Abstract
The uncoupling protein homologue UCP2 is expressed in a variety of mammalian cells. It is thought to be an uncoupler of oxidative phosphorylation. Uncoupling proteins previously have been shown to be capable of translocating protons across phospholipid bilayers in proteoliposome systems. Furthermore, studies in mitochondria from yeast overexpressing the proteins have led to suggestions that they may act as uncouplers in cells. However, this issue is controversial, and to date, definitive experimental evidence is lacking as to whether UCP2 mediates part or all of the basal mitochondrial proton leak in mammalian cells in situ. In the present study, by using thymocytes isolated from UCP2-deficient and wild-type (WT) mice, we addressed the question whether UCP2 is directly involved in catalyzing proton leak in intact cells. Over a range of mitochondrial membrane potentials (ΔΨm), proton leak activity was lower in thymocytes from UCP2-deficient mice compared with WT mice. At physiological levels of ΔΨm, a significant portion (50%) of basal proton leak in resting cells depended on UCP2. Of note, proton leak in whole cells from WT mice, but not UCP2-deficient mice, responded to stimulation by 4-[(E)-2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-napthalenyl)-1-propenyl]benzoic acid (TTNPB), a known activator of UCP2 activity. Consistent with the observed changes in proton leak, ΔΨm and ATP levels were increased in untreated thymocytes from UCP2-deficient mice. Interestingly, resting respiration was unaltered, suggesting that UCP2 function in resting cells may be concerned with the control of ATP production rather than substrate oxidation. This study establishes that UCP2, expressed at endogenous levels, mediates proton leak in intact cells.
UCP2 is a recently cloned homologue of uncoupling protein 1 (UCP1) (see ref. 1). UCP1 catalyzes the net translocation of protons from the intermembrane space of mitochondria into the matrix. The role of UCP1 as an uncoupler of oxidative phosphorylation in brown adipose tissue mitochondria is well established (see ref. 2 for review). UCP1 dissipates the proton gradient across the inner mitochondrial membrane generated in oxidative phosphorylation, which would normally be used by ATP synthase to generate ATP from ADP and Pi. Thus, it uncouples substrate oxidation and electron transport from ATP synthesis. In brown adipose tissue, dissipation of the proton motive force has a thermogenic role. However, uncoupling of oxidative phosphorylation (also termed mitochondrial proton leak or proton conductance) may serve a wide variety of purposes (3, 4), including the regulation of substrate oxidation, free radical production, and ATP production and turnover. Therefore, it is not surprising that proton leak is seen in mitochondria and cells from many different organs and species.
Proton leak activity is not an artifact of isolation, as it can be detected in mitochondria as well as intact cells (3). It is a major contributor to standard metabolic rate (5). Studies in phospholipid liposomes suggest that the proton leak phenomenon requires the presence of proteins in the membrane, as proton flux through the lipid bilayer portion may account only for about 5% of all proton leak observed in intact mitochondria (6). It seems that proton leak activity in mitochondria may depend on a number of factors, including interactions between phospholipids and proteins in the inner mitochondrial membrane and the existence of specific proteins that catalyze some of the leak (7). However, proton leak even occurs in cells from which uncoupling proteins are thought to be absent, e.g., in hepatocytes and yeast. Studies in proteoliposomes (8, 9) and yeast mitochondria (1) suggested that UCP2 may catalyze proton translocation. It is debated whether and how much UCP2 and other UCP homologues actually contribute to proton leak in a physiological context in intact mammalian cells (7). It should be noted that these proteins have been termed “uncoupling proteins” first and foremost because of their high sequence homology with UCP1. Their role in metabolism as well as their in vivo regulation have only started to become elucidated.
Based on its tissue distribution, it had been suggested that UCP2 probably does not have a thermogenic role. This idea has been confirmed in loss-of-function studies (10, 11), which demonstrated that body temperature during cold exposure was similar in control and UCP2-deficient mice. These studies also showed that body weights were equivalent in control and UCP2-deficient mice. The involvement of UCP1 homologues in mediating proton leak in a physiological context remains unclear. It has been argued that the UCP1 homologues may not account for or contribute to the ubiquitous basal proton conductance of mitochondria, considering their varied distribution and expression patterns (7). However, no study has, as yet, accurately assessed the possible role of UCP1 homologues in intact cells. To date, there is no published evidence that UCP2 is involved in proton leak in cells that naturally express the UCP homologue at endogenous levels. The present study addresses the putative role of UCP2 in intact mammalian cells with respect to (i) proton leak activity and (ii) the possible role of UCP2 in the control of energy flow and associated processes. Thymocytes prepared from wild-type (WT) and Ucp2 gene knockout (KO) mice (11) were used to screen for differences in basic bioenergetic parameters and to establish unambiguously the role of UCP2 in mediating proton leak in situ. We also addressed the question whether proton leak activity may be altered by an activator, as recently demonstrated in isolated mitochondria from yeast expressing UCP2 (12).
