Abstract
We cloned, expressed, and characterized a hemeprotein from Deinococcus radiodurans (D. radiodurans NO synthase, deiNOS) whose sequence is 34% identical to the oxygenase domain of mammalian NO synthases (NOSoxys). deiNOS was dimeric, bound substrate Arg and cofactor tetrahydrobiopterin, and had a normal heme environment, despite its missing N-terminal structures that in NOSoxy bind Zn2+ and tetrahydrobiopterin and help form an active dimer. The deiNOS heme accepted electrons from a mammalian NOS reductase and generated NO at rates that met or exceeded NOSoxy. Activity required bound tetrahydrobiopterin or tetrahydrofolate and was linked to formation and disappearance of a typical heme-dioxy catalytic intermediate. Thus, bacterial NOS-like proteins are surprisingly similar to mammalian NOSs and broaden our perspective of NO biochemistry and function.
Genes coding for NO synthases (NOSs, EC 4.14.23) are present throughout the plant and animal kingdom. NOS activities are present in plants and lower eukaryotes (1–5). Their primary structures and activities are strikingly similar to the mammalian NOSs, suggesting that NO has been important throughout evolution. All eukaryotic NOSs catalyze the NADPH- and O2-dependent oxidation of l-arginine (Arg) to citrulline and NO, with N-hydroxy-l-arginine (NOHA) formed as an enzyme-bound intermediate (6). Structurally, all animal NOSs are bi-domain proteins containing an N-terminal oxygenase domain (NOSoxy) that binds protoporphyrin IX (heme), 6R-tetrahydrobiopterin (H4B), and Arg and is linked to a C-terminal flavoprotein domain (NOS reductase domain, NOSred) by a central calmodulin (CaM) binding sequence (7, 8). NOSred bears strong sequence and functional similarity to NADPH-cytochrome P450 reductase and related electron transfer flavoproteins (9, 10), and function to transfer NADPH-derived electrons to the ferric heme for O2 activation during NO synthesis. In contrast, NOSoxy and cytochromes P450 have completely different primary, secondary, and tertiary structures, even though both enzyme families use a thiolate-ligated heme for O2 activation (11, 12). Moreover, unlike P450s, NOSoxy must dimerize to become active (13, 14). Dimerization produces functional binding sites for Arg and H4B and sequesters the heme catalytic center from solvent. These distinguishing features imply that NOSs evolved separately from other heme-thiolate enzymes and can so provide unique perspectives on their structure-function relationships.
Although oxidative (nitrification) or reductive (denitrification) pathways for prokaryote NO biosynthesis are well established (15), there have been few reports on NOS-like proteins in bacteria (16, 17). To date no prokaryotic NOS proteins have been completely sequenced, purified, or cloned. However, some have been shown to have nitrite-forming activity that depended on Arg, NADPH, and H4B. More recent genome sequencing of Bacillus subtilis, Deinococcus radiodurans, and other bacteria (18–20) confirm that a subset contain ORFs that code for proteins homologous to NOSoxy. To better understand structure-function aspects of NOS proteins, we cloned, expressed, and characterized the NOSoxy-like protein in D. radiodurans (deiNOS). Here we determine physical and catalytic features of deiNOS and compare them to mammalian NOSoxy. The results reveal striking similarities and differences regarding primary structure, heme environment, utilization of H4B or tetrahydrofolate (THF), electron transfer interactions, formation and disappearance of a heme FeIIO2 catalytic intermediate, and activity. We also show that NO synthesis is possible for deiNOS. Together these findings give an important perspective on NOS biochemistry and biological function.
Materials and Methods
Materials.
All regents and materials were obtained from Sigma or sources previously reported (9–11, 21).
Molecular Biology.
The NOS gene of D. radiodurans (ATCC strain number 13939) was amplified by PCR from genomic DNA. PCR primers generated a NdeI site before the 5′ start codon and a BamHI site after the 3′ stop codon, and the amplified fragment was cloned into a pET15B expression vector. The reductase domain of neuronal NOS (nNOS) was created by using PCR to introduce a NdeI site, start codon at Met-695, and a SpeI site at nucleotide 3040 of nNOS. This PCR product was cloned into the NdeI/SpeI-digested nNOS pCWori. The cDNA constructs were confirmed by DNA sequencing. Protein expression was carried out in Escherichia coli strain BL21 (DE3).
Expression and Purification of deiNOS, nNOSoxy, and nNOSred.
They were purified by using on Ni2+-NTA resin for nNOSoxy and deiNOS and 2′,5′-ADP-Sepharose for nNOSred (9, 22). The ferrous-CO adduct absorbing at 444 nm was used to quantitate heme protein content by using an extinction coefficient of 74 mM−1⋅cm−1 (A444–A500). The nNOSred concentration was calculated by using an extinction coefficient of 18 mM−1⋅cm−1 (A457–A800).
Arg Binding.
Arg binding affinity was studied at 25°C by perturbation difference spectroscopy using 10 mM imidazole using methods as described (23).
NO Synthesis, NADPH Oxidation, Nitrite Formation, and Citrulline Production.
The steady-state activities in the reconstituted system containing deiNOS (or nNOSoxy) and nNOSred in a 1:1.5 molar ratio or in the heterodimer system containing deiNOS (or nNOSoxy) monomer and G671A nNOS full-length monomer (5:1) were both determined at 25°C by using spectroscopic and HPLC fluorometric assays as described in detail (8, 24, 25).
Measurement of Apparent km for H4B or THF.
Apparent km values were determined by double reciprocal analysis of the NADPH-dependent nitrite formation against various concentrations of H4B or THF in a reconstitution system containing nNOSred and either deiNOS or nNOSoxy.
