Abstract
p53 can adopt two forms in vitro, a latent form that binds naked DNA poorly and an active form that binds DNA well. Conversion of the latent form to the active form is thought to occur by an allosteric mechanism induced by phosphorylation and acetylation. Despite the large differences in affinity produced by regulatory modifications in vitro, mutation of putative regulatory sites has not produced correspondingly large effects on transcription of p53 target genes in vivo. To determine whether genotoxic stress regulates DNA binding by p53 in vivo, we have performed quantitative chromatin immunoprecipitation (ChIP) assays on tumor and normal cell lines containing wild-type p53. ChIP recovers several hundredfold more p21 and MDM2 promoter DNA from p53 wild-type than p53-null cells, indicating that the assay is specific for p53. Genotoxic stress induces much smaller increases in chromatin precipitation, which are matched by changes in the p53 protein level. Thus, in the experimental systems tested, allosteric regulation of DNA binding is not a major level of regulation of p53 activity. The p53 target genes tested can be divided into a group showing high promoter occupancy in vivo (p21, MDM2, and PUMA) and a group giving substantially weaker or background p53 binding (bax, AIP1, and PIG3). Neither group shows selective recruitment of p53 to the promoter in cells undergoing apoptosis, indicating that the decision to undergo apoptosis or cell cycle arrest depends on other changes in the cell.
The p53 tumor-suppressor gene encodes a transcription factor that induces cell cycle arrest and apoptosis in response to oncogenic transformation and DNA damage. Several models have been proposed to explain why p53 induces cell cycle arrest in some conditions and apoptosis in others. The favored model postulates that p53 binds with high affinity to the promoters of cell cycle arrest genes and binds to the promoters of apoptosis-inducing genes with a lower affinity. This model is based on the observation that the p53-binding site in the p21 gene is a much better match to the consensus p53-binding site than are the sites in putative apoptotic target genes, that some low-affinity mutants retain the ability to induce cell cycle arrest, and that the level of p53 expression determines the outcome in some experimental systems (1, 2).
There is evidence for regulation of p53 activity at the level of protein stability, interaction with basal transcription factors, and affinity for DNA. Regulation of DNA binding is thought to occur through a concerted allosteric mechanism induced by posttranslational modification (3). The induction of DNA binding in vitro is a very striking effect, and led to the proposal that in the absence of genotoxic stress, p53 should not be able to bind to the promoters of its target genes. Despite the impressive changes in DNA binding seen after in vitro modification of p53 and the fact that these modifications have been shown to occur in vivo after DNA damage (4), several reports have cast doubt on whether allosteric regulation occurs in vivo. Transfection of p53 mutated at sites expected to play an important role in regulating DNA binding generally fails to produce the expected changes in activity (5–7), and DNA binding by p53 in cell lysates is sometimes not induced by irradiation before harvesting the cells (8). Furthermore, recent in vitro studies on p53 binding to DNA packed into nucleosomes suggest that allosteric regulation may be an artifact of studying short DNA fragments or naked DNA (9).
To test whether p53 DNA binding is regulated in vivo, and to determine whether DNA binding to the promoters of putative apoptotic target genes is correlated with the induction of apoptosis, we have performed quantitative chromatin immunoprecipitation assays (ChIP; refs. 10 and 11). We can find no evidence that p53 exists in a non-DNA binding state in unstressed cells. We suggest that rather than reflecting a physiological regulatory process, allosteric regulation of DNA binding by p53 indicates that the conditions used previously for in vitro DNA-binding assays do not faithfully reproduce the nuclear environment. We further show that p53 binding to the published target sites in the bax, PIG3, AIP1, and PUMA promoters does not correlate with apoptosis induction.
Materials and Methods
HCT 116 human colorectal cancer cells and p53-null derivatives (12) were supplied by B. Vogelstein (Johns Hopkins Univ., Baltimore). Human NT2, WI-38, and U-2 OS cells were supplied by the American Type Culture Collection. DO7 murine hybridoma cells (13) were supplied by D. Lane (Univ. of Dundee, Dundee, Scotland, U.K.). The stably transfected human H1299 cells expressing p53 from the tet promoter were described by Saller et al. (14). p53 expression was induced in H1299 cells as described (14). UV irradiation was performed with a Stratagene Stratalinker. γ-Irradiation was performed with a cesium-137 source. Flow cytometry was performed as described (14). The primary antibodies used for immunoblotting were DO7 for p53, SMP-14 (Santa Cruz Biotechnology; ref. 15) for MDM2, and C19 for p21 (Santa Cruz Biotechnology). The loading control in Figs. 1–3 is a nonspecific band recognized by C19; in Figs. 4 and 5, a band stained by Ponceau S. Fold induction of p53 protein level was estimated by comparing Western blots of uninduced samples with dilutions of induced samples.
