Abstract
Uracil phosphoribosyltransferase (UPRT) is a member of a large family of salvage and biosynthetic enzymes, the phosphoribosyltransferases, and catalyzes the transfer of ribose 5-phosphate from α-d-5-phosphoribosyl-1-pyrophosphate (PRPP) to the N1 nitrogen of uracil. The UPRT from the opportunistic pathogen Toxoplasma gondii represents a promising target for rational drug design, because it can create intracellular, lethal nucleotides from subversive substrates. However, the development of such compounds requires a detailed understanding of the catalytic mechanism. Toward this end we determined the crystal structure of the T. gondii UPRT bound to uracil and cPRPP, a nonhydrolyzable PRPP analogue, to 2.5-Å resolution. The structure suggests that the catalytic mechanism is substrate-assisted, and a tetramer would be the more active oligomeric form of the enzyme. Subsequent biochemical studies revealed that GTP binding, which has been suggested to play a role in catalysis by other UPRTs, causes a 6-fold activation of the T. gondii enzyme and strikingly stabilizes the tetramer form. The basis for stabilization was revealed in the 2.45-Å resolution structure of the UPRT–GTP complex, whereby residues from three subunits contributed to GTP binding. Thus, our studies reveal an allosteric mechanism involving nucleotide stabilization of a more active, higher order oligomer. Such regulation of UPRT could play a role in the balance of purine and pyrimidine nucleotide pools in the cell.
Toxoplasma gondii is a ubiquitous protozoan parasite that infects approximately one third of Americans and up to 90% of certain European countries. T. gondii is the causative agent of congenital toxoplasmosis, the leading cause of neurological birth defects, and toxoplasmic encephalitis, a devastating opportunistic infection of people with acquired immunodeficiency syndrome (1, 2). The current drugs used against T. gondii are not ideal, and therapeutic intervention and prophylaxis must often be aborted because of toxic side effects (3). Thus, the need for better drugs is acute. The T. gondii pyrimidine salvage enzyme uracil phosphoribosyltransferase (UPRT), which is absent in humans, offers a promising target for the design of specific antitoxoplasmal subversive substrates, and several uracil analogues have been identified as substrates of the enzyme (4). One analogue, 5-fluorouracil, is effective in eradicating T. gondii in vivo, a process that depends on its UPRT activity (5). However, 5-fluorouracil is also toxic to mammalian cells, thus limiting its usefulness. The effective design of more selective prodrugs that use the T. gondii UPRT depends on a detailed understanding of the structure and catalytic mechanism of this enzyme.
T. gondii UPRT (EC 2.4.2.9) is a 244-aa protein (molecular mass, 27 kDa; ref. 6) that belongs to the PRT family of enzymes. Members of this large family carry out the biosynthesis and salvage of pyrimidines, pyridines, and purines as well as the synthesis of tryptophan and histidine in lower eukaryotes and bacteria (7). These enzymes catalyze chemically similar reactions involving phosphoribosyl transfer from the substrate α-D-5-phosphoribosyl-1-pyrophosphate (PRPP) to an acceptor molecule with inversion of configuration about the ribose C1′ atom. For UPRT, the PRPP ribosyl phosphate group is transferred to the N1 nitrogen of uracil to form UMP and PPi (Fig. 1A). The Km values are 3.5 μM for uracil and 243 μM for PRPP (6). The three-dimensional structures of several PRTs (8–28) including the T. gondii UPRT (12) have been determined and reveal two structural classes (8). Most PRTs, including UPRT, belong to the “class I” family, which shares a core region that consists of a four- or five-stranded parallel β-sheet surrounded by three α-helices.
Figure 1.
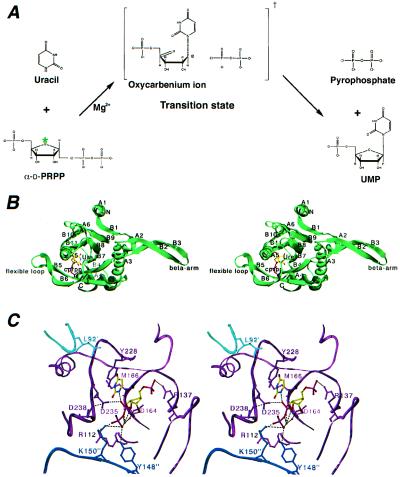
The UPRT–uracil–1-α-pyrophosphoryl-2-α,3-α-dihydroxy-4-β-cyclopentanemethanol-5-phosphate (cPRPP) structure. (A) Chemical reaction catalyzed by UPRT. The green asterisk denotes the oxygen atom that is replaced by a carbon in the cPRPP analogue. (B) Stereo view of the UPRT monomer with bound uracil and cPRPP. The secondary structural elements are labeled as in ref. 12. Uracil (Ura) and cPRPP are depicted as balls and sticks. This protomer is shown in the same orientation as the dark blue subunit in Fig. 5B. This and Fig. 4A were generated with SWISS-PDBVIEWER (47) and rendered in POVRAY (www.povray.org; ref. 48). (C) Stereo view of the active site of the UPRT–uracil–cPRPP structure. Uracil and cPRPP are shown as balls and sticks, and solvent molecules, Mg2+ and Wat-1, are shown as green and red spheres. This and Fig. 4B were made with O (37).