Heme Reduction.
Optical spectra and kinetics were recorded on a Hitachi 3010 UV-visible spectrophotometer at 25°C. Reduction of heme groups was followed anaerobically in a reconstitution system containing deiNOS (or nNOSoxy) plus nNOSred in a 1:1.5 molar ratio in the presence or absence of CaM under CO gas as described (21). The heme reduction rate was monitored at 444 nm. Anaerobic reaction solutions contained ferric deiNOS or nNOSoxy (2.8 μM) and nNOSred (4.2 μM), 40 mM 4-(2-hydroxyethyl)-1-piperazine-propanesulfonic acid (EPPS) buffer (pH 7.6), 300 mM NaCl, 0.9 mM EDTA, 1 mM DTT, 1 mM Arg, 20 μM H4B, with or without 6 μM CaM plus 1 mM Ca2+. An aliquot of NADPH solution was added to give 50 μM to initiate heme reduction.
Optical Spectroscopy of Heme FeIIO2 Formation and Decay.
Oxygen binding spectra were recorded in a stopped-flow instrument equipped with a rapid scanning diode array device (Hi-Tech MG-6000, Salisbury, UK). Rapid scanning experiments involved mixing anaerobic solutions containing dithionite-reduced deiNOS or nNOSoxy, 40 mM EPPS buffer (pH 7.6), 0.5 mM DTT, 300 μM NaCl, and 1 mM Arg with air-saturated buffer solutions at 10°C in the presence or absence of 1 mM H4B. Formation and decay of the enzyme heme complexes were followed at 410 nm (407 nm for deiNOS) or 440 nm (22). Signal-to-noise ratios were improved by averaging 10 individual traces. The time courses of FeIIO2 complex formation and decay were best-fit to a single exponential equation with a nonlinear least-squares method as described (22).
Results
Primary Structure Analysis.
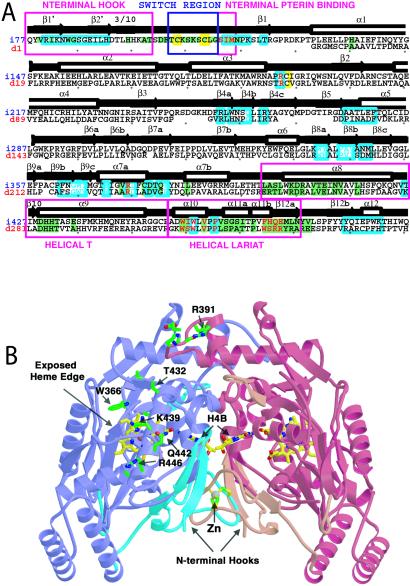
Cloning identified a 1-kb cDNA for deiNOS, coding for a 359-aa protein that has 34% identity and 52% conservation with rat nNOSoxy, and 34% identity and 50% conservation with mouse inducible NOSoxy (iNOSoxy). Similarities between primary sequences of deiNOS and mouse iNOSoxy are striking (Fig. 1A). Structural elements that make up the iNOS catalytic core are well conserved in deiNOS. This includes residues that contact the heme, bind the pteridine ring of H4B, and position substrate Arg above the heme. Residues that participate in the dimer interface or substrate binding channel are somewhat less conserved between deiNOS and iNOS. Sequence analysis predicts that the overall fold of deiNOS is very similar to iNOSoxy, and threading algorithms predict that the entire deiNOS sequence is highly compatible with structures of mammalian NOSoxy proteins. [The loopp threading algorithm (J. Mellar and R. Elber, Cornell Theory Center) indicated with high confidence that the deiNOS primary sequence adopts a topology similar to that of the mammalian NOSoxy dimers. The highest match (score = 15.3, global Z = 3.6, local Z = 3.6) occurred between the deiNOS primary sequence and the structure of mouse delta 114 iNOSoxy subunit (Protein Data Bank code 2nos).] However, a notable difference is that deiNOS lacks an extended portion of N-terminal sequence found in the mammalian enzymes. In eukaryotic NOSs this region codes for a N-terminal hook and metal binding site and contains residues that participate in forming the dimer interface and binding the dihydroxypropyl side chain of H4B (Fig. 1). Two smaller deletions downstream in the deiNOS sequence correspond to surface-exposed loops in the eukaryotic NOS structure (Fig. 1B). Thus, deiNOS likely has a topology similar to eukaryotic NOSs and contains residues likely to generate functional Arg and H4B binding sites.
Figure 1.
(A) Aligned iNOSoxy (above) and deiNOS (below) sequences with mapped residue function, secondary structure, and contributions to the dimer interface determined from iNOSoxy structures. NOS sequences are color-coded to highlight zinc ligands Cys-104 and Cys-109 and proximal heme ligand Cys-194 (yellow background), Arg-binding residues (cyan letters), H4B-binding residues (red letters), and residues that form the active-center channel leading to the heme (cyan boxed). Dimer interface residues that contribute at least 5 Å2 of buried surface area in the unswapped state of the inducible NOSoxy domain (34) are shown with green background. Above, black arrows show β-strands, white boxes show α-helices. Key sequence stretches involved in forming the dimer interface and cofactor binding sites are boxed in magenta and denoted, N-terminal hook, switch region (zinc loop), N-terminal pterin binding, helical T, and helical lariat. Seven additional C-terminal deiNOS residues (TGHAPTG) do not have analogs in the NOSoxy structures and are thus not shown. (B) Ribbon representation of the unswapped iNOSoxy dimer (34) highlighting the structural elements absent in deiNOS and surface residues conserved among NOSs. Most of the NOSoxy core (purple and red subunits) is conserved by deiNOS with the exception of the N-terminal hook, switch region, N-terminal binding pterin segments, and two peripheral loops (cyan and orange). Notably, the intermolecular zinc binding site (gray, bottom center) is not found in deiNOS, although regions that bind heme (yellow bonds) and H4B (yellow, center, on edge) are conserved. Residues that form a conserved surface patch above the active center channel and surrounding an exposed heme edge in mammalian NOSoxys (34) and are also conserved in deiNOS are highlighted (green side chains) and numbered.