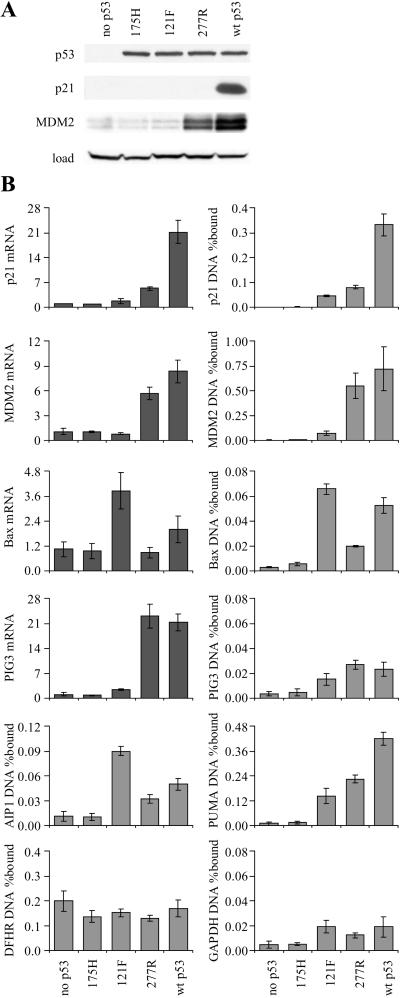
Figure 1.
ChIP from H1299 lung cancer cells expressing p53 from a tetracycline-responsive promoter. (A) Western blot for p53, p21, and MDM2. (B) Quantitative PCR for ChIP and mRNA expression. p53 binding to the p21, MDM2, bax, PIG3, AIP, and PUMA promoters was tested by ChIP. p53 binding to exon 4 of the GAPDH gene and Sp1 binding to the DHFR promoter were used as negative and positive controls, respectively. The level of p21, MDM2, bax, and PIG3 mRNA is expressed as fold-induction relative to cells lacking p53. wt, wild type.
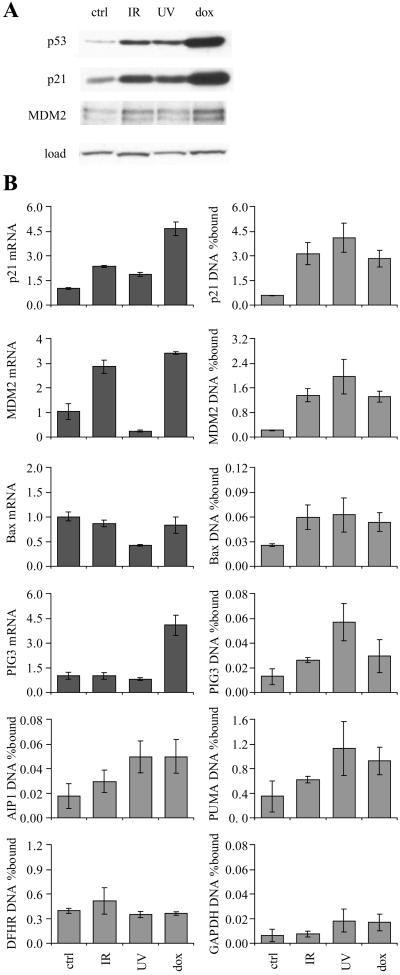
Figure 3.
ChIP from WI-38 normal diploid fibroblasts exposed to γ-rays (IR, 10 Gy), UV-C (UV, 20 J/m2), or doxorubicin (dox, 200 ng/ml). (A) Western blot. (B) Quantitative PCR assays for ChIP and mRNA expression. Cells were harvested at the time of peak p53 response (4, 12, and 24 h for IR, UV, and dox, respectively).
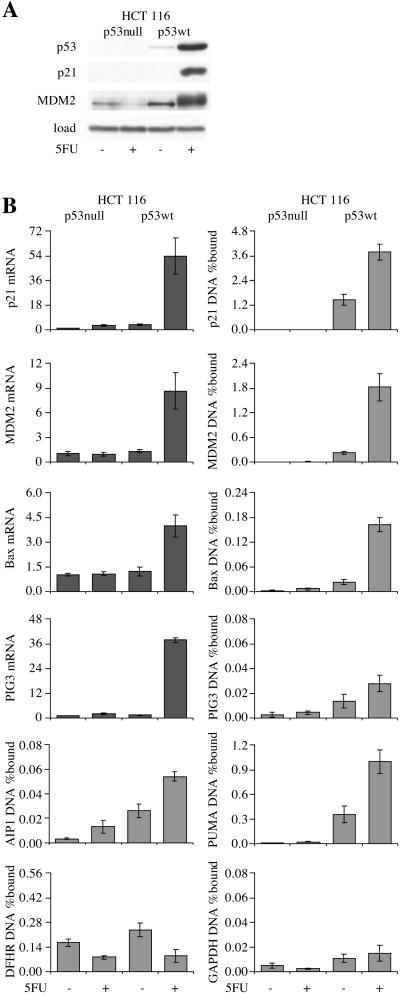
Figure 4.