UPRT has been identified and characterized to varying extents in a variety of organisms including Escherichia coli, thermophilic bacteria, Saccharomyces cerevisiae, archea, and a number of pathogenic bacteria and protozoa (29–33). The sequences of these enzymes are fairly dissimilar with identities ranging from 20 to 45%, but there is strong conservation of residues in the active site pocket (12). Most of these enzymes are homodimers including the Acholeplasma laidlawii (30), Crithidia luciliae (29), and Giardia intestinalis (33) UPRTs, whereas the E. coli (31) and S. cerevisiae (32) enzymes seem to be a homotrimer and heterooligomer, respectively.
Kinetic studies have shown that many UPRTs, including the E. coli and Sulfolobus shibatae enzymes, are activated by GTP (29, 31–34). How GTP regulates these enzymes is unknown, although it does affect the oligomerization state of the E. coli enzyme (31). Also uncertain is the catalytic mechanism of UPRT. Studies on the E. coli UPRT suggest a sequential mechanism with PRPP binding before uracil, whereas the A. laidlawii, S. cerevisiae, and G. intestinalis enzymes seem to use a random order reaction mechanism (30, 32–33). A random order mechanism for the T. gondii enzyme is supported by the structure of its UPRT–uracil complex, which reveals a complete binding site for the base in the absence of PRPP (12). The structure also demonstrated that a dimer is essential for uracil binding. A puzzling feature of this structure was the presence of a second dimer that led to the creation of a dimer of dimers, the functional significance of which was unclear.
To understand more fully the catalytic mechanism of the T. gondii UPRT, we determined the crystal structure of this enzyme bound to uracil and a nonhydrolyzable PRPP analogue, cPRPP. Further biochemical and structural experiments were carried out to address the role of GTP in catalysis by this enzyme and revealed its allosteric mechanism.
Materials and Methods
Mutagenesis, Expression, and Purification of T. gondii UPRT.
For mutagenesis, the UPRT cDNA containing the C128V mutation that is a requisite for structural studies was isolated as described (6, 12). New mutations verified by automated sequencing were introduced by using the Gene Editor site-directed mutagenesis system. The mutants were transformed into JM109 E. coli, expression was induced at 37°C for 16 h, and all proteins were purified as described (6).
Enzyme Assays.
Radiometric assays for determining UPRT activity were performed essentially as described (6). In experiments that studied the kinetic effects of GTP, 2 mM GTP was included in the assay buffer. All reagents were purchased from Sigma unless otherwise indicated. 2-14C-Uracil [54 mCi⋅mmol−1 (1 Ci = 37 GBq)] and [γ-32P]GTP (30 Ci·mmol) were purchased from DuPont-NEN. DE-53 anion exchange matrix was obtained from Whatman.
Ligand Binding Studies.
GTP binding to UPRT was determined by using a variation of the nitrocellulose filtration method (35). [γ-32P]GTP was diluted with nonlabeled GTP (final specific activity of ≈12,000 cpm/nmol) and mixed with UPRT (≈120 nM) in solution A (50 mM Tris⋅HCl, pH 7.5/10 mM MgCl2/5 mM DTT/1 mM EDTA/0.1% BSA). The mixture was incubated at 4°C for 30 min. Then 10-μl aliquots were withdrawn and filtered rapidly through 0.45-μm nitrocellulose filter discs. The filters were washed with 20 ml of solution A and allowed to dry for 1 h at 27°C. Retained filter radioactivity was quantitated by liquid scintillation counting and used to calculate values for bound GTP. Total GTP was obtained from counting unfiltered 10-μl samples. Free GTP was calculated from the difference between total and bound GTP. Kd values for GTP binding were determined by Scatchard analysis using ENZFITTER.
UPRT Sedimentation Studies.
UPRT sedimentation properties in the presence or absence of GTP were determined by using sucrose density gradients (5–25%, 13 ml total volume) formed in Ultra-Clear tubes in 50 mM Tris⋅HCl/10 mM MgCl2/10 mM DTT/2 mM PRPP. GTP was included in gradients as indicated. Samples (200 μl) containing UPRT and BSA (at concentrations of ≈100 μM) were loaded on top of the gradients. The samples were centrifuged at 39,000 rpm for 30 h in a Beckman SW41 rotor. After completion, 200-μl fractions were collected from the bottom of the gradients, and samples from each fraction were run on 15% Tris-N-tris(hydroxymethyl)glycine gels. Gels were stained with Coomassie brilliant blue R-250 dye. Distribution of UPRT and the marker proteins along the gradients was quantified by using a Bio-Rad GS-700 imaging densitometer.
Crystallization and Data Collection.
The UPRT–uracil–cPRPP complex was obtained by adding 10 mM cPRPP and saturating amounts of uracil to protein at 20 mg/ml. Crystals of the ternary complex were grown via hanging drop-vapor diffusion by mixing equal volumes of the complex with 0.2 M NaCl/15 mM sodium citrate/phosphate, pH 4.7/10% polyethylene glycol 3400. The crystals are monoclinic, space group P21, with the unit cell dimensions a = 71.4 Å, b = 111.4 Å, c = 71.9 Å, and β = 97.7°. The UPRT–GTP complex was obtained by either soaking apo UPRT crystals (12) or cocrystallization with 1 mM GTP. The crystals are monoclinic, space group P21, with the unit cell dimensions a = 60.6 Å, b = 141.8 Å, c = 71.4 Å, and β = 115.0°. X-ray intensity data were collected at room temperature with an R-AXIS IV imaging plate system and processed with BIOTEX. Selected statistics are presented in Table 1 for each complex.
Table 1.
