.012 (0.043) | 0.561 | 5.445E-07 | |
| NAD | −0.035 (0.032) | 0.070 | 5.231E-07 | |
| Urea | −0.015 (0.040) | 0.459 | 5.304E-07 | |
| Guanidine | −0.005 (0.013) | 0.395 | 5.460E-07 | |
| GSSH | −0.024 (0.028) | 0.130 | 5.310E-07 | |
| Hydroxylamine | 0.476 (0.235) | 0.011 | 5.084E-07 | |
| N(5)-Hydroxy-L-ornithine | 7.03 (1.47) | 5.504E-07 | − | |
| N(5)-Hydroxy-L-ornithine | SidF N-ter | −0.014 (0.054) | 0.660 | 5.371E-07 |
| N(5)-Hydroxy-L-ornithine | SidF C-trim | 2.15 (0.613) | 0.001 | 2.123E-06 |
Note1Standard deviation; 2One-Sample t-Test; 3Welch's t-test (two-sample t-test with unequal variances); NAD: Nicotinamide Adenine Dinucleotide; GSSH: Oxidized Glutathione. Data points represent the mean of n = 5 replicates, except for SidF WT tested with N(5)-Hydroxy-L-ornithine, where n = 9.
Functional characterization of SidF
GNAT Broad-Substrate screen
GNAT family enzymes exhibit a generally conserved donor site and a more variable acceptor site. This versatility allows them to acetylate diverse metabolites, antibiotics, and proteins, potentially leading to multiple functionalities (Kuhn et al., 2013). To evaluate the substrate range of SidF, a DTNB assay was employed with AcCoA as the donor substrate and a variety of potential acceptor substrates (Fig. 5B and Table 2). SidF exhibited no activity on L-ornithine, but activity on the natural substrate N5-hydroxy-L-ornithine notably increased to 7.03 ± 1.47 µmol/min*mg (p-value < 0.001). The sole distinction between L-ornithine and N5-hydroxy-L-ornithine is the presence of the hydroxyl group, which suggests that the hydroxyl group may play a significant role in substrate recognition. To further investigate, hydroxylamine, which contains a hydroxyl group (OH) and an ammonia group (NH2), was used to assess the impact of the hydroxyl group. SidF demonstrated the ability to process hydroxylamine, but with a relatively low activity of 0.476 ± 0.235 µmol/min*mg (p-value = 0.011). This suggests that the hydroxyl group influences SidF acceptor recognition to a limited extent. SidF may employ a substrate recognition mode analogous to MbtK/IucB-like proteins (InterPro entry IPR019432), which 82.6 % of the proteins of this group are bacterial and specifically recognize hydroxyl groups (Card et al., 2005). Cadaverine showed a detectable signal as acceptor substrate (0.65 ± 0.745 µmol/min*mg), but the high standard deviation rendered the difference from background non-significant. Conversely, SidF did not utilize lysine, glutamic acid, ornithine, glycine, kanamycin A, ampicillin, chloramphenicol, NAD, urea, guanidine, or GSSH as acceptor substrates, highlighting its substrate specificity.
Functional characterization of SidF N-terminal domain and C-trim
DTNB assay employing AcCoA as the donor substrate and N5-hydroxyornithine acceptor was used to assess the enzymatic activity of the SidF N-terminal domain and SidF C-trim. The N-terminal domain exhibited no detectable activity compared to the background control (p-value = 0.660) (Table 2). This observation aligns with the predicted absence of enzymatic activity within the N-terminus due to the lack of the conserved GNAT motif.
The dimeric form of SidF C-trim, displayed a significant reduction (69.95 %) in activity compared to the full-length protein (p-value < 0.001) (Table 2). This observation underscores the importance of tetramerization for optimal catalytic efficiency of SidF. Furthermore, the C-terminal tail’s proximity to the acyl-CoA binding site (Fig. 2D) suggests a potential regulatory role by influencing the accessibility of the donor substrate.