ChIP from HCT 116 colon cancer cells treated with 5FU for 12 h. (A) Western blot. (B) Quantitative PCR for ChIP and mRNA expression. The p53-null cells were derived from parental HCT 116 by gene targeting (12).
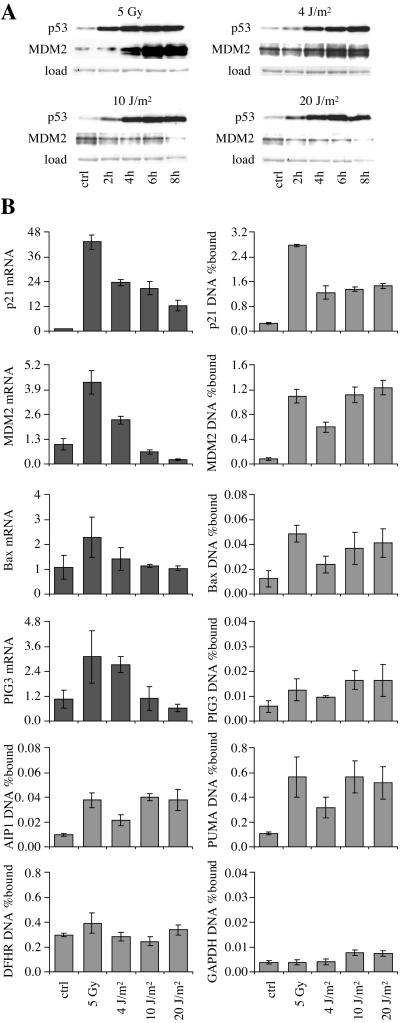
Figure 5.
ChIP from NT2 teratocarcinoma cells exposed to γ-rays (5 Gy) and UV-C (4, 10, or 20 J/m2). (A) Western blot. (B) Quantitative PCR for ChIP and mRNA expression. Cells were harvested for PCR 6 h after irradiation.
For ChIP, a 9-cm dish of subconfluent cells (1–15 × 106 cells, depending on the cell line) was fixed with 1% formaldehyde for 10 min at room temperature. Formaldehyde was neutralized by addition of 125 mM glycine for 5 min. Cells were washed twice in ice-cold PBS and collected by scraping in 1 ml of 1% SDS, 100 mg/liter sonicated salmon sperm DNA, and protease inhibitors (15 mg/liter aprotinin, 2 mg/liter leupeptin and 0.2 g/liter PMSF). To eliminate free p53, lysates were vortexed and insoluble material was collected by centrifugation at 14,000 × g at 4°C for 5 min. Pellets were resuspended in 1 ml 0.25% SDS/200 mM NaCl/100 mg/liter sonicated salmon sperm DNA and protease inhibitors and were sonicated to an average fragment size of 1 kb by using a microtip on a Branson sonicator. Remaining insoluble material was removed by centrifugation at 14,000 × g at 4°C for 5 min. The supernatant was diluted with 2 vol of 1% Nonidet P-40/350 mM NaCl and incubated for 12 h at 4°C with 10 μl of antibody-coated paramagnetic protein G beads (Dynal). One microgram of DO7 and 1 μg of Pep-2 (anti-Sp1, Santa Cruz Biotechnology) were used per ChIP. Immune complexes were collected with a magnet, washed four times in 1% Nonidet P-40/350 mM NaCl/100 mg/liter sonicated salmon sperm DNA, resuspended in 125 μl of 1% SDS/16 mg/liter salmon sperm DNA, and eluted by heating to 85°C for 10 min. Crosslinking was reversed by incubation of the eluate for 6 h at 65°C. Samples were diluted with 125 μl of water containing 160 mg/liter proteinase K, and incubated for 1 h at 50°C. DNA was purified by extraction with phenol/chloroform and precipitated with isopropyl alcohol and glycogen. The amount of input DNA per ChIP was 0.5 μg for WI-38, 5 μg for U-2 OS, 12 μg for COS-7, 25 μg for H1299 and NT2, and 50 μg for HCT 116.