Materials and Methods
Animals. Genotyping of Mice.
Ucp2 gene KO mice were generated as described (11). Multiplex PCR was used to genotype mice. Mice were housed three to four per cage in a temperature-controlled room with a 12-h light/12-h dark cycle. The animals had ad libitum access to chow (Rodent Diet no. 8664, Harlan Teklad, Madison, WI) and water and were handled in accordance with the principles and guidelines established by the National Institutes of Health.
Materials.
[3H]Triphenylmethylphosphonium (TPMP) iodide was obtained from NEN, and myxothiazol was obtained from Fluka. Antibodies for fluorescence-activated cell sorter (FACS) analysis were obtained from PharMingen. All other chemicals were obtained from Sigma.
Isolation of Thymocytes.
Thymocytes were isolated and incubated in RPMI medium 1640 (without glucose, with 2 mM glutamine) essentially as described (13). Cells were kept in plastic vials under 95% air/5% CO2 atmosphere in a shaking water bath at 37°C until used in the experiment. The viability of cells was greater than 95%, as determined by trypan blue exclusion.
Electron Microscopy and Analysis of Thymocyte Morphology and Mitochondrial:Cytosolic Volume Ratios.
Cells were prepared for electron microscopy in fixing buffer [1.25% (vol/vol) formaldehyde/2.5% (vol/vol) glutaraldehyde/0.003% picric acid] and washed with 0.1 M cacodylate buffer, pH 7.4. From the electron micrographs, mitochondrial:cytosolic volume ratios were estimated based on real fraction determinations using a transparent lattice grid.
Measurement of Oxygen Consumption and Mitochondrial Membrane Potential (ΔΨm).
Oxygen consumption was determined polarographically by using a Clark-type oxygen electrode (Digital Model 10, Rank Brothers, Cambridge, U.K.). All oxygen consumption rates were corrected for background and drift in the electrode. ΔΨm was determined concomitantly with oxygen consumption rates using 3H-labeled TPMP as a probe, essentially as described (14).
Proton leak curves were determined by concomitantly measuring nonphosphorylating oxygen consumption at different levels of ΔΨm. To this end, 0.5 ml of thymocyte suspension (≈3.5 × 107 cells) were incubated in the oxygen electrode chamber for 20 min with 160 ng/ml oligomycin, and either 0, 40, 80, or 480 nM myxothiazol (to inhibit electron transport). [3H]TPMP and carriers (TPMP and tetraphenylboron) were added as described (13). After 20 min, oxygen consumption was measured for 10 min, and then 0.4 ml of cell suspension was taken to determine potential-dependent distribution of [3H]TPMP. ΔΨm was calculated by using the equation and binding corrections given in Buttgereit et al. (14).
ATP Measurements.
Approximately 1.5 × 106 cells were extracted with 0.1 M NaOH/0.5 mM EDTA and assayed for ATP in a Wallac Victor2 multilabel plate-reader system using a luciferase-based assay (15).
UCP2 Immunoblotting and Northern Studies.
A full-length rat Ucp2 cDNA hybridization probe was used for Northern analysis. For detection of UCP2 protein in mitochondria isolated from thymocytes, immunoblotting was performed by using 50 μg of mitochondrial protein, goat anti-UCP2 IgG [UCP2(C-20):sc6525, Santa Cruz Biotechnology] as the first antibody (1:100) and donkey anti-goat IgG (PA42005, Amersham Pharmacia) as the second antibody (1:2,000).
FACS Analysis of Thymocytes: Cell Preparations, Antibodies.
Viable suspensions of thymocytes were prepared as described (13). Cells (≈1 × 106) in PBS/1% (vol/vol) FCS/0.1% NaN3 were labeled with anti-CD3e-PE, anti-CD8a-PerCP, and anti-CD4-FITC antibodies and analyzed on a FACSort (Becton Dickinson) by using forward and side-scatter gating to exclude nonviable cells.