Conclusions
This study presents a comprehensive investigation of the structure and function of A. fumigatus SidF. X-ray crystallography, SAXS analysis, and enzyme activity assay support the biological relevance of the tetrameric assembly of the full-length protein. The SidF subunits possess a repetitive structure, resulting in a size larger than that typically observed for GNAT family enzymes. This characteristic is common to homologs from other Aspergillus species and numerous other ascomycetes. Notably, SidF structure represents the first structural characterization of this group. The absence of GNAT activity in the SidF N-terminal domain, consistent with the lack of a conserved GNAT motif, raises questions about its functional necessity. Our findings suggest that this region might be essential for solubility and proper tetramer assembly of SidF. The C-terminal domain is essential for tetramer formation with its 19-residue tail which may specifically contribute to the dimerization. Our analysis further suggests that SidF recognizes the hydroxyl group in the acceptor substrate. Based on structural similarities and shared donor substrate utilization verified in this study, SidF is proposed to be a functional counterpart of SidL supporting the production of both fusarinine- and ferrichrome-type siderophores.
Our results provide insights into the structure of SidF and propose a novel function for SidF in siderophore biosynthesis. The study of this uncharacterized GNAT protein enhances our understanding of A. fumigatus siderophore biosynthesis and has promising implications for disease treatment.
CRediT authorship contribution statement
Thanalai Poonsiri: Writing – review & editing, Writing – original draft, Methodology, Investigation, Conceptualization. Jan Stransky: Writing – review & editing, Methodology, Investigation. Nicola Demitri: Writing – review & editing, Methodology, Investigation. Hubertus Haas: Writing – review & editing, Supervision, Conceptualization. Michele Cianci: Writing – review & editing, Methodology, Investigation. Stefano Benini: Writing – review & editing, Supervision, Methodology, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We gratefully acknowledge Elettra and XRD2 beamline for providing beamtime and support under proposal 0220565. CIISB, Instruct-CZ Centre of Instruct-ERIC EU consortium, funded by MEYS CR infrastructure project LM2023042 and European Regional Development Fund-Project „UP CIISB“ (No. CZ.02.1.01/0.0/0.0/18_046/0015974), is gratefully acknowledged for the financial support of the measurements at the CF Diffraction techniques. Instruct/MOSBRI visit For MOSBRI: This project has received funding from the European Unions Horizon 2020 Research and Innovation Program under grant no. 101004806, MOSBRI-2021-28. We extend our sincere gratitude to the staff of the IBT CAS, particularly to Dr. Tatsiana Chernovets, for their invaluable cooperation and assistance during the MOSBRI visit.
Funding
This work was supported by IPN95 project funded by the EGCT within the 3rd call for the Euregio Fund for Scientific Research. The publication of this work was supported by the Open Access Publishing Fund of the Free University of Bozen-Bolzano.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.yjsbx.2024.100119.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Aguiar M., Orasch T., Shadkchan Y., Caballero P., Pfister J., Sastré-Velásquez L.E., Gsaller F., Decristoforo C., Osherov N., Haas H. Uptake of the Siderophore Triacetylfusarinine c, but Not Fusarinine c, Is Crucial for Virulence of Aspergillus Fumigatus. 2022;mBio 13 doi: 10.1128/mbio.02192-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson G.J., Frazer D.M. Current understanding of iron homeostasis. Am J Clin Nutr. 2017;106:1559S–S1566. doi: 10.3945/ajcn.117.155804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arputhanantham, S.S., Raja, K., Shanmugam, L., Raman, V., 2021. Basics of Fungal Siderophores: Classification, Iron Transport and Storage, Chemistry and Biosynthesis, Application, and More.
- Baumgartner, J.T., Habeeb Mohammad, T.S., Czub, M.P., Majorek, K.A., Arolli, X., Variot, C., Anonick, M., Minor, W., Ballicora, M.A., Becker, D.P., Kuhn, M.L., 2021. Gcn5-Related N-Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too. Frontiers in Molecular Biosciences 8. [DOI] [PMC free article] [PubMed]
- Blatzer M., Schrettl M., Sarg B., Lindner H.H., Pfaller K., Haas H. SidL, an Aspergillus fumigatus transacetylase involved in biosynthesis of the siderophores ferricrocin and hydroxyferricrocin. Applied and Environmental Microbiology. 2011;77:4959–4966. doi: 10.1128/AEM.00182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond C.S. TopDraw: a sketchpad for protein structure topology cartoons. Bioinformatics. 2003;19:311–312. doi: 10.1093/bioinformatics/19.2.311. [DOI] [PubMed] [Google Scholar]
- Card G.L., Peterson N.A., Smith C.A., Rupp B., Schick B.M., Baker E.N. The crystal structure of Rv1347c, a putative antibiotic resistance protein from Mycobacterium tuberculosis, reveals a GCN5-related fold and suggests an alternative function in siderophore biosynthesis. Journal of Biological Chemistry. 2005;280:13978–13986. doi: 10.1074/jbc.M413904200. [DOI] [PubMed] [Google Scholar]
- Cdc. 2019. Antibiotic resistance threats in the United States, 2019, Atlanta, Georgia.