Quantitative PCR was performed on a PE5700 PCR machine using a TaqMan Master Mix (Perkin–Elmer). The primers and probe for measuring 18S ribosomal RNA was purchased from Perkin–Elmer. The PCR cycles were 50°C for 2′ to digest dUTP-containing DNA, 95°C for 10 min to activate Taq Gold polymerase, then cycles of 95°C for 15 s and 60°C for 1 min repeated 40 times, except for the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers, where the annealing temperature was 55°C. The DNA from two experiments was pooled, and the error bars show the standard deviation of triplicate measurements, except for p21, MDM2, and DHFR in Fig. 2, where the error bars show the range of values for two experiments. Total RNA was extracted by using Trizol (Invitrogen), and 2 μg were reverse transcribed by using SuperScript II (Invitrogen). The level of mRNA was normalized to that of ribosomal RNA, and the results are expressed on an arbitrary scale, where the uninduced value is set to 1. The probe and primer sequences are given below. Primers are in italics, TaqMan probes are in bold, and p53-binding sites (p21, MDM2, bax, PIG3, AIP, and PUMA) and Sp1 binding sites (dihydrofolate reductase; DHFR) are underlined.
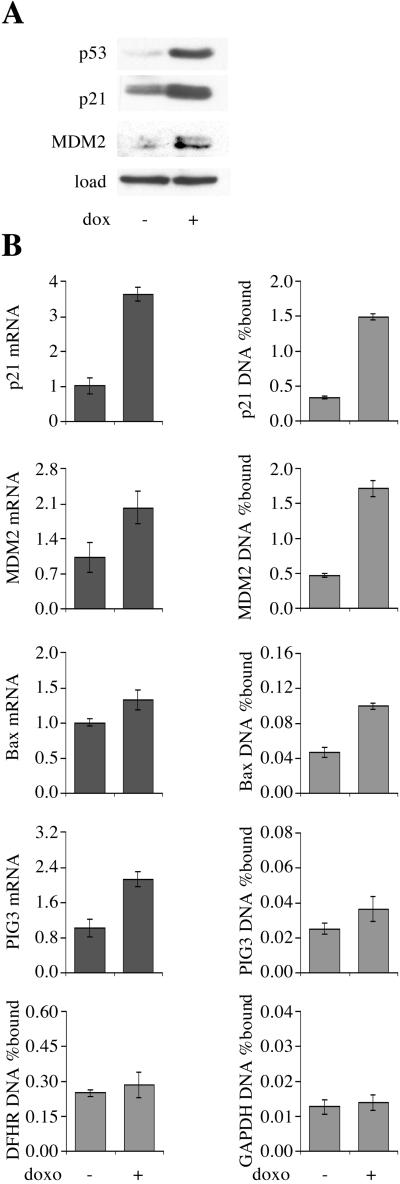
Figure 2.
ChIP from U-2 OS osteosarcoma cells treated with doxorubicin (dox) for 8 h (+) or untreated (−). (A) Western blot. (B) Quantitative PCR for ChIP and mRNA expression.
p21: GTGGCTCTGATTGGCTTTCTGGCCATCAGGAACATGTCCCAACATGTTGAGCTCTGGCATAGAAGAGGCTGGTGGCTATTTTGTCCTTGGGCTGCCTGTTTTCAG.
MDM2: GGTTGACTCAGCTTTTCCTCTTGAGCTGGTCAAGTTCAGACACGTTCCGAAACTGCAGTAAAAGGAGTTAAGTCCTGACTTGTCTCCAGCTGGGGCTATTTAAACCATGCATTTTCC.
Bax: TAATCCCAGCGCTTTGGAAGGCTGAGACGGGGTTATCTCTTGGGCTCACAAGTTAGAGACAAGCCTGGGCGTGGGCTATATTGCTAGATCCAGGTCTCTGCA.
PIG3: CACTCCCAACGGCTCCTTTCTCTTCTCTTAGCAGCACCCAGCTTGCCCACCCATGCTCAAGATGGGC.
AIP1: GCTGCCCTCCCTTCTCCTAGCTCTGTCCCCTCTCACTTCAGGAGTCTCAAGTCCTTCAGACTACTCCAAAGTCGGGG-38-TCTCTTGCCCGGGCTTGTCG.
PUMA: GCGAGACTGTGGCCTTGTGTCTGTGAGTACATCCTCTGGGCTCTGCCTGCACGTGACTTTGTGGACCCTGGAACG-76-CTCCTTGCCTTGGGCTAGGCC-63-CTGCAAGTCCTGACTTG.
DHFR: GGGGCGGGGCCTCGCCTGCACAAATAGGGACGAGGGGGCGGGGCGGCCACAATTTCGCGCCAAACTTGACCGCGCGTTCT.