Physical and Spectral Characteristics of deiNOS.
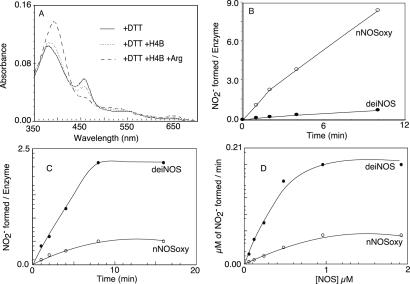
Recombinant deiNOS migrated in a denaturing SDS/PAGE gel at a molecular mass of ≈40 kDa, identical to its cDNA calculated molecular mass. Its migration in low-temperature PAGE and column filtration both indicated that deiNOS was predominantly dimeric in its native form (data not shown). Spectral changes obtained upon Arg or H4B binding are shown in Fig. 2A. Ferric deiNOS showed characteristic absorbance maxima at 460 and 380 nm, indicating its heme bound DTT to form a bis-thiolate species that is indistinguishable from nNOSoxy (24). Adding Arg caused the Soret maximum to shift from 460 to 395 nm, indicating Arg displaced DTT and caused a shift toward five-coordinate high-spin heme. Adding H4B caused a similar spectral transition and generated a broad Soret peak at 400 nm. The data indicate that these molecules bind to deiNOS in a similar manner to eukaryotic NOSs.
Figure 2.
(A) Spectrum of DTT-bound deiNOS and displacement of DTT upon H4B and Arg binding after incubating for 1 h at 25°C. (B) Kinetics of H2O2-dependent NOHA oxidation by nNOSoxy and deiNOS at 25°C was measured as described in Materials and Methods. (C and D) Time-dependent and enzyme concentration-dependent nitrite formation from NOHA by nNOSoxy or deiNOS in a reconstitution assay with nNOSred at 25°C in the presence of H4B and CaM and 1 mM NADPH.
We next monitored the ability of Arg to displace heme-bound imidazole in deiNOS and nNOSoxy to determine and compare their Arg binding affinity. Upon addition of Arg there was a concentration-dependent spectral shift that indicated Arg could achieve a complete displacement of heme-bound imidazole in both proteins (data not shown). The apparent kd value for Arg in the presence of 10 mM imidazole and 20 μM H4B was derived by double reciprocal analysis and was 97 ± 10 μM and 55 ± 4 μM in deiNOS and nNOSoxy, respectively. We conclude that their Arg binding affinities are similar.
Spectra of ferric or ferrous deiNOS with small heme ligands like NO, CO, and O2 also were obtained, and their features are compared with nNOSoxy in Table 1. In general, spectra of deiNOS complexed with these ligands were identical to those previously reported for mammalian NOSs. This similarity extended to deiNOS forming stable ferrous heme-CO or heme-NO complexes in the presence of Arg (26). The spectral data suggest that deiNOS has a similar, if not identical, heme environment as nNOSoxy.
Table 1.
UV-visible spectral properties of deiNOS and nNOSoxy with Arg or various heme ligands
| Enzyme-ligand complex | deiNOS, nm
|
nNOSoxy, nm
|
||
|---|---|---|---|---|
| Soret | Visible | Soret | Visible | |
| Fe3+-Arg | 393 | 650 | 393 | 650 |
| Fe3+-imidazole | 427 | 550 | 427 | 550 |
| Fe3+-DTT | 380, 460 | 650 | 380, 460 | 650 |
| Fe3+-NO | 440 | 549, 589 | 440 | 549, 589 |
| Fe3+-Arg | 410 | 560 | 412 | 560 |
| Fe2+-NO | 436 | 560 | 436 | 560 |
| Fe2+-CO | 444 | 540 | 444 | 540 |
| Fe2+-O2 | 427 | 560 | 427 | 560 |
H2O2-Supported NOHA Oxidation.
We next examined whether deiNOS would oxidize NOHA to nitrite in a H2O2-driven reaction. H2O2 binds to the NOS ferric heme to form reactive heme-oxy species that can react with Arg or NOHA in the presence or absence of H4B (25). In this reaction the heme need not acquire electrons from the NOSred or H4B. In Fig. 2B, the turnover number for deiNOS was 0.7 nitrite min−1, ≈12-fold slower than nNOSoxy tested in the same assay. Including 0.1 mM H4B in the assay failed to stimulate nitrite formation by deiNOS but stimulated nNOSoxy activity about 3-fold. This finding suggests that the deiNOS heme may have lower affinity toward H2O2 or may generate a heme-oxy species that is less reactive toward NOHA than nNOSoxy.
Catalytic Activity in Two Reconstituted Systems.
NO synthesis by animal NOSs involves electron transfer between reductase and oxygenase domains in a NOS dimer. Because deiNOS has no attached reductase domain, its heme can receive electrons only from an externally added reductase protein. We therefore examined its catalytic ability in two types of reconstitution systems. One involved mixing deiNOS with purified nNOSred that contained a functional CaM binding site, whereas the other involved generation of a heterodimer comprised of one deiNOS oxygenase subunit and one nNOS full-length subunit. The nNOS subunit contained a G671A mutation to prevent its forming a homodimer (24). In similar heterodimers of mammalian NOSs, functional electron transfer occurs only between the reductase domain of the full-length subunit and the surrogate oxygenase domain (13, 24, 27). We also repeated all reconstitution reactions by using nNOSoxy for direct comparison.