Crystallographic data
| Structure | UPRT–uracil–cPRPP | UPRT–GTP |
|---|---|---|
| Date collection | ||
| Resolution range, Å | 60.0–2.50 | 16.1–2.45 |
| Highest resolution range, Å | 2.66–2.50 | 2.60–2.40 |
| Unique/total reflections, no. | 32,692/99,823 | 34,646/77,473 |
| Completeness, % | 84.6 | 86.6 |
| I/(σ)I | 13.5 (1.5)* | 10.6 (3.8) |
| Rsym† | 10.1 (26.5) | 3.9 (14.3) |
| Refinement | ||
| Resolution range, Å | 60.0–2.50 | 16.1–2.45 |
| Rwork, %‡ | 22.5 | 18.4 |
| Rfree, %§ | 26.5 | 24.2 |
| rms deviations | ||
| Bond lengths, Å | 0.017 | 0.007 |
| Bond angles, ° | 2.18 | 1.50 |
| B value, Å2 | 1.5 | 2.1 |
| Ave B, Å2 | 58.1 | 25.1 |
Data for high resolution shell in parentheses.
Rsym = ∑/Io − 〈I〉/Io, where Io = observed intensity, and 〈I〉 = average intensity obtained from multiple observations of symmetry-related reflections.
Rwork = ∑∥Fobs| − |Fcalc∥/∑|Fobs|.
Rfree = ∑∥Fobs| − |Fcalc∥/∑|Fobs|, where all reflections belong to a test set of 10% randomly selected data.
Structure Determination and Refinement.
The tetrameric T. gondii UPRT–uracil–cPRPP structure was solved by molecular replacement (36) using the T. gondii apo UPRT dimer structure as a search model. Initial difference Fourier maps revealed a uracil in each of the four subunits. Only one subunit showed unambiguous density for a cPRPP molecule, which along with four uracil molecules was built into the electron density (37). The structure was subjected to simulated annealing and positional and thermal parameter refinement by using CNS (38). The Rwork is 22.5% to 2.5-Å resolution, and the Rfree is 26.5% (Table 1). The structure includes residues 21–244 of the subunit with bound uracil and cPRPP, residues 21–244 of a second subunit, residues 21–136 and 142–244 of the other two subunits, 9 phosphates, 4 uracils, 1 cPRPP, and 117 water molecules. PROCHECK (39) reveals 597 (77.6%) residues in the most favored regions of the Ramachandran plot, 146 (19.0%) in additionally allowed regions, 17 (2.2%) in generously allowed regions, and 9 (1.2%) in disallowed regions. The outlier residues include Arg-112 from each subunit, which plays a role in PRPP binding, and flexible loop residues.
The UPRT–GTP tetramer was solved by molecular modification using the apo UPRT structure as the starting model. Excellent density for the four GTP molecules was evident after rigid body refinement (37). The coordinates were refined by a combination of simulated annealing/positional/thermal parameter refinement (38). After several rounds of refinement, the density for N-terminal residues 10–21 became clear. These residues were added, and the structure was refined to an Rwork of 18.4% and Rfree of 24.2% (Table 1). The structure consists of residues 10–244 of each subunit, 8 phosphates (in the active site pockets), 169 solvent atoms, and 4 GTPs. PROCHECK reveals 726 (86.8%) residues in the most favored regions of the Ramachandran plot, 106 (12.7%) in additionally favored regions, none in generously allowed regions, and 4 (0.5%) outliers, again residue Arg-112 from each subunit. The coordinates and structure factors for the UPRT–GTP and UPRT–uracil–cPRPP complexes have been deposited in the RCSB database under the accession codes 1JLR and 1JLS, respectively.
Dynamic Light Scattering.
Dynamic light-scattering experiments were done using a 2001 DynaPro dynamic light-scattering instrument and UPRT at a concentration of 1–3 mg/ml in 20 mM Tris, pH 7.5/1 mM DTT (buffer A). All solutions were filtered through 0.1-μm Anotop filters (Whatman) to remove particulates. Multiple runs were made in which 20–40 scans of 30 s each were taken at 22°C. The data were analyzed by the DYNAMICS 3.30 software. The UPRT-alone sample was fit using a monomodal analysis, giving a molecular mass of 63 ± 5 kDa, concordant with a dimer. To test the effect of the phosphate addition, sodium/potassium phosphate, pH 7.5, was added to buffer A such that its final concentration was 150 mM. Analysis of these data revealed a UPRT molecular mass of 62 ± 7 kDa, again consistent with a dimer. When GTP was added to buffer A to a final concentration of 1 mM, the resultant molecular mass was 110 ± 10 kDa, i.e., a tetramer. Previous dynamic light-scattering experiments on UPRT at 6 mg/ml (or higher) in the presence of 1 M ammonium sulfate resulted in molecular masses of 108 ± 20 kDa, indicating that UPRT can form tetramers at very high concentrations.
Results and Discussion
UPRT Interactions with Uracil and cPRPP.
The structure of the UPRT–uracil–cPRPP complex reveals the same tetramer (a dimer of dimers) as taken by apo UPRT (12). Each subunit contains a “core” region (A1-B1-A2-A3-B4-A4-B5-B6-B7-A5-B8-A6-B9, where As are α-helices and Bs are β-strands) and a “hood” (B10-B11-A7), which provides residues critical for pyrimidine binding and specificity (Fig. 1B). The structure also contains an antiparallel β-arm (B2-B3) inserted within the canonical PRT fold between A3 and B4. This β-arm forms part of the dimerization interface and caps the hood of the uracil binding site, preventing access to bulk solvent.