Calculations and Statistical Analyses.
Data are presented as averages ± SEM. Where applicable, mean values were compared by using a two-tailed Student's t test.
Results
UCP2 Expression Levels, Thymocyte Morphology, and Cell Population Composition.
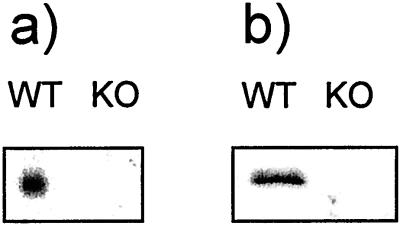
UCP2 is expressed in thymocytes and is absent in KO mice, as shown by Northern and Western analyses (Fig. 1). It has been shown that UCP2 is expressed in spleen (11, 16); expression levels in thymocytes are comparable to those observed in spleen mitochondria (data not shown).
Figure 1.
(a) RNA was extracted from thymocytes, and Northern blot analysis was performed with a full-length rat Ucp2 cDNA hybridization probe. (b) Mitochondria were isolated from thymus and immunoblotted for UCP2 protein.
Cellular and mitochondrial morphology affect the distribution of the potential-sensitive probe [3H]TPMP, which was used to determine ΔΨm in situ, as described (14). As assessed by electron microscopy, thymocyte morphology as well as mitochondrial content and distribution in UCP2 KO mice appeared normal (data not shown). Mitochondrial:cytosolic volume ratios (which appear in the equation used to calculate potentials from [3H]TPMP distribution) were 0.05 ± 0.01 for both UCP2 WT (n = 14) and KO (n = 16) thymocytes. Thus, the same protocol and calculation procedures could be used to determine potentials in WT and UCP2 KO mice.
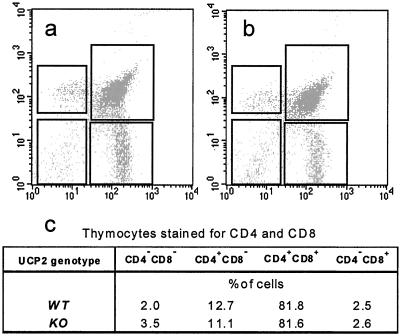
Direct comparison of bioenergetic parameters between thymocyte populations from WT and UCP2 KO mice also required that the populations did not differ with respect to their cell composition. The majority of cells retrieved in the preparation were thymocytes. To see whether UCP2 expression levels affected thymic selection of lymphocytes, FACS analyses of thymocyte preparations were performed. The analyses revealed comparable distributions of different cell types [lymphocytes, red blood cells, peripheral blood mononuclear cells, natural killer cells and macrophages] in individual thymocyte preparations; among the T cells, CD4/8 distribution was similar (Fig. 2).
Figure 2.
FACS analysis of thymocytes from (a) WT and (b) UCP2-deficient mice. (c) Quantitative assessment of CD4/CD8 distributions shown in a and b.
Proton Leak in Thymocytes.
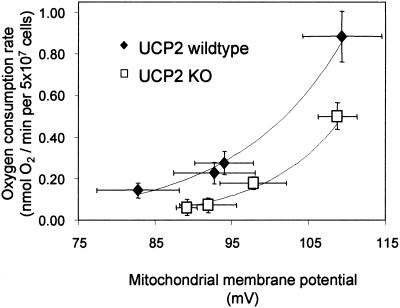
Mitochondrial proton leak was assayed in intact cells by using an established assay (17). Proton leak rate was measured in units of oxygen consumption under nonphosphorylating conditions (state 4 respiration, i.e., with saturating amounts of the ATP synthase inhibitor oligomycin). Under these conditions, in steady state, oxygen consumption reflects proton pumping by the respiratory chain to drive proton leak. However, because ATP synthase is inhibited, ΔΨm assumes an unphysiologically high value. To determine proton leak activity around the ΔΨm of resting cells, ΔΨm needs to be lowered to values below the resting level, with the proton leak rate determined concomitantly. To this end, different concentrations were added of myxothiazol, an inhibitor of the ubiquinol-cytochrome c oxidoreductase of the electron-transport chain. The resulting curves representing proton leak kinetics are shown in Fig. 3. As expected, proton leak increases with ΔΨm (e.g., see ref. 17). This increase is due to the fact that the membrane potential is the driving force for proton leak. At any value of ΔΨm, the oxygen consumption rate that drives proton leak is lower in thymocytes that lack UCP2 protein. The value of the ΔΨm in resting cells is 98 mV (see Fig. 4a). From Fig. 3 it can be seen that, at this level of ΔΨm, the proton leak in UCP2-deficient thymocytes is at least 50% less than that observed in WT thymocytes. Thus, endogenous levels of UCP2 mediate at least 50% of proton leak activity in resting thymocytes.