GAPDH: GTATTCCCCCAGGTTTACATGTTCCAATATGATTCCACCCATGGCAAATTCCATGGCACCGTCAAGGCTGAGAACGGGAAGCTTGTCATCAATGGAAATCCCATCACCATCTTCCAGGAGTGAGTGGAAGACAGAA.
MDM2 mRNA: GTGAATCTACAGGGACGCCATCGAATCCGGATCTTGATGCTGGTGTAAGTGAACATTCAGGTGATTGGTTGGATCAG.
p21 mRNA: CTGGAGACTCTCAGGGTCGAAAACGGCGGCAGACCAGCATGACAGATTTCTACCACTCCAAACGCCGGCTGATCTTCTCCAAGAGGAAGCCCTAATC.
Bax mRNA: GGGTTGTCGCCCTTTTCTACTTTGCCAGCAAACTGGTGCTCAAGGCCCTGTGCACCAAGGTGCCGGAACTGATCAGAACCATCATGGGCT.
PIG3 mRNA: TCTCTGAAGCAACGCTGAAATTCACCAAAGGTGCTGGAGTTAATCTTATTCTAGACTGCATAGGCGGATCCTACTGGGAGAAGAACGT.
Results
To demonstrate that the ChIP assay detects sequence-specific DNA binding by p53 in vivo, p53-null H1299 lung cancer cells expressing p53 mutants were tested (14). The Ser-121 → Phe (121F) and Cys-277 → Arg (277R) p53 mutants have an altered sequence specificity in vitro and differentially activate transcription of p53 target genes in vivo (14, 16). The mutations alter the sequence specificity of p53 through effects on hydrogen bonding of p53 to the bases in the major groove (Lys-120 and Cys-277 are DNA contact residues; ref. 17). Hence, differences in chromatin binding by the mutants would provide strong evidence that the ChIP assay measures direct DNA binding. The Arg → His (175H) mutant, which does not bind to DNA, was used as a negative control. Because it retains the ability to interact with basal transcription factors, lack of chromatin precipitation by 175H would provide further evidence that the ChIP assay measures direct DNA binding rather than interactions with chromatin proteins. Sp1 binding to the DHFR promoter was used as a positive control for the efficiency of crosslinking and sonication. Amplification of exon 4 of the GAPDH gene was used as a negative control. To allow accurate measurement of the amount of DNA precipitated, quantitative PCR was performed by using TaqMan probes. The amount of target DNA precipitated is expressed as a percentage of the input target DNA. The antibody used for immunoprecipitation of p53 was DO7, which binds with high affinity to the amino terminus of p53 in a manner shown not to be influenced by regulatory phosphorylation (18).
Western blotting showed that the cells expressed the same amount of the different p53 alleles and confirmed that the p53 mutants are defective in induction of p21 (175H, 121F, and 277R) and MDM2 (175H and 121F; Fig. 1A). mRNA induction and ChIP by the 175H mutant gave background values for all genes tested. ChIP analysis allowed division of the p53 target genes into those giving strong binding (p21, MDM2, PUMA), and those giving weak (bax, AIP1) or background binding (PIG3 and the GAPDH control). Compared with parental p53-null cells, wild-type p53 expression increased the amount of DNA precipitated 350-, 300-, and 45-fold at the p21, MDM2, and PUMA promoters, respectively. The amount of DNA precipitated was 0.4–0.7% for these genes. Anti-Sp1 antibody precipitated 0.1–0.2% of the input DHFR promoter, a value unchanged by p53 expression. Nonspecific precipitation of GAPDH DNA by DO7 was increased 3- to 4-fold by wild-type p53 expression, possibly through nonspecific DNA binding by the p53 carboxyl terminus.
The pattern of DNA binding to the p21, MDM2, and bax promoters by the 121F and 277R mutants closely matched the pattern of mRNA expression (Fig. 1B). The simplest interpretation of this result is that the ChIP assay can detect exogenous p53 at the promoters of p53 target genes in vivo. Because the amount of PIG3 promoter precipitated was similar to the amount of GAPDH exon 4 precipitated, it is unlikely that the PIG3 gene is regulated by direct p53 binding to the published p53-binding site (Fig. 1B). Despite this, PIG3 mRNA was induced 20-fold by wild-type p53 and the 277R mutant, consistent with the original identification of PIG3 as a p53-induced gene. These results confirm observations by Szak et al. (11), who detected weak PIG3 ChIP that was temporally distinct from induction of PIG3 mRNA expression. The amount of bax and AIP1 DNA precipitated by DO7 was intermediate between that of the negative control and the strong binding group. The variation in binding seen with the specificity mutants suggests that this binding is direct. Furthermore, the pattern of binding is consistent with these genes playing a role in p53-dependent apoptosis, because the 121F mutant preferentially induces apoptosis (14).
