Abstract
Detectable splicing by the Saccharomyces cerevisiae mitochondrial bI3 group I intron RNA in vitro is shown to require both an intron-encoded protein, the bI3 maturase, and the nuclear-encoded protein, Mrs1. Both proteins bind independently to the bI3 RNA. The bI3 maturase binds as a monomer, whereas Mrs1 is a dimer in solution that assembles as two dimers, cooperatively, on the RNA. The active six-subunit complex has a molecular mass of 420 kDa, splices with a kcat of 0.3 min−1, and binds the guanosine nucleophile with an affinity comparable to other group I introns. The functional bI3 maturase domain is translated from within the RNA that encodes the intron, has evolved a high-affinity RNA-binding activity, and is a member of the LAGLIDADG family of DNA endonucleases, but appears to have lost DNA cleavage activity. Mrs1 is a divergent member of the RNase H fold superfamily of dimeric DNA junction-resolving enzymes that also appears to have lost its nuclease activity and now functions as a tetramer in RNA binding. Thus, the bI3 ribonucleoprotein is the product of a process in which a once-catalytically active RNA now obligatorily requires two facilitating protein cofactors, both of which are compromised in their original functions.
Many small ribozymes, both those that occur naturally (1) and those that are products of in vitro selection experiments (2), display an impressive array of catalytic activities. Although these simple structures are capable of performing catalysis, most cellular ribozymes have undergone a process of elaboration in which potentially simple RNA active sites are bolstered by additional RNA structural domains (3, 4) or protein cofactors (5–9).
One example of this process is the group I introns. The group I intron RNA core consists of two extended and roughly coaxially stacked helices that form a catalytic cleft, which binds, in turn, the 5′ and 3′ splice site helices and the guanosine cofactor (guanosine 5′-monophosphate, pG) (4, 5, 10). Although the catalytic core is relatively compact and can independently form a functional active site (11), most group I introns have acquired additional peripheral structures that function to stabilize the RNA core (4, 12, 13). Moreover, whereas a large number of group I introns have been identified by sequence and structural analysis (4, 14), both anecdotal and experimental evidence suggests that many have lost their self-splicing activity (15). It appears that group I intron RNAs commonly recruit protein cofactors and now function as obligatory ribonucleoproteins (8, 16).
The Saccharomyces cerevisiae mitochondrial cytochrome b bI3 group I intron represents an especially intriguing case of elaboration by accretion of peripheral RNA domains and protein cofactors. First, the bI3 RNA potentially spans two group I intron subgroups, IA2 and IB4, characterized by RNA sequence insertions between helices P7 and P3 and adjacent to P9 and by an extension of the P5 helix, respectively (4) (Fig. 1A, boxed structures). The bI3 intron also contains two tertiary interactions common to many catalytic RNAs, the L2-P8 and L9-P5 tetraloop–minor groove receptor motifs (4, 17, 18) (Fig. 1A, dashed lines).
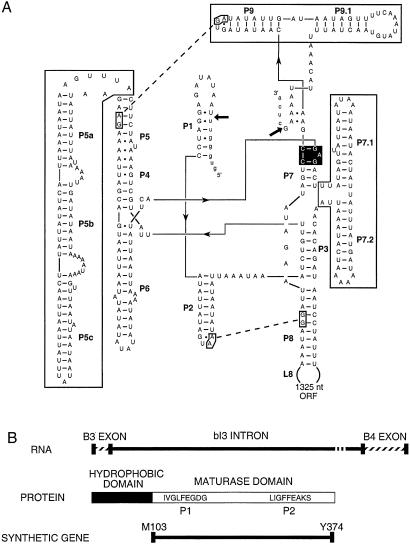
Figure 1.
Organization of the bI3 group I intron. (A) Secondary structure of the bI3 RNA (4). 5′ and 3′ splice sites are marked with arrows. Peripheral domains are boxed, the guanosine cofactor-binding site is in black, and the L2-P8 and L9-P5 interactions are shown with dashed lines. Px identifies base paired helices. (B) The bI3 maturase RNA is translated in frame with the cyt b B3 exon to yield the bI3 maturase protein, consisting of hydrophobic (potentially membrane bound) and maturase domains. The ′LAGLIDADG′ motifs, P1 and P2, reside within the “soluble” portion of the protein, for which a synthetic gene was constructed.
Second, genetic evidence indicates that at least two genes are required for bI3 RNA splicing in vivo. The first encodes the bI3 maturase, most of whose coding sequence is inserted within a peripheral domain of the intron, in L8 (Fig. 1A) and is translated in frame with the native cytochrome b B3 exon (19) (Fig. 1B). The maturase is a member of a large family of DNA endonucleases identifiable, in part, by conserved LAGLIDADG motifs (20). The presence of the bI3 maturase ORF within the intron probably represents an early colonization event by a LAGLIDADG DNA endonuclease precursor (21–24). The RNA-directed maturase, or splicing facilitation, activity is likely a subsequent innovation. Because the bI3 maturase coding sequence lies within the intron, this function is potentially linked with evolution of the intron RNA.
The second gene is MRS1 (25, 26), whose protein product is encoded in the nucleus, targeted to the mitochondria, and homologous with the nuclear-encoded mitochondrial DNA-resolving enzyme, Cce1 (27, 28). Cce1 recognizes, manipulates, and cleaves four-way DNA junctions (27) and is a member of the RNase H fold family of DNA junction-resolving enzymes (29, 30). Mrs1 is highly diverged from the RNase H fold superfamily and lacks several residues implicated in DNA cleavage.