In the UPRT–uracil complex uracil and cPRPP are juxtaposed (Fig. 1C). The uracil is bound as described previously (12) in a cleft that is formed by residues from the PRPP binding motif and the hood (Fig. 1C). Specifically, the side chains of the residues Met-166 and Tyr-228 sandwich the base, and its exocyclic O2 and N3 ring atoms are anchored in β-strand fashion against the backbone of the hood residues Gly-234 and Phe-236. The cPRPP molecule makes extensive interactions with residues from the PRPP binding motif, the flexible loop, and the turn between B4 and A4. Contacts to the 5′-phosphate include hydrogen bonds from conserved flexible loop residue Arg-137, which secures the loop over the active site in a manner analogous to the stabilizing contacts of the SY dipeptide residues in hypoxanthine-guanine-(xanthine) phosphoribosyltransferase (23–26). In the apo structure of UPRT this loop is poised over the active site, already in its active conformation (12). This conformation results from the interaction of the side chain of residue Arg-137 and a phosphate ion bound in the active site, which acts as a structural surrogate for the 5′-phosphate of the substrate PRPP (Fig. 1B). Arg-137 seems to be the critical residue in this loop among the UPRTs, because it is the only conserved residue in this region aside from Thr-141, which appears to be important structurally. Although all PRTases contain a flexible loop, each enzyme has a structurally distinct hood that is used in nucleobase recognition and is likely responsible for some of the mechanistic distinctions among PRTases. In UPRT, instead of the flexible loop, its larger hood region appears to play the major role in substrate-solvent exclusion, because the hood covers the uracil N1 nitrogen and the cPRPP C1′ carbon.
The 5′-phosphate of cPRPP is stabilized also by the helix dipole of A5 and contacts with the backbone amide nitrogens of residues 168–172 (A5) and the side chain Oγ oxygens of Thr-169 and Ser-172. Both the O3′ and O2′ hydroxyls of the cPRPP contact a solvent molecule (Wat-1), which contacts Asp-164 and a second solvent molecule. The distorted 6-fold coordination of the second solvent molecule, involving three oxygens of the β-phosphate, Wat-1 and the O2′ and O3′ hydroxyls, and its relatively low B-factor (45 Å2) suggest it might be an Mg2+ ion, which accompanies the protein through purification. A similarly located ion has been observed in the structures of the glutamine phosphoribosylpyrophosphate amidotransferase–cPRPP complex and several HGPRT-(substrate/substrate analogue/product) complexes (21, 23–26). In UPRT the PPi moiety of the cPRPP is tethered to the active site by interactions with the side chains of residues Arg-112, Tyr-148", and Lys-150" (where the double prime indicates contacts from a third subunit of the tetramer). The strained conformation of Arg-112, which is a Ramachandran outlier and takes a cis peptide conformation, allows its backbone to form a pocket for β-phosphate binding and possibly to participate in catalysis. A similar close approach of the peptide to the β-phosphate is observed in other PRTs when bound to pyrophosphate, cPRPP, or PRPP (21, 23–26). The putative divalent cation in the UPRT–uracil–cPRPP structure appears to stabilize a strained cPRPP conformation in which the PPi tail curls back toward the sugar (Fig. 1C). Interestingly, this conformation of the cPRPP is essentially identical to that taken by the cPRPP and PRPP molecules complexed with other PRTs (21, 23–26), indicating this structure as a potentially key feature of PRT catalysis.
The structure of this ternary complex reveals three critical features of the active site pocket. First, when cPRPP is bound, the “bottom” of the uracil pocket is shielded completely, thus occluding uracil from the bulk solvent. Second, the binding pocket anchors uracil optimally for an in-line attack on the PRPP molecule that inverts the stereochemistry of the C1′ carbon atom (Fig. 1C). Third, the interactions from Tyr-148" and Lys-150" to the PPi were unexpected, because they can occur only after tetramerization.
Substrate-Assisted Catalysis.
A notable feature of the T. gondii UPRT–uracil–cPRPP ternary structure is the proximity of the uracil N1 nitrogen to an α-phosphate oxygen of the cPRPP analogue [2.4 Å, N1(Ura)-O2A(cPRPP)]. This contact and the active site architecture suggests that UPRT may employ substrate-assisted catalysis (40, 41), whereby the PRPP α-phosphate abstracts the uracil N1 proton and thus serves as the general base in the reaction. Such a mechanism would contrast with that suggested for hypoxanthine-guanine-(xanthine) phosphoribosyltransferases (42), in which an aspartate in proximity to the purine N7 atom seems to play this role (23–25). Examination of the uracil binding site of UPRT reveals that the only other candidate for the general base is Asp-235, which is conserved in all UPRTs. However, the Oδ of Asp-235 is too far from the uracil N1 (3.8 Å). Yet, the importance of Asp-235 in catalysis is underscored by our kinetic analyses, which revealed that replacement of Asp-235 with asparagine or alanine results in a catalytically inactive enzyme. Thus, Asp-235 might serve as the general base upon a shift in its side chain orientation. Alternatively, the position of its side chain is such that it could act as a proton shuffle in which its carboxyl group, which is 2.7 Å from an α-phosphate oxygen, would act as a proton acceptor (in the forward reaction) from the scissile PPi. Moreover, the pKa of this side chain likely is increased because of its proximity to the side chain of Asp-238 (3.6 Å). After proton abstraction the resulting negative charge of the uracil pyrimidine ring and the strained conformation of the bound PRPP would induce the formation of a positively charged oxocarbenium-like state. The reaction between the uracil N1 nucleophile and the PRPP C1′ could then proceed via an SN1 (complete oxocarbenium formation) or SN2 (glycosidic bond formation before dissociation of the PPi leaving group) mechanism or a mixture of these and lead to UMP and PPi formation.