Doxorubicin treatment of U-2 OS cells, which contain wild-type p53, led to induction of p21, MDM2, and PIG3 expression (Fig. 2). This was accompanied by a 3- to 4-fold increase in p53 binding to the p21 and MDM2 promoters (Fig. 2B). Bax showed a 2-fold increase in ChIP but only a small increase in mRNA level. Compared with the p21 and MDM2 promoters, the amount of bax promoter precipitated was 10-fold lower. There was no significant change in DHFR ChIP by Sp1 or PIG3 ChIP by p53, which was again close to the GAPDH value. The 3- to 4-fold increase in p21 and MDM2 promoter precipitation was matched by a commensurate increase in p53 protein level determined by Western blotting (Fig. 2A). We conclude that p53 is present at the p21 and MDM2 promoters of unstressed U-2 OS cells, and there is no evidence that doxorubicin regulates the affinity of p53 for DNA.
Because transformed cells containing wild-type p53 frequently have defects in upstream regulators of p53 such as p14ARF, we performed ChIP assays in WI-38 normal human diploid fibroblasts. To rule out the possibility that p53 binding to chromatin is regulated by only a subset of stresses, we tested three classic activators of p53-dependent transcriptional responses: γ-irradiation, UV-C irradiation, and doxorubicin treatment. All three treatments led to increased p53 protein expression (Fig. 3A). The increase in p21 and MDM2 promoter ChIP was comparable to the increase in p53 protein level; for doxorubicin, the measured increase was 6-fold for p53 level, 5-fold for p21 ChIP, and 6-fold for MDM2 ChIP. The increase in p53 level with UV-C and γ-irradiation was lower, but still broadly similar to the increase in p21 and MDM2 ChIP. In contrast with p21 mRNA expression, which increased after all three genotoxic stresses, MDM2 mRNA level fell after UV-C irradiation (Fig. 3B). MDM2 promoter binding and mRNA expression thus show opposite responses to UV-C irradiation. The other promoters tested again gave intermediate results. PUMA gave relatively strong ChIP signals (up to 1% of input), with modest induction by all three treatments. AIP1, bax and PIG3 gave weaker ChIP signals which for bax and PIG3 did not correlate with the mRNA levels (AIP1 was not tested for mRNA level). The Sp1/DHFR control showed that the efficiency of crosslinking and chromatin precipitation was not affected by genotoxic stress. The GAPDH control showed a 3-fold increase after UV-C treatment, but it should be noted that the p21 and MDM2 ChIP was 100 times more efficient than the GAPDH ChIP.
The low amount of p53 at the promoters of putative apoptotic target genes in U-2 OS and WI-38 cells is consistent with the lack of apoptosis induction by genotoxic stress at the doses used in these experiments. Two experimental systems were tested in which p53 strongly induces apoptosis. First, wild-type p53-containing HCT 116 colon carcinoma cells and p53-null derivatives were treated with 5 fluorouracil (5FU). Consistent with a published report (19), apoptosis was induced by 5FU only in the parental cells (28% sub-G1 by flow cytometry at 24 h, vs. 1% in the absence of 5FU, and 1% in the null cells in the presence or absence of 5FU). The p53 protein level rose 10-fold after 5FU treatment (Fig. 4A). This rise was accompanied by induction of p21 and MDM2 protein expression (Fig. 4A) and induction of p21, MDM2, bax, and PIG3 mRNA expression, which were not seen in the p53-null cells (Fig. 4B). 5FU treatment increased p53 binding to all of the putative target genes, but the increases never exceeded the increase in p53 protein level. The absolute amount of DNA precipitated was again much greater for p21, MDM2, and PUMA than for the other target genes, despite the fact that the cells were undergoing apoptosis. PIG3 and AIP1 ChIP from 5FU-treated wild-type cells gave signals only slightly above the background in untreated p53-null cells (4- and 6-fold increases for AIP1 and PIG3, respectively). This finding contrasts with the figures for the equivalent comparisons at the p21, MDM2, and PUMA promoters (430-, 240-, and 49-fold increases, respectively). Bax again gave an intermediate result; after 5FU treatment, the amount of promoter precipitated was 10-fold higher than the GAPDH value but 10- to 20-fold lower than the p21 and MDM2 values (Fig. 4B). The 7-fold increase in bax ChIP after 5FU treatment was accompanied by a 3-fold increase in bax mRNA level. The Sp1 control showed a decrease in DHFR ChIP after 5FU treatment, but the effect was identical in the parental and p53-null cells, effectively ruling out a role for p53 in the decline (Fig. 4B).