Our initial work revealed that mixtures of nNOSred plus nNOSoxy or deiNOS could catalyze conversion of either Arg or NOHA to products, but activity was greater using NOHA as substrate, consistent with previous NOS reconstitution systems (28, 29). We chose an assay condition where deiNOS or nNOSoxy dimers were present in a 0.7 molar ratio with added nNOSred. Varying the ratio of deiNOS or nNOSoxy to reductase protein from 1:1.5 to 3:1 had no effect on activity (S.A. and D.J.S., unpublished data). As shown in Fig. 2 C and D, nitrite formation from NOHA was time-dependent and enzyme concentration-dependent in reconstitution reactions that contained H4B and either deiNOS or nNOSoxy, with deiNOS being about four times more active than nNOSoxy. Compared with intact nNOS, the amount of NADPH consumed per nitrite generated by deiNOS or nNOSoxy was considerably higher in the reconstitution assay (data not shown), consistent with NADPH depletion occurring within 10 min (Fig. 2C). The nitrite synthesis activities of deiNOS and nNOSoxy in the reconstitution assay were ≈200 times less than intact nNOS (21), consistent with the uncoupled NADPH oxidation.
Results from reconstitution assays that mixed nNOSred with deiNOS or nNOSoxy dimers under various conditions are summarized in Table 2. Nitrite and citrulline formation were undetectable in reactions that contained Arg without H4B, but were detected in identical assays that contained NOHA as substrate. Adding H4B or CaM was clearly not obligatory. However, in reactions that contained CaM and NOHA, H4B stimulated the activity of deiNOS and nNOSoxy by 20-fold and 10-fold, respectively. In comparison to nNOSoxy, deiNOS activities ranged from being equivalent to four times more active in the presence of CaM, H4B, and NOHA. Despite their nitrite and citrulline production, none of the assays generated detectable NO in the oxyhemoglobin assay, possibly because of uncoupled NADPH oxidation generating superoxide. Our results imply that deiNOS can accept NADPH-derived electrons from free nNOSred, albeit in an uncoupled manner, to convert Arg or NOHA to citrulline and nitrite in a reaction that is further stimulated by H4B. Remarkably, under these circumstances the activity of deiNOS was equivalent or greater than nNOSoxy.
Table 2.
Catalytic activities of deiNOS versus nNOSoxy when supported by nNOSred
| Reaction system | NO production,
10−2 min−1 production,
10−2 min−1
|
Citrulline
production, 10−2 min−1
|
NO synthesis +CaM | ||
|---|---|---|---|---|---|
| −CaM | +CaM | −CaM | +CaM | ||
| nNOSoxy + Arg | ND | ND | ND | ND | ND |
| nNOSoxy + NOHA | 0.6 ± 0.1 | 1.2 ± 0.1 | 0.9 ± 0.1 | 1.3 ± 0.2 | ND |
| deiNOS + Arg | ND | ND | ND | ND | ND |
| deiNOS + NOHA | 1 ± 0.2 | 1.5 ± 0.2 | 1 ± 0.2 | 1.5 ± 0.2 | ND |
| nNOSoxy + Arg + H4B | 1.2 ± 0.1 | 1.2 ± 0.2 | 1.2 ± 0.1 | 1.6 ± 0.2 | ND |
| nNOSoxy + NOHA + H4B | 6.2 ± 1 | 10 ± 1 | 8.7 ± 1 | 13 ± 1 | ND |
| deiNOS + Arg + H4B | 1.2 ± 0.1 | 1.9 ± 0.1 | 1.2 ± 0.1 | 2.1 ± 0.1 | ND |
| deiNOS + NOHA + H4B | 10 ± 1 | 31 ± 3 | 10.7 ± 1 | 37 ± 3 | ND |
Values represent the mean and SD for three measurements. ND means not detectable.
Results from our heterodimer reconstitution experiments are summarized in Table 3. Product formation by both deiNOS and nNOSoxy heterodimer depended on CaM binding, consistent with CaM triggering interdomain electron transfer in the nNOS heterodimer (24, 27). The deiNOS and nNOSoxy heterodimers displayed nearly equivalent activities under all circumstances. Moreover, we detected similar rates of NO formation in the oxyhemoglobin assay for both proteins, demonstrating that deiNOS does make NO under this circumstance.
Table 3.
Catalytic activities of deiNOS and nNOSoxy heterodimers
| Reaction system | NO production,
10−2 min−1 production,
10−2 min−1
|
Citrulline
production, 10−2 min−1
|
NO synthesis +CaM | ||
|---|---|---|---|---|---|
| −CaM | +CaM | −CaM | +CaM | ||
| G671A nNOS/nNOSoxy | ND | 160 ± 4 | ND | 320 ± 3 | ND |
| G671A nNOS/deiNOS | ND | 210 ± 20 | ND | 320 ± 40 | ND |
| G671A nNOS/nNOSoxy + H4B | ND | 1,010 ± 40 | 120 ± 10 | 1,200 ± 60 | 10.4 ± 0.3 |
| G671A nNOS/deiNOS + H4B | ND | 820 ± 20 | 110 ± 10 | 1,000 ± 100 | 8.1 ± 0.4 |
ND, not detectable.
Affinity Toward H4B and THF.