T. gondii UPRT Is a Physiologically Active Tetramer.
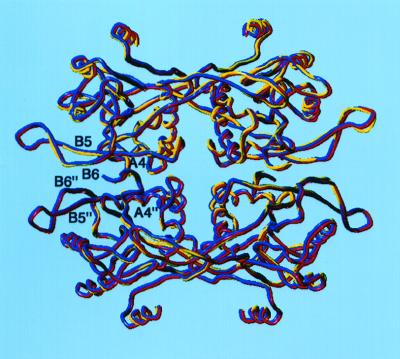
The tetramer found in the UPRT–uracil–cPRPP complex is identical to that observed in the apo UPRT, and superimposition of all Cα atoms of these tetramers results in an rms deviation of 0.44 Å despite their different crystal forms and crystallization conditions (Fig. 2). The tetramer is stabilized by interactions between residues from the turn before A4, A4, B5, and B6 from each subunit, which result in the burial of 1,100-Å2 surface area per subunit within the dimer of dimers interface (43, 44). Further, the UPRT–uracil–cPRPP structure reveals interactions between the β-phosphate oxygen of cPRPP and residues Tyr-148 and Lys-150 from a third subunit that belongs to the “other” dimer (Fig. 1C). These contacts simultaneously stabilize the tetramer and anchor the β-phosphate in the active site, thus properly orienting the PRPP for catalysis. The role of Lys-150 in PRPP binding explains its universal conservation among UPRTs (12). These findings strongly suggest that the tetramer is important in the physiological function of UPRT.
Figure 2.
The superposition of the apo (red), GTP complex (blue), and uracil–cPRPP complex (yellow) UPRT tetramers. The interacting elements of the dimer of dimers interface are labeled.
GTP Binding Stabilizes UPRT Tetramerization and Activates Catalysis.
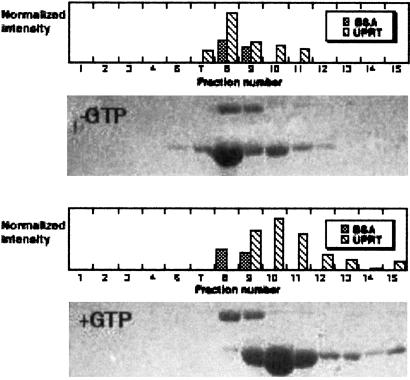
The addition of GTP to E. coli UPRT reduces the Km of the enzyme for PRPP by 7-fold without affecting Vmax (34, 45). Moreover, the oligomerization state of the enzyme changes from a dimer or trimer to a pentamer or hexamer (34). The mechanisms by which GTP increases PRPP affinity or induces and stabilizes higher order oligomers are unknown. Given our finding that T. gondii UPRT is in equilibrium between a dimer and tetramer (12) and our structural data, which reveals the functional importance of the tetramer, we tested whether GTP influences the oligomeric state and activity of the T. gondii enzyme. Sucrose density gradient sedimentation experiments revealed that in the absence of GTP, UPRT sedimented as a dimer, whereas in the presence of 2 mM GTP, it sedimented as a tetramer (Fig. 3).
Figure 3.
Sedimentation analyses of T. gondii UPRT oligomerization state in the absence (−GTP) and presence (+GTP) of GTP. In both gels, the upper band is BSA (64 kDa, fraction 8), and the lower band is UPRT (54 kDa, fraction 8, or 108 kDa, fraction 10).
Any physiological importance of GTP-stabilized UPRT tetramerization is tied to the intracellular levels of GTP. Although unknown in T. gondii, intracellular GTP concentrations in eukaryotic cells typically range from 0.2 to 0.5 mM (46). Therefore, depending on its affinity for UPRT, GTP could serve either as a constitutively bound cofactor or an allosteric regulator. To discern the specific role of GTP in UPRT function, the Kd of UPRT for GTP was determined by a rapid membrane filter-binding technique in the presence and absence of PRPP. In the absence of PRPP the Kd(GTP) is 465 ± 52 μM. In the presence of 3 mM PRPP the Kd(GTP) drops to 169 ± 7 μM. These data suggest that GTP plays a regulatory role, possibly functioning to sense and balance purine and pyrimidine nucleotide levels in the T. gondii cell. To demonstrate a correlation between the apparent GTP-induced change in oligomerization and the kinetic behavior of UPRT, steady-state kinetic constants were measured in both the presence and absence of GTP (Table 2). GTP addition lowered the Km for PRPP by nearly 6-fold but only marginally affecting the kcat. This result demonstrates that the kinetic consequence of GTP addition is an increased affinity of the enzyme for PRPP with little or no effect on subsequent rate-limiting steps of catalysis. This effect is specific for the purine nucleotide GTP, because the addition of ATP or other nucleotides did not alter the Km for PRPP (data not shown).
Table 2.