ChIP analysis was then performed on NT2 teratocarcinoma cells induced to undergo cell cycle arrest by ionizing radiation and apoptosis by UV-C. The sub-G1 fraction was 2%, 2%, 1%, 3%, and 10% at 9 h in untreated cells, γ-irradiated cells, and cells treated with 4, 10, and 20 J/m2 UV-C, respectively. ChIP was performed at 6 h, when the p53 protein level was 6-fold higher in cells treated with 5 Gy of ionizing radiation and 7-fold higher in cells treated with 20 J/m2 UV-C than in control cells (Fig. 5A). p53 binding measured by ChIP showed the same general pattern of binding as in cells not undergoing apoptosis: strong binding to p21, MDM2, and PUMA DNA, but weak or background binding to AIP1, PIG3, and bax DNA. There was no evidence for preferential binding to promoters of putative apoptotic target genes in cells undergoing apoptosis. With the exception of p21 ChIP after ionizing radiation, there was no difference between the magnitude of ChIP on the p53 target promoters treated with ionizing radiation or high doses of UV-C. Ionizing radiation produced the strongest increase in target gene mRNA expression. Bax mRNA level was not increased by UV-C, despite the fact that this was the treatment that induced apoptosis. High-dose UV-C reversed the induction of MDM2 mRNA seen at 4 J/m2, despite the continued presence of p53 at the promoter, as was seen in WI-38.
Discussion
Many studies have shown very large differences in DNA binding in vitro after phosphorylation, acetylation, and deletion of the p53 carboxyl terminus. The model that emerged from those studies was that the carboxyl-terminal basic domain in p53 is an allosteric regulator of DNA binding. The difference in affinity typically observed is so great that it should have been easily detectable with the ChIP assay. The most plausible explanation for the disparity with the ChIP data is that bandshift assays do not recapitulate a key property of the nuclear environment, which overcomes the inhibitory effect of the p53 basic domain in vivo. This function could plausibly be provided by chromatin, because recent studies have shown that unmodified p53 binds actively to its target sites in the p21 promoter in chromatin assembled in vitro (9).
The fact that the 121F and 277R mutants showed the expected alterations in chromatin precipitation strongly suggests that the assay measures direct DNA binding because the altered sequence specificity of these mutants is caused by subtle changes in hydrogen bonding to the bases in the major groove of the DNA (14, 17). Precipitation of the negative control GAPDH DNA increased slightly in the presence of p53. This increase could be caused by weak p53 binding to chromatin proteins or nonspecific DNA binding by the p53 carboxyl terminus (20). In contrast, the amount of p21 and MDM2 promoter precipitated increased several hundredfold in the presence of wild-type p53. The increases in chromatin precipitation after genotoxic stress were much lower, maximally 15-fold on the MDM2 promoter after high-dose UV-C treatment of NT2 cells. These changes were always matched by changes in p53 protein level, strongly suggesting that the affinity of p53 for DNA was not changed. The level of mRNA showed much more variation, as expected given the multiple levels of regulation of promoter activity and mRNA processing. The most striking disparity was seen after UV-C irradiation, where p53 binding to the MDM2 promoter increased with the p53 level, but the level of MDM2 mRNA fell substantially. The decrease in MDM2 mRNA level is caused, at least in part, by a decrease in transcription initiation (21). This decrease is presumably caused by the activation of DNA damage and stress signaling pathways, which repress MDM2 transcription, despite the continued presence of p53 at the promoter. This repression could be mediated by recruitment of corepressors to p53 or by mechanisms entirely independent of p53.
The genes tested can be divided into two groups: p21, MDM2, and PUMA, which bind p53 well, and bax, AIP1, and PIG3, which give substantially weaker ChIP signals. For the latter group, the amount of DNA precipitated was frequently comparable to that of the GAPDH control. Szak et al. (11) also saw weak p53 binding to the PIG3 promoter, which was temporally distinct from the PIG3 transcriptional response. These data are compatible with the known low affinity of p53 for its binding sites in the promoters of apoptotic p53 target genes. Lower promoter occupancy measured by ChIP could be caused by transient DNA binding by p53 or incorrect identification of the p53-binding sites in the promoter, both of which are still compatible with biologically meaningful regulation of these genes by p53, or could indicate that these genes are not directly transactivated by p53 in vivo. Indirect transactivation has been described for the gadd45 gene, which responds to DNA damage through a WT1 site (22). Determining whether a candidate gene is genuinely involved in mediating p53-dependent apoptosis is a very difficult problem, because the normal criteria of necessity and sufficiency cannot easily be applied to complex redundant responses. We suggest that the criteria for classifying putative apoptotic p53 target genes should include an assessment of whether credible amounts of p53 can be detected at the promoter in apoptotic conditions. By this measure, PUMA is a much better candidate for mediating p53-dependent apoptosis than AIP or PIG3. Our data suggest that the distinction between cell cycle arrest and apoptosis induction is not taken at the level of p53 binding to the promoter of any of the putative apoptotic target genes tested, because there was no consistent relationship between promoter binding and apoptosis induction. This finding could indicate that bax, AIP1, and PUMA are not involved in induction of p53-dependent apoptosis. Alternatively, the decision to invoke the apoptotic program may depend on differences in the interaction of p53 with coactivators or on changes in the activity of other pathways in the cell.