Figure 3.
Proton leak kinetics in thymocytes from WT and UCP2-deficient mice. Titrations were performed as described in Materials and Methods. State 4 respiration (i.e., the cells do not synthesize ATP) was induced by incubation with 160 ng/ml oligomycin (top right-hand points of each curve). ΔΨm was altered by titrations with different concentrations of myxothiazol (40, 80, and 480 nM). Data are means ± SEM (n = 6 and n = 5 cell preparations for WT and UCP2-deficient thymocytes, respectively). At steady state, the number of protons pumped by the complexes of the electron-transport chain equals the number of protons that leak back into the mitochondrial matrix. Because proton pumping and oxygen consumption are stoichiometrically linked, the oxygen consumption rates at any given potential reflect proton leak across the inner mitochondrial membrane.
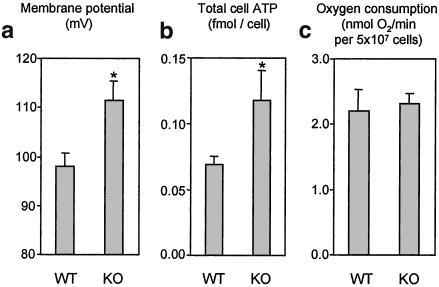
Figure 4.
Comparison of ΔΨm, total cell ATP content, and oxygen consumption in resting thymocytes. ATP data are means ± SEM for 9- and 13-cell preparations from WT and UCP2-deficient mice, respectively. Respiration and ΔΨm data are means ± SEM for 7- and 5-cell preparations from WT and UCP2-deficient mice, respectively. *, P < 0.05.
ATP Levels, Respiration, and ΔΨm in Resting Cells.
Given that UCP2 catalyzes proton leak in resting cells, it can be hypothesized that UCP2 deficiency also would affect ΔΨm and steady-state ATP levels. Thymocytes from UCP2 KO mice had a ΔΨm of 98 mV (Fig. 4a). This potential was increased by about 15% to a value of 112 mV in cells that lack UCP2. Deficiency in UCP2 also increases total cell ATP levels by about 70%, from 0.069 to 0.118 fmol per cell (Fig. 4b). Interestingly, UCP2 deficiency did not alter oxygen consumption in resting thymocytes (Fig. 4c).
Effects of the Retinoic Acid Analog TTNPB on State 4 Respiration in Thymocytes.
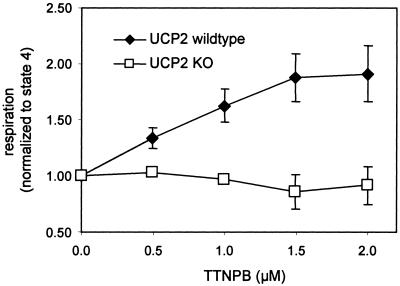
To see whether the proton leak activity mediated by UCP2 in thymocytes could be modulated at the cellular level, the effect of 4-[(E)-2-(5, 6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-napthalenyl)-1-propenyl]benzoic acid (TTNPB) on thymocyte state 4 respiration was assessed. Rial et al. (12) reported that TTNPB is an activator of UCP2-mediated proton leak. In the low micromolar range, TTNPB indeed was able to stimulate state 4 respiration in WT thymocytes by almost 2-fold (Fig. 5). In comparison, TTNPB had no effect on state 4 respiration in thymocytes isolated from UCP2-deficient mice, indicating that TTNPB acts on proton leak activity in a UCP2-dependent fashion.
Figure 5.
Effect of TTNPB on state 4 (i.e., nonphosphorylating) respiration in intact thymocytes. Thymocytes (at ≈7 × 107 cells per ml) were incubated with 160 ng/ml oligomycin, and respiration was measured. TTNPB was added in 0.5 μM increments, and respiration was measured after steady state was established.