Steady-state kinetic parameters* for wild-type and mutant UPRTs
|
kcat, μmol
min−1 mg
protein−1
|
KmPRPP,
μM
|
Fold activation† | |||
|---|---|---|---|---|---|
| −GTP | 2 mM GTP | −GTP | 2 mM GTP | ||
| Wild type | 1.64 ± 0.41 | 1.50 ± 0.25 | 216 ± 22 | 37.4 ± 12.1 | 5.8 |
| K150A | 1.12 ± 0.33 | 1.35 ± 0.21 | 254 ± 35 | 195 ± 41 | 1.3 |
| R68A | 1.62 ± 0.55 | 1.32 ± 0.19 | 251 ± 45 | 77.4 ± 22.1 | 3.2 |
| K59A | 1.44 ± 0.34 | 1.78 ± 0.34 | 223 ± 25 | 171 ± 10 | 1.3 |
Data from three experimental determinations were fitted using the computer program enzfitter. Errors represent standard error calculated for each fit.
Activation is defined as the ratio of Km PRPP(−GTP) to Km PRPP(+GTP).
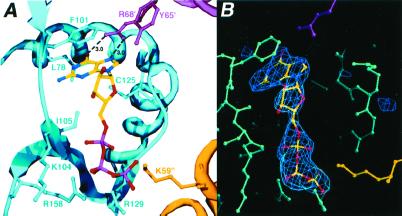
To ascertain directly how GTP binding stabilizes the UPRT tetramer, we determined the structure of the UPRT–GTP complex. Electron density for four GTP molecules, which adopt anti conformations, was observed at the interfaces between three subunits of the tetramer (Fig. 4 A and B). The conformation of the GTP-bound UPRT tetramer is identical to the apo tetramer with an rms deviation of 0.33 Å between all Cα atoms (Fig. 2). The GTP binding pockets are 25 Å from the nearest active site and correspond to a phosphate/sulfate binding site described previously in the apo UPRT structure (Fig. 5 A and B; ref. 12). Specificity for the guanine nucleobase is provided by residue Arg-68′, whereby the NH1 and NH2 atoms of the guanidium group read the base by hydrogen bonding to its O6 and N7 atoms. van der Waals contacts to the guanine are provided by Tyr-65′, Leu-78, Phe-101, Ile-105, and Cys-125 (Fig. 4A). At the other end of the binding pocket, the GTP phosphates engage in multiple electrostatic contacts and hydrogen bonds with the side chains of the “basic patch” residues Lys-104, Arg-129, Arg-158, and Lys-59′′ (Figs. 4A and 5A). The formation of the GTP binding pocket from three subunits explains the role of this nucleotide in tetramer stabilization and thus catalytic activation.
Figure 4.
GTP stabilized tetramerization of the T. gondii UPRT. (A) Closeup view of the GTP binding site showing key contacts to the nucleotide. GTP is contacted by residues from three subunits of the tetramer, which are colored magenta, cyan, and yellow. The GTP and side chains are depicted as balls and sticks. (B) Simulated annealing omit the Fo − Fc map (blue mesh contoured at 3 σ, calculated with a starting temperature of 1,500 K) in which GTP was omitted from refinement.
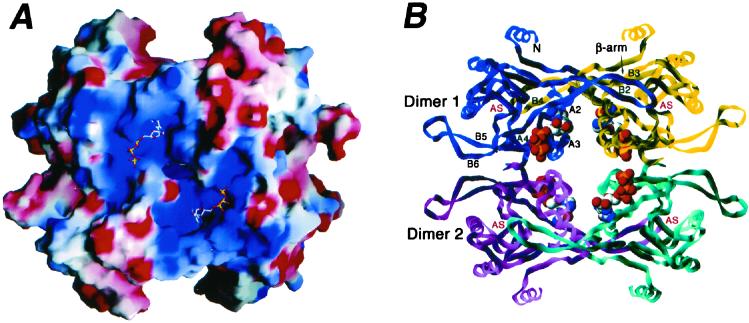
Figure 5.
The tetrameric UPRT–GTP complex. (A) Electrostatic surface of the UPRT tetramer illustrating the striking electropositive surface around the GTP binding site. Positive regions are blue, negative regions are red, and the GTP molecules are shown as sticks. This figure was generated in GRASP (48). (B) Ribbon diagram of the overall T. gondii UPRT–GTP tetramer structure with the GTP molecules shown as Corey–Pauling–Koltun models. Secondary structure elements involved in oligomerization are labeled, as is the active site (AS) of each subunit.
The entire complement of interactions between UPRT and GTP seem critical for stabilization of the tetramer. Indeed, even high phosphate concentrations of 150 mM, which far exceed in vivo phosphate concentrations (4 mM; ref. 46), do not promote UPRT tetramerization. Dynamic light-scattering experiments carried out on the protein alone, in the presence of 150 mM sodium/potassium phosphate, and 1 mM GTP reveal molecular masses of ≈63, 62, and 110 kDa, respectively, indicative of tetramerization only in the presence of GTP. Moreover, the addition of 50 mM sodium sulfate does not result in catalytic activation of UPRT (Km for PRPP of 237 μM in the presence of sulfate vs. 216 μM for the protein alone and 37 μM for the protein plus GTP).
The importance of Lys-59, Arg-68, and Lys-150 in GTP binding and catalytic enhancement was assessed further by their substitution with alanine. As compared with wild-type enzyme, each mutant protein exhibited diminished GTP-induced catalytic activation (5.8- vs. 1.3–3.2-fold) as a result of its decreased affinity for PRPP (Table 2). Lys-59 and Lys-150 are particularly crucial, because their substitution with alanine results in the loss of GTP activation, which suggests that phosphate neutralization is more important to catalytic function than guanine recognition per se.