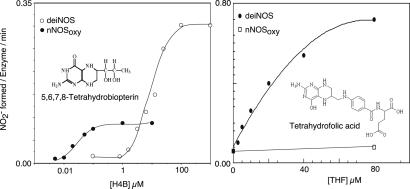
We used the mixed protein reconstitution system to determine an apparent Km for H4B. Fig. 3 shows that nitrite formation by deiNOS approached Vmax conditions near 100 μM H4B. Reciprocal analysis gave an apparent Km for H4B of 10 ± 2 μM for deiNOS versus 30 ± 10 nM for nNOSoxy. Clearly, deiNOS exhibits poorer affinity toward H4B than nNOSoxy or other mammalian NOSs, whose apparent Km values range between 0.05 μM and 1 μM.
Figure 3.
Affinities and activities of deiNOS and nNOSoxy toward H4B and THF. NADPH-dependent nitrite formation was measured as a function of H4B (Left) or THF concentration (Right) in a reconstitution assay containing NOHA, CaM, and nNOSred.
Although D. radiodurans does not biosynthesize H4B, it does contain THF. Because THF is a tetrahydropteridine like H4B, we determined apparent Km values for THF in the mixed protein reconstitution assay (Fig. 3). Only deiNOS was found to productively bind THF, with apparent Km value of 20 ± 5 μM. The estimated Vmax for deiNOS with THF was twice the value obtained with H4B. Thus, deiNOS differs from nNOSoxy in using THF in place of H4B to support catalysis. This is also true for other bacterial NOS-like proteins (C. S. Raman, personal communication).
Heme Reduction.
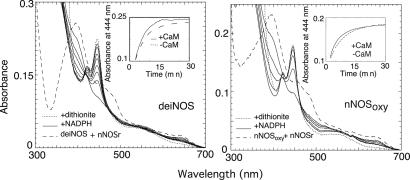
We compared the ability of added nNOSred to catalyze NADPH-dependent heme reduction in deiNOS or nNOSoxy as determined by CO binding under an anaerobic atmosphere. As shown in Fig. 4, about 98% of deiNOS or nNOSoxy heme iron became reduced after NADPH addition to the mixed protein reconstitution system relative to the value obtained by using dithionite, which completely reduces the heme iron independent of nNOSred. Time courses for heme reduction are shown in Fig. 4 Insets. Heme reduction was monophasic and slightly faster in the presence of CaM in both cases and was faster in deiNOS compared with nNOSoxy. The apparent rates are listed in Table 4. Faster heme reduction in deiNOS is consistent with its greater activity in this reconstitution assay (see Table 2). However, heme reduction in the reconstituted system was about 1,000 times slower than in intact nNOS when determined under identical conditions (30). This finding is consistent with slower catalytic activity in the reconstituted system.
Figure 4.
Heme iron reduction by nNOSred in deiNOS (Left) versus nNOSoxy (Right) under anaerobic conditions. Spectra were collected before (dashed line) and after addition of 50 μM NADPH (solid lines), and then after addition of 100 μM dithionite (dotted line). (Insets) The kinetics of heme iron reduction as measured by CO binding in the presence or absence of Ca2+ and CaM.
Table 4.
Rates of NADPH-dependent ferric heme reduction and FeIIO2 complex formation and decay in deiNOS and nNOSoxy
| Enzyme | Heme reduction,
10−3 s−1
|
FeIIO2
formation, s−1
|
FeIIO2 decay,
s−1
|
|||
|---|---|---|---|---|---|---|
| −CaM | +CaM | −H4B | +H4B | −H4B | +H4B | |
| nNOSoxy | 1.5 ± 0.2 | 3.0 ± 0.4 | 200* | 163* | 0.14* | 10* |
| deiNOS | 2.3 ± 0.3 | 5.7 ± 0.7 | 72 ± 4 | 60 ± 6 | 1.37 ± 0.04 | 3.9 ± 0.2 |
Values were taken from ref. 22.
Reaction of Ferrous deiNOS with O2.
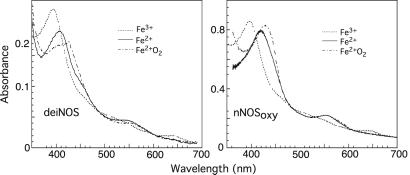
During NO synthesis, formation of a transient FeIIO2 intermediate is prerequisite for catalysis (22, 31, 32). We used rapid-scanning stopped-flow spectroscopy to examine spectral and kinetic properties of the FeIIO2 intermediate in deiNOS and to observe whether H4B would affect its properties. An anaerobic solution of dithionite-reduced deiNOS (or nNOSoxy) containing Arg with or without H4B was rapid-mixed with air-saturated buffer at 10°C. Groups of spectra were recorded to follow formation and decay of the FeIIO2 intermediate (Fig. 5) The initial ferrous species had a Soret peak at 407 nm and converted to a transient species with Soret peak at 427 nm in both deiNOS and nNOSoxy. In deiNOS the transient species Soret peak was less well defined and built up to a lesser extent. Both transient species converted to stable ferric enzymes that displayed a Soret peak at 395 nm and visible absorbance band at 650 nm (Fig. 5). Thus, deiNOS formed a FeIIO2 intermediate that is quite similar to nNOSoxy.
Figure 5.
Spectrum of a transient FeIIO2 catalytic intermediate formed in deiNOS (Left) and nNOSoxy (Right). The initial ferrous spectra shown were collected before mixing. Spectra indicating the FeIIO2 intermediate were collected at ≈7 ms and 28 ms after mixing for the nNOSoxy and deiNOS reactions, respectively, whereas spectra of the ferric species were collected 3 s after mixing. Other collected spectra were omitted for clarity.