We conclude that in the classic models for regulation of p53 function commonly used in the literature, there is no evidence for allosteric regulation of DNA binding. Our data force a reassessment of many published conclusions on the regulation of p53 activity. We cannot exclude the possibility that p53 DNA binding is regulated in models we did not test. In particular, we cannot exclude that culture shock may have activated p53 in all of the cell lines tested (23). Nevertheless, rather than assuming that posttranslational modifications such as phosphorylation and acetylation of the p53 carboxyl terminus act through regulation of DNA binding, it may be instructive to look for effects of these modifications on other aspects of p53 function.
Acknowledgments
We thank Drs. David Lane and Bert Vogelstein for antibodies and cell lines, Drs. Alan Bridge and Matthias Peter for constructive advice, and the Swiss National Science Foundation and Institut Suisse de Recherches Experimentales sur le Cancer (Lausanne, Switzerland) for financial support.
Abbreviations
- ChIP
chromatin immunoprecipitation
- 5FU
5 fluorouracil
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- DHFR
dihydrofolate reductase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ludwig R L, Bates S, Vousden K H. Mol Cell Biol. 1996;16:4952–4960. doi: 10.1128/mcb.16.9.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X, Ko L J, Jayaraman L, Prives C. Genes Dev. 1996;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- 3.Hupp T R, Lane D P. Curr Biol. 1994;4:865–875. doi: 10.1016/s0960-9822(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 4.Meek D W. Oncogene. 1999;18:7666–7675. doi: 10.1038/sj.onc.1202951. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs B, O'Connor D, Fallis L, Scheidtmann K H, Lu X. Oncogene. 1995;10:789–793. [PubMed] [Google Scholar]
- 6.Blattner C, Tobiasch E, Litfen M, Rahmsdorf H J, Herrlich P. Oncogene. 1999;18:1723–1732. doi: 10.1038/sj.onc.1202480. [DOI] [PubMed] [Google Scholar]
- 7.Ashcroft M, Kubbutat M H G, Vousden K H. Mol Cell Biol. 1999;19:1751–1758. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siliciano J D, Canman C E, Taya Y, Sakaguchi K, Appella E, Kastan M B. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espinosa J M, Emerson B M. Mol Cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- 10.Murphy M, Ahn J, Walker K K, Hoffman W H, Evans R M, Levine A J, George D L. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szak S T, Mays D, Pietenpol J A. Mol Cell Biol. 2001;21:3375–3386. doi: 10.1128/MCB.21.10.3375-3386.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown J P, Sedivy J M, Kinzler K W, Vogelstein B. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 13.Vojtesek B, Bartek J, Midgley C A, Lane D P. J Immunol Methods. 1992;151:237–244. doi: 10.1016/0022-1759(92)90122-a. [DOI] [PubMed] [Google Scholar]
- 14.Saller E, Tom E, Brunori M, Otter M, Estreicher A, Mack D, Iggo R. EMBO J. 1999;18:4424–4437. doi: 10.1093/emboj/18.16.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Picksley S M, Vojtesek B, Sparks A, Lane D P. Oncogene. 1994;9:2523–2529. [PubMed] [Google Scholar]
- 16.Freeman J, Schmidt S, Scharer E, Iggo R. EMBO J. 1994;13:5393–5400. doi: 10.1002/j.1460-2075.1994.tb06874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho Y, Gorina S, Jeffrey P D, Pavletich N P. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 18.Chehab N H, Malikzay A, Stavridi E S, Halazonetis T D. Proc Natl Acad Sci USA. 1999;96:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bunz F, Hwang P M, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler K W, Vogelstein B. J Clin Invest. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pavletich N P, Chambers K A, Pabo C O. Genes Dev. 1993;7:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- 21.Wu L, Levine A J. Mol Med. 1997;3:441–451. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhan Q, Chen I T, Antinore M J, Fornace A J., Jr Mol Cell Biol. 1998;18:2768–2778. doi: 10.1128/mcb.18.5.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez R D, Morales C P, Herbert B S, Rohde J M, Passons C, Shay J W, Wright W E. Genes Dev. 2001;15:398–403. doi: 10.1101/gad.859201. [DOI] [PMC free article] [PubMed] [Google Scholar]