- Cianci M., Bourenkov G., Pompidor G., Karpics I., Kallio J., Bento I., Roessle M., Cipriani F., Fiedler S., Schneider T.R. P13, the EMBL macromolecular crystallography beamline at the low-emittance PETRA III ring for high- and low-energy phasing with variable beam focusing. J Synchrotron Radiat. 2017;24:323–332. doi: 10.1107/S1600577516016465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D.W., Cadranel J., Beigelman-Aubry C., Ader F., Chakrabarti A., Blot S., Ullmann A.J., Dimopoulos G., Lange C. Chronic pulmonary aspergillosis: Rationale and clinical guidelines for diagnosis and management. European Respiratory Journal 47. 2016 doi: 10.1183/13993003.00583-2015. [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, M.C., Alastruey-Izquierdo, A., Berman, J., Bicanic, T., Bignell, E.M., Bowyer, P., Bromley, M., Brüggemann, R., Garber, G., Cornely, O.A., Gurr, S.J., Harrison, T.S., Kuijper, E., Rhodes, J., Sheppard, D.C., Warris, A., White, P.L., Xu, J., Zwaan, B., Verweij, P.E. 2022. Tackling the emerging threat of antifungal resistance to human health, pp. 557-571 Nature Reviews Microbiology, Vol. 20. Nature Research. [DOI] [PMC free article] [PubMed]
- Gerwien, F., Skrahina, V., Kasper, L., Hube, B., Brunke, S. 2018. Metals in fungal virulence, pp. 1-21 FEMS Microbiology Reviews, Vol. 42. Oxford University Press. [DOI] [PMC free article] [PubMed]
- Gründlinger M., Yasmin S., Lechner B.E., Geley S., Schrettl M., Hynes M., Haas H. Fungal siderophore biosynthesis is partially localized in peroxisomes. Molecular Microbiology. 2013;88:862–875. doi: 10.1111/mmi.12225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge, J.M.C., Halliwell, B. 2018. Mini-Review: Oxidative stress, redox stress or redox success?, pp. 183-186 Biochemical and Biophysical Research Communications, Vol. 502. Elsevier B.V. [DOI] [PubMed]
- Happacher I., Aguiar M., Alilou M., Abt B., Baltussen T.J.H., Decristoforo C., Melchers W.J.G., Haas H. The Siderophore Ferricrocin Mediates Iron Acquisition in Aspergillus fumigatus. Microbiol Spectr. 2023;11:e0049623. doi: 10.1128/spectrum.00496-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hissen A.H.T., Wan A.N.C., Warwas M.L., Pinto L.J., Moore M.M. Infection and Immunity 73. 2005. The Aspergillus fumigatus siderophore biosynthetic gene sidA, encoding L-ornithine N5-oxygenase, is required for virulence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins J.B., Gillilan R.E., Skou S. BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J Appl Crystallogr. 2017;50:1545–1553. doi: 10.1107/S1600576717011438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen, N.A.F., Nyga, R., Vanderbeke, L., Jacobs, C., Ergün, M., Buil, J.B., Van Dijk, K., Altenburg, J., Bouman, C.S.C., Van der Spoel, H.I., Rijnders, B.J.A., Dunbar, A., Schouten, J.A., Lagrou, K., Bourgeois, M., Reynders, M., Van Regenmortel, N., Rutsaert, L., Lormans, P., Feys, S., Debavaye, Y., Tamion, F., Costa, D., Maizel, J., Dupont, H., Chouaki, T., Nseir, S., Sendid, B., Brüggemann, R.J.M., Van de Veerdonk, F.L., Wauters, J., Verweij, P.E., 2021. Multinational observational cohort study of covid-19-associated pulmonary aspergillosis1. Emerging Infectious Diseases 27. [DOI] [PMC free article] [PubMed]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsarou A., Pantopoulos K. Vol. 75. Elsevier Ltd.; 2020. (Basics and Principles of Cellular and Systemic Iron Homeostasis Molecular Aspects of Medicine). [DOI] [PubMed] [Google Scholar]