Formation and decay kinetics of the FeIIO2 intermediate were determined by monitoring absorbance change at 407 or 440 nm versus time. The direction of absorbance change at these two wavelengths was reversed as expected (see Fig. 5), but otherwise proceeded with identical kinetics (data not shown), as found previously for nNOSoxy (22). Spectral change during the O2 binding or FeIIO2 decay phases was best described by a single exponential equation in all cases, suggesting both transitions are monophasic. FeIIO2 formation was somewhat slower in deiNOS compared with nNOSoxy at the O2 concentration used here (Table 4) and was unaffected by added H4B in both proteins, consistent with H4B not significantly affecting O2 binding kinetics in animal NOSs (22). FeIIO2 disappearance in H4B-free deiNOS was three times faster compared with nNOSoxy. Adding H4B to deiNOS increased this rate by about 3-fold, whereas H4B addition to nNOSoxy caused a 60-fold acceleration in its FeIIO2 disappearance, as reported (22). Thus, in the presence of H4B, FeIIO2 disappearance in deiNOS was three times slower compared with nNOSoxy (Table 4).
Discussion
The NOS-like protein of D. radiodurans provides unique perspective on NOS structure-function. For example, deiNOS was dimeric and catalytically active despite its missing an N-terminal segment that is conserved among most animal NOSs. This segment contains a N-terminal hook that helps form the dimer interface, two cysteine residues that form a metal ion binding site (Zn2+) at the dimer interface, and several residues that bind the dihydroxypropyl side chain of H4B (refs. 11, 12, 33, and 34; see Fig. 1). Apparently, these elements are not essential for deiNOS dimerization, H4B and Arg binding, proper heme complex formation with NO, CO, or O2, or electron import from a separate reductase domain for catalysis. This finding differs from how loss of these N-terminal elements affect certain animal NOSs. In iNOSoxy, deletion of the first 117 aa removes its N-terminal hook, metal binding site, and residues interacting with the H4B side chain. This deletion generated a thiol-ligated heme protein that was monomeric and catalytically inactive and had no capacity to bind H4B or Arg or form a homodimer (35). In the case of endothelial NOS, deletion of the first 105 residues also removed the three N-terminal elements, but did not completely abolish dimerization, substrate and H4B binding, and catalysis (36). Regarding nNOS, two alternatively spliced variants that are missing the N-terminal hook and metal binding site (37), or missing both of these elements plus residues that bind the H4B side chain (38), displayed normal or minimal activity, respectively, when expressed in animal cells. Thus, a picture is emerging where NOS isoforms exhibit a range of dependence on the three structural elements contained in the N terminal, with iNOS at one extreme and deiNOS at the other.
Our current work defines two reconstitution methods (added nNOSred, and heterodimer formation) for studying prokaryotic NOSs regarding catalysis and structure-function. Remarkably, deiNOS oxidized Arg or NOHA when supported by nNOSred and was more active than nNOSoxy in this setting. Its greater activity correlated with a faster rate of heme reduction. DeiNOS must have a competent docking site for the nNOSred to allow electron transfer and catalysis. However, heme reduction was still very slow and was associated with uncoupled NADPH consumption in this particular reconstitution system. Similar results were reported in reconstitution studies with separate iNOS oxygenase and reductase domains (28). This finding is consistent with heme reduction in animal NOSs requiring specific interactions in an intact dimer rather than collisions between free reductase and oxygenase domains in solution. However, because deiNOS has no attached reductase domain, it must normally rely on interactions with a separate redox partner to receive electrons. Our current results suggest that its native redox partner probably differs significantly from nNOSred. Indeed, the genome of D. radiodurans codes for no flavoproteins that match cytochrome P450 reductase or nNOSred, although several alternative electron transfer proteins are present (20).
deiNOS also oxidized Arg when it was part of a heterodimer that contained full-length G671A nNOS as the partner subunit. The oxyhemoglobin assay directly demonstrated that deiNOS can generate NO. Remarkably, heterodimers containing deiNOS or nNOSoxy displayed equivalent rates of NO synthesis that were well coupled to their NADPH consumption. Thus, interactions between the deiNOS subunit and the mammalian reductase domain were fully functional in a heterodimer, despite their distant evolutionary relationship and the fact that a 350-aa N-terminal segment is missing in deiNOS. Studies with NOS chimeras have shown that electrons can transfer between domains from different isoforms (i.e., nNOS and endothelial NOS reductase domains transferred electrons at their native rates to either nNOS or endothelial NOS oxygenase domains) (30). Thus, an interaction surface may be conserved that is independent of the N-terminal elements. The overall conservation of surface residues is predicted to be small between deiNOS and mammalian NOSoxy proteins (for example, 14% identity and 23% homology with mouse iNOSoxy). However, one conserved cluster of surface residues formed from helix α9 on one subunit and the C-terminal end of helix α7a on another (Fig. 1), corresponds to a region identified as a potential interaction site for the mammalian NOSred. This patch, which is adjacent to an exposed heme edge, was implicated in flavoprotein interactions based on conservation of surface residues among eukaryotic NOSs and the electrostatic character of NOSoxy and NOSred surfaces (34). Thus, electron transfer between deiNOS and its native redox partner may rely on similar interactions as those between the mammalian oxygenase and reductase domains. On the other hand, specific surface features may matter little once a foreign oxygenase subunit becomes part of a heterodimer (i.e., electron transfer may be made possible primarily by reductase domain proximity). These possibilities can now be addressed.