Discussion
Based on distribution and expression patterns of the UCP1 homologues, it has been suggested that they may not account for the ubiquitous basal proton conductance of mitochondria (see ref. 7 for a more detailed review). Studies on proton-translocating activities of UCP2 (and UCP3) have, so far, been limited to mitochondrial and proteoliposome systems (9, 18–20). Thus, the potential of UCP2 to uncouple respiration in intact mammalian cells has not been addressed. In this article, it is shown that, expressed at endogenous levels, UCP2 contributes importantly to mitochondrial proton leak in resting thymocytes. From the proton leak titration curve shown in Fig. 3, it is clear that UCP2-deficient cells, compared with WT cells, have markedly decreased proton leak over a wide range of ΔΨms. Comparing proton leak activity in cells from UCP2 WT and KO mice at the resting ΔΨm level (about 98 mV, as reported in Fig. 4a), it is evident that UCP2-dependent uncoupling activity accounts for at least 50% of all proton leak observed at that potential (see Fig. 3). It can be concluded that in thymocytes, UCP2 at endogenous levels is required for catalyzing mitochondrial proton leak.
This result adds significantly to previous studies that used isolated mitochondria or proteoliposomes to define UCP1 homologue function. Those systems have been instrumental in establishing the proton-translocation activity of UCP2 (8) or the modulation of UCP2 activity by regulatory molecules (12). However, it is not possible from these studies to know the role of UCP2 in mediating proton leak in vivo because conditions under which isolated mitochondria are studied are unlikely to reflect the physiological environment in the cell, and this fact is even more true for proteoliposome studies. Two points are relevant: (i) cells exert many levels of control over substrate supply, and (ii) the identity and concentration of metabolites that might posttranslationally regulate UCP2 activity are virtually unknown. The present results indicate that UCP2 is required for at least 50% of the leak observed in the thymocyte model; however, it is also true that ≈50% of proton leak is mediated by UCP2-independent mechanisms. Whether this UCP2-independent proton leak is mediated by other uncoupling proteins or is independent of uncoupling proteins remains to be determined.
The difference seen in proton leak between cells from WT and UCP2 KO mice could depend on UCP2 in an indirect fashion. There are two possibilities. (i) UCP2 deficiency may affect thymic selection of lymphocytes and the composition of the thymocyte population, and (ii) proton leak could change because of UCP2-unrelated mechanisms, e.g., through chronic adaptation after deletion of the Ucp2 gene. Great care was taken to exclude the possibility that deletion of the Ucp2 gene affects either the composition of cell populations in primary cultures or the morphological appearance that would require adaptation of the method to determine membrane potentials in intact cells. Electron microscopy and FACS analysis showed that morphology of thymocytes or cell composition in suspensions were not changed in UCP2-deficient cells. Thus, it is justified to ascribe the difference in leak between cells from WT and UCP2 KO mice to the expression and activity of UCP2. To address ii, TTNPB experiments were conducted (Fig. 5). TTNPB stimulates state 4 respiration rate in WT mice but does not affect state 4 respiration in UCP2-deficient thymocytes. Because TTNPB acts in a UCP2-dependent fashion (12), this fact supports the finding that differences in proton leak between WT and UCP2 KO mice are directly dependent on UCP2.
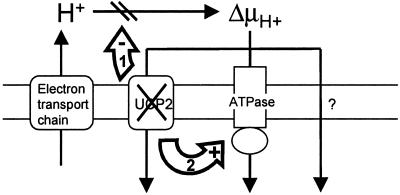
It is known that proton leak can control other pathways of oxidative phosphorylation by altering the level of proton motive force (e.g., see refs. 13 and 21). That is to say, a small change in proton leak activity may lead to significant changes in activities of other pathways, such as substrate oxidation or ATP production, because proton leak changes proton motive force, and the affected pathways are sensitive to these changes. Given that at least 50% of the leak in resting thymocytes is UCP2-dependent, UCP2 may share control over substrate oxidation or ATP production. Thus, UCP2-deficiency can have three possible consequences: the resulting increase in ΔΨm may inhibit respiration, or increase ATP production, or both (Fig. 6). From data reported in this and other studies, there is evidence that UCP2 is involved in controlling ATP production and not substrate oxidation. It was shown previously that, upon exposure to a given glucose load, ATP in pancreatic islets from UCP2 KO mice was increased compared with islets from WT mice (11). Here, it is shown that total cell ATP in thymocytes from UCP2 KO mice is increased by 70% (Fig. 4b). Interestingly, Cline et al. (22) reported an increased ATP/ADP ratio in muscle from UCP3 KO mice, along with an increased rate of ATP synthesis from Pi, but no noticeable change in tricarboxylic acid cycle (i.e., substrate oxidation) rate. Consistent with the view that UCP2 does not regulate substrate oxidation is the finding that resting respiration was not altered in UCP2-deficient thymocytes (Fig. 4c). Taken together, these findings suggest that UCP2 controls ATP production rather than substrate oxidation. Thus, modulation of UCP2 activity and the subsequent change in ΔΨm is likely to divert flux to and from ATP synthase (route 2 in Fig. 6), but it does not affect substrate oxidation rates significantly.