- Keegan R.M., Winn M.D. MrBUMP: an automated pipeline for molecular replacement. Acta Crystallogr D Biol Crystallogr. 2008;64:119–124. doi: 10.1107/S0907444907037195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler P., Bassetti M., Chakrabarti A., Chen S.C.A., Colombo A.L., Hoenigl M., Klimko N., Lass-Flörl C., Oladele R.O., Vinh D.C., Zhu L.P., Böll B., Brüggemann R., Gangneux J.P., Perfect J.R., Patterson T.F., Persigehl T., Meis J.F., Ostrosky-Zeichner L., White P.L., Verweij P.E., Cornely O.A. Defining and managing COVID-19-associated pulmonary aspergillosis: the 2020 ECMM/ISHAM consensus criteria for research and clinical guidance. The Lancet Infectious Diseases. 2021;Vol. 21 doi: 10.1016/S1473-3099(20)30847-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E. On the relationship between sequence and structure similarities in proteomics. Bioinformatics. 2007;23:717–723. doi: 10.1093/bioinformatics/btm006. [DOI] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Krissinel E., Lebedev A.A., Uski V., Ballard C.B., Keegan R.M., Kovalevskiy O., Nicholls R.A., Pannu N.S., Skubak P., Berrisford J., Fando M., Lohkamp B., Wojdyr M., Simpkin A.J., Thomas J.M.H., Oliver C., Vonrhein C., Chojnowski G., Basle A., Purkiss A., Isupov M.N., McNicholas S., Lowe E., Trivino J., Cowtan K., Agirre J., Rigden D.J., Uson I., Lamzin V., Tews I., Bricogne G., Leslie A.G.W., Brown D.G. CCP4 Cloud for structure determination and project management in macromolecular crystallography. Acta Crystallogr D Struct Biol. 2022;78:1079–1089. doi: 10.1107/S2059798322007987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M.L., Majorek K.A., Minor W., Anderson W.F. Broad-substrate screen as a tool to identify substrates for bacterial Gcn5-related N-acetyltransferases with unknown substrate specificity. Protein Science. 2013;22:222–230. doi: 10.1002/pro.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latgé J.P. Aspergillus fumigatus and Aspergillosis. Clinical Microbiology Reviews. 1999;Vol. 12 doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lausi A., Polentarutti M., Onesti S., Plaisier J.R., Busetto E., Bais G., Barba L., Cassetta A., Campi G., Lamba D., Pifferi A., Mande S.C., Sarma D.D., Sharma S.M., Paolucci G. Status of the crystallography beamlines at Elettra. The European Physical Journal plus. 2015;130:43. [Google Scholar]
- Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Marchler G.H., Song J.S., Thanki N., Yamashita R.A., Yang M., Zhang D., Zheng C., Lanczycki C.J., Marchler-Bauer A. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48:D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes M.L., Vetting M.W., Gao F., Freiburger L., Auclair K., Blanchard J.S. Kinetic and structural analysis of bisubstrate inhibition of the Salmonella enterica aminoglycoside 6'-N-acetyltransferase. Biochemistry. 2008;47:579–584. doi: 10.1021/bi701957c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M., Schutze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misslinger, M., Hortschansky, P., Brakhage, A.A., Haas, H. 2021. Fungal iron homeostasis with a focus on Aspergillus fumigatus Biochimica et Biophysica Acta - Molecular Cell Research, Vol. 1868. Elsevier B.V. [DOI] [PubMed]
- Moore S.D., Prevelige P.E., Jr. A P22 scaffold protein mutation increases the robustness of head assembly in the presence of excess portal protein. J. Virol. 2002 doi: 10.1128/JVI.76.20.10245-10255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M., Gorba C., Mertens H.D., Konarev P.V., Svergun D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Science. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton L., Agirre J., Ballard C., Cowtan K., Dodson E., Evans P.R., Jenkins H.T., Keegan R., Krissinel E., Stevenson K., Lebedev A., McNicholas S.J., Nicholls R.A., Noble M., Pannu N.S., Roth C., Sheldrick G., Skubak P., Turkenburg J., Uski V., von Delft F., Waterman D., Wilson K., Winn M., Wojdyr M. CCP4i2: the new graphical user interface to the CCP4 program suite. Acta Crystallogr D Struct Biol. 2018;74:68–84. doi: 10.1107/S2059798317016035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebowski G., Boczkowska M., Drazic A., Ree R., Goris M., Arnesen T., Dominguez R. Mechanism of actin N-terminal acetylation. Sci Adv. 2020;6:eaay8793. doi: 10.1126/sciadv.aay8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomone-Stagni M., Bartho J.D., Polsinelli I., Bellini D., Walsh M.A., Demitri N., Benini S. A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora. J Struct Biol. 2018;202:236–249. doi: 10.1016/j.jsb.2018.02.002. [DOI] [PubMed] [Google Scholar]
- Schrettl M., Bignell E., Kragl C., Joechl C., Rogers T., Arst H.N., Haynes K., Haas H. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. Journal of Experimental Medicine. 2004;200 doi: 10.1084/jem.20041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., Bignell E., Kragl C., Sabiha Y., Loss O., Eisendle M., Wallner A., Arst H.N., Haynes K., Haas H. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathogens. 2007;3:1195–1207. doi: 10.1371/journal.ppat.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S., Lin J., Cai Y., Yu J., Hong H., Ji K., Downey J.S., Lu X., Chen R., Han J., Han A. Dimeric structure of p300/CBP associated factor. BMC Struct Biol. 2014;14:2. doi: 10.1186/1472-6807-14-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tria G., Mertens H.D., Kachala M., Svergun D.I. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ. 2015;2:207–217. doi: 10.1107/S205225251500202X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ud-Din A.I.M.S., Tikhomirova A., Roujeinikova A. Structure and functional diversity of GCN5-related n-acetyltransferases (GNAT) International Journal of Molecular Sciences. 2016;17 doi: 10.3390/ijms17071018. MDPI AG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A., Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- Vetting, M.W., Luiz, L.P., Yu, M., Hegde, S.S., Magnet, S., Roderick, S.L., Blanchard, J.S. 2005. Structure and functions of the GNAT superfamily of acetyltransferases, pp. 212-226 Archives of Biochemistry and Biophysics, Vol. 433. [DOI] [PubMed]
- Wallner A., Blatzer M., Schrettl M., Sarg B., Lindner H., Haas H. Ferricrocin, a siderophore involved in intra- and transcellular iron distribution in Aspergillus fumigatus. Appl Environ Microbiol. 2009;75:4194–4196. doi: 10.1128/AEM.00479-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Pantopoulos K. Regulation of cellular iron metabolism. Biochemical Journal. 2011;434:365–381. doi: 10.1042/BJ20101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg E.D. Nutritional immunity. Host's attempt to withold iron from microbial invaders. JAMA. 1975;231:39–41. doi: 10.1001/jama.231.1.39. [DOI] [PubMed] [Google Scholar]
- Weston S.A., Camble R., Colls J., Rosenbrock G., Taylor I., Egerton M., Tucker A.D., Tunnicliffe A., Mistry A., Mancia F., de la Fortelle E., Irwin J., Bricogne G., Pauptit R.A. Crystal structure of the anti-fungal target N-myristoyl transferase. Nat Struct Biol. 1998;5:213–221. doi: 10.1038/nsb0398-213. [DOI] [PubMed] [Google Scholar]
- Yasmin, S., Alcazar-Fuoli, L., Grun̈dlinger, M., Puempel, T., Cairns, T., Blatzer, M., Lopez, J.F., Grimalt, J.O., Bignell, E., Haas, H., 2012. Mevalonate governs interdependency of ergosterol and siderophore biosyntheses in the fungal pathogen Aspergillus fumigatus. Proceedings of the National Academy of Sciences of the United States of America 109. [DOI] [PMC free article] [PubMed]
- Zhang C., Shine M., Pyle A.M., Zhang Y. US-align: universal structure alignments of proteins, nucleic acids, and macromolecular complexes. Nature Methods. 2022;19:1109–1115. doi: 10.1038/s41592-022-01585-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.