The ability of a deiNOS subunit to function so well in the heterodimer could also involve its being “rescued” by a phenomenon known as N-terminal hook swapping. Our previous work with iNOS showed that N-terminal hook swapping occurs in both homodimers and heterodimers (34, 35). Swapping within a heterodimer explains how iNOSoxy subunits can display good activity despite their having N-terminal mutations that debilitate catalysis in a homodimer. For example, a heterodimer containing an iNOSoxy subunit missing the first 117 aa (and thus similar to deiNOS, see Fig. 1) catalyzed NO synthesis at 50% the rate of wild type, apparently because its partner subunit provided an intact N-terminal hook to the mutant subunit (35). In our current study, the nNOS G671A full-length subunit could have provided its N-terminal hook to deiNOS through a swapping interaction, and in this way made deiNOS more like nNOSoxy regarding structure and catalysis in the heterodimer. On the other hand, it is still possible that deiNOS is constructed so as to not require a N-terminal hook for good catalytic function.
Reduced pteridines bound to deiNOS and participated in its catalysis. This is evidenced by H4B stimulating deiNOS citrulline formation in the mixed protein reconstitution system and being required for heterodimer NO synthesis. The primary structure of deiNOS is entirely consistent with its behavior toward H4B. Residues that surround the H4B ring in mammalian NOSs and position it near the heme are conserved in deiNOS. Conversely, the absence of N-terminal residues that in animal NOSs bind the 6-dihydroxypropyl side chain of H4B may explain why deiNOS has poorer affinity toward H4B. The missing N-terminal region probably also allows deiNOS to bind THF, which has a bulkier group in place of the 6-dihydroxypropyl side chain. This characteristic may be biologically important because it suggests how D. radiodurans could generate NO despite its lack of H4B.
In mammalian NOSs, H4B is implicated as an electron donor (32, 39) and provides an electron to the heme FeIIO2 intermediate for Arg hydroxylation in a single turnover reaction (31). In this circumstance electron transfer from H4B speeds disappearance of the FeIIO2 intermediate (22). Remarkably, H4B also increased the rate of FeIIO2 disappearance in deiNOS under identical circumstances. This behavior, together with deiNOS requiring H4B for NO synthesis and its conserving residues important for H4B function, strongly suggest that a native reduced pteridine may perform an identical redox function in D. radiodurans. Our current work sets the stage for detailed biochemical and biological investigations of bacterial NO synthesis and should provide a deeper understanding of all NOSs.
Acknowledgments
We thank Kartikeya Pant for help with loopp calculations. This work was supported by National Institutes of Health Grants CA53914 (to D.J.S.) and HL58883 (to E.D.G.), and grants from The American Heart Association (to S.A.) and The Camille and Henry Dreyfus Foundation (to B.R.C.).
Abbreviations
- NOS
NO synthase
- NOSoxy
NOS oxygenase domain
- NOSred
NOS reductase domain
- deiNOS
D. radiodurans NOS
- nNOS
neuronal NOS
- iNOS
inducible NOS
- CaM
calmodulin
- H4B
6R-tetrahydrobiopterin
- NOHA
N-hydroxy-l-arginine
- THF
tetrahydrofolate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Muller U. Prog Neurobiol. 1997;51:363–381. doi: 10.1016/s0301-0082(96)00067-6. [DOI] [PubMed] [Google Scholar]
- 2.Klesig D F, Durner J, Noad R, Navarre D A, Wendehenne D, Kumar D, Zhou J M, Shah J, Zhang S, Kachroo P, et al. Proc Natl Acad Sci USA. 2000;97:8849–8855. doi: 10.1073/pnas.97.16.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golderer G, Werner E R, Leitner S, Grobner P, Werner-Felmayer G. Genes Dev. 2001;15:1299–1209. doi: 10.1101/gad.890501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ninnemann H, Maier J. Photochem Photobiol. 1996;64:393–398. doi: 10.1111/j.1751-1097.1996.tb02477.x. [DOI] [PubMed] [Google Scholar]
- 5.Radomski M W, Martin J F, Moncada S. Philos Trans R Soc London B. 1991;334:129–133. [Google Scholar]
- 6.Griffith O W, Stuehr D J. Annu Rev Physiol. 1995;57:707–736. doi: 10.1146/annurev.ph.57.030195.003423. [DOI] [PubMed] [Google Scholar]
- 7.Sheta E A, McMillan K, Masters B S. J Biol Chem. 1994;269:15147–15153. [PubMed] [Google Scholar]
- 8.Ghosh D K, Stuehr D J. Biochemistry. 1995;34:801–807. doi: 10.1021/bi00003a013. [DOI] [PubMed] [Google Scholar]
- 9.Gachhui R, Presta A, Bentley D F, Abu-Soud H M, McArthur R, Brudvig G, Ghosh D K, Stuehr D J. J Biol Chem. 1996;271:20594–20602. doi: 10.1074/jbc.271.34.20594. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Martasek P, Paschke R, Shea T, Masters B S, Kim J J. J Biol Chem. 2001;276:37506–37513. doi: 10.1074/jbc.M105503200. [DOI] [PubMed] [Google Scholar]