Figure 6.
Model of UCP2 function in resting thymocytes (see Discussion for details). Protons are pumped into the mitochondrial intermembrane space by complexes of the electron transport chain during oxidation of substrates. UCP2 mediates a significant portion of mitochondrial proton leak in resting thymocytes, thus providing one of three possible routes for protons to re-enter the matrix (the other two being ATP synthase and UCP2-independent leak). UCP2 deficiency causes an increase in ΔΨm. This increase may either put backpressure on the proton pumps in the electron-transport chain (1), thus inhibiting substrate oxidation and oxygen consumption rates, or increase ATP production (2).
We previously reported that UCP2 is a negative regulator of insulin secretion in pancreatic β-cells (11). In vivo, UCP2-deficient mice secrete more insulin in response to similar increases in blood glucose levels. In vitro experiments showed that pancreatic islets isolated from UCP2-deficient mice secreted more insulin compared with WT islets. This finding was ascribed to the uncoupling activity of UCP2. Consistent with this notion, the islets from UCP2-deficient mice had elevated ATP levels when incubated in vitro, which is the intracellular stimulus for insulin secretion. These and other results suggested that UCP2-deficiency causes higher coupling of oxidative phosphorylation, thereby up-regulating insulin secretion in an ATP-dependent mechanism. However, islets are a complex, multicellular system, and it is difficult to assess proton leak and characterize the bioenergetic consequences of UCP2-deficiency at a molecular level. Here, resting thymocytes were used to address these fundamental issues in intact cells. It is shown that proton leak activity in resting thymocytes is indeed UCP2-dependent. Consistently, UCP2-deficiency leads to increases in ΔΨm and total cell ATP levels. The control of ATP production (but not substrate oxidation) is likely to be a major function of UCP2.
The UCP2-dependent proton leak in thymocytes expressing UCP2 is sensitive to modulation by TTNPB, a nonphysiological retinoic acid analog. Thus, the efficiency of ATP production in glucose-sensing systems such as the pancreatic β-cell may be adjusted posttranslationally by regulatory molecules. If this mechanism holds in islets, it may be possible to develop new treatments for Type 2 diabetes by identifying UCP2 inhibitors that, at given glucose levels, increase ATP production and, therefore, improve insulin secretion in pancreatic β-cells.
Acknowledgments
We thank Shane Gray (Immunobiology Research Center, Beth Israel Deaconess Medical Center) for help with the FACS analyses. This research was funded by National Institutes of Health Grant RO1 DK 53477 and Ely Lilly.