The importance of these ionic interactions is underscored by the universal conservation of Lys-150 and Lys-59 (or its functional surrogate Lys-60) and divergence at residue 68 (12). However, because the E. coli enzyme is GTP-activated but lacks the corresponding arginine, either an alternative arginine or different nucleotide recognition mode must be used. Structural studies, on this and other UPRTs, will be necessary to delineate the role of GTP in oligomerization and enzyme activation, and as importantly, why some UPRTs do not undergo such activation.
As noted, the surface enveloping the GTP displays a continuous positive patch that extends beyond the nucleotide-binding site (Fig. 5A). This striking electropositive surface includes the residues Lys-104, Arg-129, and Arg-158 and suggests its role in nucleic acid binding, for which there is precedence from biochemical and structural studies on the Bacillus subtilis protein PyrR (21). The main function of PyrR is to regulate the expression of the pyrimidine nucleotide biosynthetic pathway by binding specific RNA stem-loop sequences and thus modulating the attenuation of transcription at three loci of the pyr operon (22). Remarkably, PyrR contains a PRT fold and displays weak UPRT activity. The parallels between the T. gondii UPRT and B. subtilis PyrR are intriguing, especially given our initial data that indicate the T. gondii UPRT can bind stem-loop structures with moderately high affinity (data not shown).
In conclusion, the UPRT–uracil–cPRPP structure suggests that T. gondii UPRT may use a substrate-assisted catalysis mechanism involving both substrate-assisted proton abstraction and reaction intermediate stabilization. In addition, this complex reveals the importance of the tetramer in the UPRT catalytic mechanism by the presence of a PRPP contact to the conserved residue Lys-150 from a third protomer. Our structural and biochemical data confirm that GTP binding stabilizes the more active tetrameric form of the enzyme and functions as a positive allosteric regulator. This mechanism of regulation of the UPRT oligomerization state by GTP may serve to balance purine and pyrimidine nucleotide pools in the T. gondii cell.
Acknowledgments
We thank Sao Jiralerspong and Laurie Remer for work on the cPRPP preparation. M.A.S. is a Burroughs Wellcome Career Development Awardee. This work was supported by National Institutes of Health Grants AI46416, AI23682 and GM26569.
Abbreviations
- PRT
phosphoribosyltransferase
- UPRT
uracil PRT
- PRPP
α-d-5-phosphoribosyl-1-pyrophosphate
- cPRPP
1-α-pyrophosphoryl-2-α,3-α-dihydroxy-4-β-cyclopentanemethanol-5-phosphate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors for the UPRT–GTP and UPRT–uracil–cPRPP complexes have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1JLR and 1JLS, respectively).
References
- 1.Beazley D M, Egerman R S. Semin Perinatol. 1998;22:332–338. doi: 10.1016/s0146-0005(98)80022-0. [DOI] [PubMed] [Google Scholar]
- 2.Luft B J, Remington J S. Clin Infect Dis. 1992;15:211–222. doi: 10.1093/clinids/15.2.211. [DOI] [PubMed] [Google Scholar]
- 3.Brooks R G, Remington J S, Luft B J. Antimicrob Agents Annu. 1987;2:297–306. [Google Scholar]
- 4.Iltzsch M H, Tankersley K O. Biochem Pharmacol. 1994;48:781–792. doi: 10.1016/0006-2952(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 5.Donald R G K, Roos D. Proc Natl Acad Sci USA. 1995;92:5749–5753. doi: 10.1073/pnas.92.12.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter D, Donald R G K, Roos D, Ullman B. Mol Biochem Parasitol. 1997;87:137–144. doi: 10.1016/s0166-6851(97)00058-3. [DOI] [PubMed] [Google Scholar]
- 7.Musick D L. CRC Crit Rev Biochem. 1981;11:1–33. doi: 10.3109/10409238109108698. [DOI] [PubMed] [Google Scholar]
- 8.Smith J L. Curr Opin Struct Biol. 1995;5:752–757. doi: 10.1016/0959-440x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 9.Craig S P, Eakin A E. J Biol Chem. 2000;275:20231–20234. doi: 10.1074/jbc.R000002200. [DOI] [PubMed] [Google Scholar]
- 10.Eads J C, Scapin G, Xu Y, Grubmeyer C, Sacchettini J C. Cell. 1994;78:325–334. doi: 10.1016/0092-8674(94)90301-8. [DOI] [PubMed] [Google Scholar]
- 11.Hendricksen A, Aghajari N, Jensen K F, Galhede M. Biochemistry. 1996;35:3803–3809. doi: 10.1021/bi952226y. [DOI] [PubMed] [Google Scholar]
- 12.Schumacher M A, Carter D, Scott D M, Roos D S, Ullman B, Brennan R G. EMBO J. 1998;17:3219–3232. doi: 10.1093/emboj/17.12.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vos S, de Jersey J, Martin J L. Biochemistry. 1997;36:4125–4134. doi: 10.1021/bi962640d. [DOI] [PubMed] [Google Scholar]