- 11.Crane B R, Arvai A S, Ghosh D K, Wu C, Getzoff E D, Stuehr D J, Tainer J A. Science. 1998;279:2121–2126. doi: 10.1126/science.279.5359.2121. [DOI] [PubMed] [Google Scholar]
- 12.Fischmann T O, Hruza A, Niu X D, Fossetta J D, Lunn C A, Dolphin E, Prongay A J, Reichert P, Lundell D J, Narula S K, Weber P C. Nat Struct Biol. 1999;6:233–242. doi: 10.1038/6675. [DOI] [PubMed] [Google Scholar]
- 13.Siddhanta U, Presta A, Fan B, Wolan D, Rousseau D L, Stuehr D J. J Biol Chem. 1998;273:18950–18958. doi: 10.1074/jbc.273.30.18950. [DOI] [PubMed] [Google Scholar]
- 14.Klatt P, Schmidt K, Lehner D, Glatter O, Bachinger H P, Mayer B. EMBO J. 1995;14:3687–3695. doi: 10.1002/j.1460-2075.1995.tb00038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goretski J, Hollocher T C. J Biol Chem. 1990;265:889–895. [PubMed] [Google Scholar]
- 16.Chen Y, Rosazza J P N. Biochem Biopys Res Commun. 1994;203:1251–1258. doi: 10.1006/bbrc.1994.2317. [DOI] [PubMed] [Google Scholar]
- 17.Choi W S, Chang M S, Han J W, Hong S Y, Lee H W. Biochem Biophys Res Commun. 1997;237:554–558. doi: 10.1006/bbrc.1997.7189. [DOI] [PubMed] [Google Scholar]
- 18.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V. Nature (London) 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 19.White O, Eisen J A, Heidelberg J F, Hickey E K, Peterson J D, Dodson R J, Haft D H, Gwinn M L, Nelson W C, Richardson D L. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Degtyarenko K N, Kulikova T A. Biochem Soc Trans. 2001;29:139–147. doi: 10.1042/0300-5127:0290139. [DOI] [PubMed] [Google Scholar]
- 21.Adak S, Ghosh S, Abu-Soud H M, Stuehr D J. J Biol Chem. 1999;274:22313–22320. doi: 10.1074/jbc.274.32.22313. [DOI] [PubMed] [Google Scholar]
- 22.Abu-Soud H M, Gachhui R, Raushel F M, Stuehr D J. J Biol Chem. 1997;272:17349–17353. doi: 10.1074/jbc.272.28.17349. [DOI] [PubMed] [Google Scholar]
- 23.Adak S, Stuehr D J. J Inorg Biochem. 2001;83:301–308. doi: 10.1016/s0162-0134(00)00176-8. [DOI] [PubMed] [Google Scholar]
- 24.Panda K, Ghosh S, Stuehr D J. J Biol Chem. 2001;276:23349–23356. doi: 10.1074/jbc.M100687200. [DOI] [PubMed] [Google Scholar]
- 25.Adak S, Wang Q, Stuehr D J. J Biol Chem. 2000;275:33554–33561. doi: 10.1074/jbc.M004337200. [DOI] [PubMed] [Google Scholar]
- 26.Abu-Soud H M, Wu C, Ghosh D K, Stuehr D J. Biochemistry. 1998;37:3777–3786. doi: 10.1021/bi972398q. [DOI] [PubMed] [Google Scholar]
- 27.Sagami I, Daff S, Shimizu T. J Biol Chem. 2001;276:30036–30042. doi: 10.1074/jbc.M104123200. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh D K, Abu-Soud H M, Stuehr D J. Biochemistry. 1995;34:11316–11320. doi: 10.1021/bi00036a003. [DOI] [PubMed] [Google Scholar]
- 29.Chen P F, Tsai A L, Berka V, Wu K K. J Biol Chem. 1996;271:14631–14635. [PubMed] [Google Scholar]
- 30.Adak S, Aulak K S, Stuehr D J. J Biol Chem. 2001;276:23246–23252. doi: 10.1074/jbc.M102509200. [DOI] [PubMed] [Google Scholar]
- 31.Wei C C, Wang Z Q, Wang Q, Meade A L, Hemann C, Hille R, Stuehr D J. J Biol Chem. 2001;276:315–319. doi: 10.1074/jbc.M008441200. [DOI] [PubMed] [Google Scholar]
- 32.Bec N, Gorren A C, Voelker C, Mayer B, Lange R. J Biol Chem. 1998;273:13502–13508. doi: 10.1074/jbc.273.22.13502. [DOI] [PubMed] [Google Scholar]
- 33.Raman C S, Li H, Martasek P, Kral V, Masters B S, Poulos T L. Cell. 1998;95:939–950. doi: 10.1016/s0092-8674(00)81718-3. [DOI] [PubMed] [Google Scholar]
- 34.Crane B R, Rosenfeld R J, Arvai A S, Ghosh D K, Ghosh S, Tainer J A, Stuehr D J, Getzoff E D. EMBO J. 1999;18:6271–6281. doi: 10.1093/emboj/18.22.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh D K, Crane B R, Ghosh S, Wolan D, Gachhui R, Crooks C, Presta A, Tainer J A, Getzoff E D, Stuehr D J. EMBO J. 1999;18:6260–6270. doi: 10.1093/emboj/18.22.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez-Crespo I, Moenne-Loccoz P, Loehr T M, Ortiz de Montellano P R. Biochemistry. 1997;36:8530–8538. doi: 10.1021/bi970192j. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Newton D C, Robb G B, Kau C-L, Miller T L, Cheung A H, Hall A V, VanDamme S, Wilcox J N, Marsden P A. Proc Natl Acad Sci USA. 1999;96:12150–12155. doi: 10.1073/pnas.96.21.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brenman J E, Xia H, Chao D S, Black S M, Bredt D S. Dev Neurosci. 1997;19:224–231. doi: 10.1159/000111211. [DOI] [PubMed] [Google Scholar]
- 39.Hurshman A R, Krebs C, Edmondson D E, Huynh B H, Marletta M A. Biochemistry. 1999;38:15689–15696. doi: 10.1021/bi992026c. [DOI] [PubMed] [Google Scholar]