Abbreviations
- TPMP
triphenylmethylphosphonium
- FACS
fluorescence-activated cell sorter
- ΔΨm
mitochondrial membrane potential
- KO
knockout
- TTNPB
4-[(E)-2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-napthalenyl)-1-propenyl]benzoic acid
- WT
wild type
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fleury C, Neverova M, Collins S, Raimbault S, Champigny O, Levi-Meyrueis C, Bouillaud F, Seldin M F, Surwit R S, Ricquier D, Warden C H. Nat Genet. 1997;15:269–272. doi: 10.1038/ng0397-269. [DOI] [PubMed] [Google Scholar]
- 2.Klingenberg M. Trends Biochem Sci. 1990;15:108–112. doi: 10.1016/0968-0004(90)90194-g. [DOI] [PubMed] [Google Scholar]
- 3.Rolfe D F, Brand M D. Biosci Rep. 1997;17:9–16. doi: 10.1023/a:1027327015957. [DOI] [PubMed] [Google Scholar]
- 4.Skulachev V P. Biochim Biophys Acta. 1998;1363:100–124. doi: 10.1016/s0005-2728(97)00091-1. [DOI] [PubMed] [Google Scholar]
- 5.Rolfe D F, Brown G C. Physiol Rev. 1997;77:731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- 6.Brookes P S, Rolfe D F, Brand M D. J Membr Biol. 1997;155:167–174. doi: 10.1007/s002329900168. [DOI] [PubMed] [Google Scholar]
- 7.Stuart J A, Cadenas S, Jekabsons M B, Roussel D, Brand M D. Biochim Biophys Acta. 2001;1504:144–158. doi: 10.1016/s0005-2728(00)00243-7. [DOI] [PubMed] [Google Scholar]
- 8.Echtay K S, Winkler E, Frischmuth K, Klingenberg M. Proc Natl Acad Sci USA. 2001;98:1416–1421. doi: 10.1073/pnas.98.4.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaburek M, Varecha M, Gimeno R E, Dembski M, Jezek P, Zhang M, Burn P, Tartaglia L A, Garlid K D. J Biol Chem. 1999;274:26003–26007. doi: 10.1074/jbc.274.37.26003. [DOI] [PubMed] [Google Scholar]
- 10.Arsenijevic D, Onuma H, Pecqueur C, Raimbault S, Manning B S, Miroux B, Couplan E, Alves-Guerra M C, Goubern M, Surwit R, et al. Nat Genet. 2000;26:435–439. doi: 10.1038/82565. [DOI] [PubMed] [Google Scholar]
- 11.Zhang C, Baffy G, Perret P, Krauss S, Peroni O, Grujic D, Hagen T, Vidal-Puig A J, Boss O, Kim Y, et al. Cell. 2001;105:745–755. doi: 10.1016/s0092-8674(01)00378-6. [DOI] [PubMed] [Google Scholar]
- 12.Rial E, Gonzalez-Barroso M, Fleury C, Iturrizaga S, Sanchis D, JimenezJimenez J, Ricquier D, Goubern M, Bouillaud F. EMBO J. 1999;18:5827–5833. doi: 10.1093/emboj/18.21.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krauss S, Buttgereit F, Brand M D. Biochim Biophys Acta. 1999;1412:129–138. doi: 10.1016/s0005-2728(99)00058-4. [DOI] [PubMed] [Google Scholar]
- 14.Buttgereit F, Grant A, Müller M, Brand M D. Eur J Biochem. 1994;223:513–519. doi: 10.1111/j.1432-1033.1994.tb19020.x. [DOI] [PubMed] [Google Scholar]
- 15.Ronner P, Friel E, Czerniawski K, Frankle S. Anal Biochem. 1999;275:208–216. doi: 10.1006/abio.1999.4317. [DOI] [PubMed] [Google Scholar]
- 16.Pecqueur C, Alves-Guerra M C, Gelly C, Levi-Meyrueis C, Couplan E, Collins S, Ricquier D, Bouillaud F, Miroux B. J Biol Chem. 2001;276:8705–8712. doi: 10.1074/jbc.M006938200. [DOI] [PubMed] [Google Scholar]
- 17.Nicholls D G, Ferguson S J. Bioenergetics 2. London: Academic; 1992. pp. 82–90. [Google Scholar]
- 18.Cadenas S, Buckingham J A, Samec S, Seydoux J, Din N, Dulloo A G, Brand M D. FEBS Lett. 1999;462:257–260. doi: 10.1016/s0014-5793(99)01540-9. [DOI] [PubMed] [Google Scholar]
- 19.Echtay K S, Liu Q, Caskey T, Winkler E, Frischmuth K, Bienengraber M, Klingenberg M. FEBS Lett. 1999;450:8–12. doi: 10.1016/s0014-5793(99)00460-3. [DOI] [PubMed] [Google Scholar]
- 20.Echtay K S, Winkler E, Klingenberg M. Nature (London) 2000;408:609–613. doi: 10.1038/35046114. [DOI] [PubMed] [Google Scholar]
- 21.Fell D A. Understanding the Control of Metabolism. London: Portland; 1997. [Google Scholar]
- 22.Cline G W, Vidal-Puig A J, Dufour S, Cadman K S, Lowell B B, Shulman G I. J Biol Chem. 2001;276:20240–20244. doi: 10.1074/jbc.M102540200. [DOI] [PubMed] [Google Scholar]