- 14.Muchmore C R, Krahn J M, Kim J H, Zalkin H, Smith J L. Protein Sci. 1998;7:39–51. doi: 10.1002/pro.5560070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozturk D, Dorfman R H, Scapin G, Sacchetini J C, Grubmeyer C. Biochemistry. 1995;34:10764–10770. doi: 10.1021/bi00034a008. [DOI] [PubMed] [Google Scholar]
- 16.Scapin G, Ozturk D H, Grubmeyer C, Sacchettini J C. Biochemsitry. 1995;34:10744–10754. doi: 10.1021/bi00034a006. [DOI] [PubMed] [Google Scholar]
- 17.Schumacher M A, Carter D, Roos D, Ullman B, Brennan RG. Nat Struct Biol. 1996;3:881–887. doi: 10.1038/nsb1096-881. [DOI] [PubMed] [Google Scholar]
- 18.Smith J L, Zaluzec E J, Wery J P, Niu L, Switzer R L, Zalkin H, Satow Y. Science. 1994;264:1427–1433. doi: 10.1126/science.8197456. [DOI] [PubMed] [Google Scholar]
- 19.Phillips C, Ullman B, Brennan R G, Hill C. EMBO J. 1999;18:3533–3545. doi: 10.1093/emboj/18.13.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Somoza J R, Chin M S, Foda P J, Wang C C, Fletterick R J. Biochemistry. 1996;35:7032–7040. doi: 10.1021/bi953072p. [DOI] [PubMed] [Google Scholar]
- 21.Krahn J M, Kim J H, Burns M R, Parry R J, Zalkin H, Smith J L. Biochemistry. 1997;36:11061–11068. doi: 10.1021/bi9714114. [DOI] [PubMed] [Google Scholar]
- 22.Tomchick D R, Turner R J, Switzer R L, Smith J L. Structure (London) 1998;6:337–350. doi: 10.1016/s0969-2126(98)00036-7. [DOI] [PubMed] [Google Scholar]
- 23.Focia P J, Craig S P, III, Eakin A E. Biochemistry. 1998;37:17120–17127. doi: 10.1021/bi9821465. [DOI] [PubMed] [Google Scholar]
- 24.Shi W, Li C M, Tyler PC, Furneaux R H, Grubmeyer C, Schramm V L, Almo S C. Nat Struct Biol. 1999;6:588–593. doi: 10.1038/9376. [DOI] [PubMed] [Google Scholar]
- 25.Shi W, Li C M, Tyler P C, Furneaux R H, Cahill S M, Girvin M E, Grubmeyer C, Schramm V L, Almo S C. Biochemistry. 1999;38:9872–9880. doi: 10.1021/bi990664p. [DOI] [PubMed] [Google Scholar]
- 26.Balendiran G K, Molina J A, Xu Y, Torres-Martinez J, Stevens R, Focia P J, Eakin A E, Sacchettini J C, Craig S P., III Protein Sci. 1999;8:1023–1031. doi: 10.1110/ps.8.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eads J C, Ozturk D, Wexler T B, Grubmeyer C, Sacchettini J C. Nat Struct Biol. 1997;5:47–58. doi: 10.1016/s0969-2126(97)00165-2. [DOI] [PubMed] [Google Scholar]
- 28.Sharma V, Grubmeyer C, Sacchettini J C. Structure (London) 1998;6:1587–1599. doi: 10.1016/s0969-2126(98)00156-7. [DOI] [PubMed] [Google Scholar]
- 29.Asai T, Lee C S, Chandler A, O'Sullivan W J. Comp Biochem Physiol B Biochem Mol Biol. 1990;95:159–163. doi: 10.1016/0305-0491(90)90264-t. [DOI] [PubMed] [Google Scholar]
- 30.McIvor R S, Wöhlhueter R M, Plagemann P G W. J Bacteriol. 1983;156:192–197. doi: 10.1128/jb.156.1.192-197.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen K F, Mygind B. Eur J Biochem. 1996;240:638–645. doi: 10.1111/j.1432-1033.1996.0637h.x. [DOI] [PubMed] [Google Scholar]
- 32.Natalini P, Ruggieri S, Santarelli I, Vita A, Magni G. J Biol Chem. 1979;254:1558–1563. [PubMed] [Google Scholar]
- 33.Dai Y P, Lee C S, O'Sullivan W J. Int J Parasitol. 1995;25:207–214. doi: 10.1016/0020-7519(94)00090-b. [DOI] [PubMed] [Google Scholar]
- 34.Linde L, Jensen K F. Biochim Biophys Acta. 1996;1296:16–22. doi: 10.1016/0167-4838(96)00045-3. [DOI] [PubMed] [Google Scholar]
- 35.Northrup J K, Smigel M D, Sternweis P C, Gilman A G. J Biol Chem. 1982;257:11416–11423. [PubMed] [Google Scholar]
- 36.Fitzgerald P M D. J Appl Crystallogr. 1988;21:273–278. [Google Scholar]
- 37.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 38.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Gross-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges N, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski R A, MacArthur M W, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 40.Dall'Acqua W, Carter P. Protein Sci. 2000;9:1–9. doi: 10.1110/ps.9.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kosloff M, Selinger Z. Trends Biochem Sci. 2001;26:161–166. doi: 10.1016/s0968-0004(00)01748-5. [DOI] [PubMed] [Google Scholar]
- 42.Xu Y, Grubmeyer C. Biochemistry. 1998;37:4114–4124. doi: 10.1021/bi972519m. [DOI] [PubMed] [Google Scholar]
- 43.Jones S, Marin A, Thornton J M. Protein Eng. 2000;13:77–82. doi: 10.1093/protein/13.2.77. [DOI] [PubMed] [Google Scholar]
- 44.Contee L L, Chothia C L, Janin J. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 45.Lundegaard C, Jensen K F. Biochemistry. 1999;38:3327–3334. doi: 10.1021/bi982279q. [DOI] [PubMed] [Google Scholar]
- 46.Traut T W. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 47.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 48.Nicholl A, Sharp K, Honig B H. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]