In this work, we find that detectable in vitro splicing by the bI3 intron has become wholly dependent on both the intron-encoded bI3 maturase and the co-opted Mrs1 proteins. The bI3 maturase and Mrs1 bind the intron independently and assemble to form an active ribonucleoprotein catalyst of ≈420 kDa. Both splicing cofactor proteins have evolved such that they appear to have compromised the original endonuclease activities of their structural homologs.
Experimental Procedures
The bI3 Maturase, Mrs1, and bI3 RNA.
To circumvent extensive codon misrepresentation, an Escherichia coli expression construct for the mitochondrial bI3 maturase gene [strain 777–3A (19)] was cloned by total gene synthesis by using recursive PCR (31). The native mRNA encodes a strongly hydrophobic N-terminal peptide of 100 amino acids [http://www.cbs.dtu.dk/services/TMHMM-2.0 (32)], derived from both 5′-exon and intron (75 amino acids) sequences. Therefore, a gene for the conserved soluble maturase domain (850 bp, 272 amino acids, starting at the sequence MNNTI) was synthesized from 12 overlapping 120-nt oligos and two 25-nt amplification oligos in a two-step PCR reaction (pfu turbo, Stratagene) (Fig. 1B). The MRS1-coding sequence was generated by PCR from total yeast genomic DNA (strain DBY746). Coding sequences were confirmed by sequencing and expressed as N-terminal (His)10 fusions (pET19b, Novagen) in E. coli BL21(DE3)pLysS or BL21(DE3) cells, respectively. Pelleted cells from 1 L cultures were resuspended in 25 ml of either Mrs1 prep buffer [40 mM Hepes (pH 7.5)/650 mM NaCl/5 mM 2-mercaptoethanol/10% (vol/vol) glycerol] or maturase prep buffer [20 mM phosphate buffer (pH 8.0)/900 mM NaCl/5 mM 2-mercaptoethanol/10% (vol/vol) glycerol]. Cells were sonicated, and the lysate was cleared by centrifugation at 35,000 rpm (Beckman 60 Ti rotor) and mixed with 1.5 ml of Ni2+-NTA agarose slurry (Qiagen, Chatsworth, CA). Maturase-bound resin was washed twice with 10 ml of maturase prep buffer plus 2 mM imidazole (pH 8.0), then both proteins were washed with Mrs1 prep buffer with up to 60 mM imidazole (pH 8.0). Proteins were eluted in 5 ml of 40 mM Hepes (pH 7.5), 600 mM NaCl, 5 mM 2-mercaptoethanol, 20% (vol/vol) glycerol, and 250 mM imidazole (pH 8.0). Eluted fractions were dialyzed against 40 mM Hepes (pH 7.5), 400 mM NaCl, 5 mM DTT, and 50% (vol/vol) glycerol and stored at −20°C. Both proteins were judged greater than 95% homogenous by SDS/PAGE analysis and Coomassie blue staining. Protein concentrations were determined by UV spectroscopy (33). Mrs1 concentrations are reported in terms of the solution dimer, unless stated otherwise.
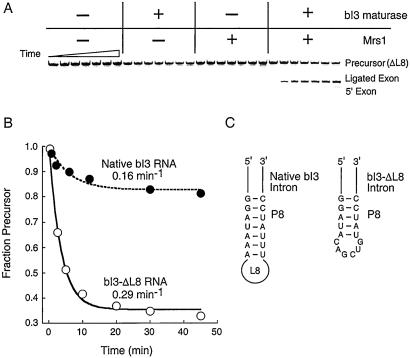
Intron precursor sequences for the native and ΔL8 constructs (15) were cloned into pTZ18U, and RNAs were generated by runoff transcription using T7 polymerase from HindIII-digested plasmid DNA (34). The bI3 RNA precursor spanned 10 nt of vector sequence, the entire 75-nt 5′-exon, the 1,752-nt intron, and 31 nt of the 3′-exon. Most experiments were performed with the bI3-ΔL8 construct (designed by R. Waring and coworkers), in which the 1,325-nt L8 loop is replaced by 6 nt (Fig. 2C).
Figure 2.
Reconstitution of an active bI3 ribonucleoprotein. (A) Detectable splicing by the bI3 intron requires the presence (+) of both the bI3 maturase and Mrs1. Reactions were monitored using a 5′-32P-end-labeled bI3-ΔL8 precursor and resolved by denaturing electrophoresis. Timepoints are 0, 2, 5, 10, 20, 30, and 45 min. (B) Comparative splicing by the native and bI3-ΔL8 intron RNAs, under conditions of saturating protein and pG. (C) Structure of the bI3-ΔL8 intron RNA. The 1,325-nt deletion lies in L8 and spans most of the ORF encoding the bI3 maturase.
bI3 RNA Splicing and Protein Binding.
Quantitative RNA splicing reactions (20–25 μl) were performed by using ≈1 nM 5′-32P-end-labeled RNA, 5 nM bI3 maturase, and 10 nM Mrs1 dimer. Mrs1 binding is coupled with maturase binding such that the K1/2 for binding is ≈2 nM in the presence of maturase. Thus, both Mrs1 and maturase were saturating in the splicing experiments reported here (Figs. 2 and 3A). Precursor RNA was incubated at 90°C for 1 min, placed on ice, and refolded at 36°C for 10 min in reaction buffer [40 mM Hepes (pH 7.5)/80 mM KOAc (pH 7.4)/20 mM MgCl2/1 mM DTT/10 μg/ml of BSA]. Proteins were added simultaneously and allowed to bind for 30 min at 36°C; control experiments showed that RNA splicing is not affected by longer preincubation times or by order of protein addition. Reactions were initiated by addition of pG to a final concentration of 3 mM, resolved by denaturing 8% polyacrylamide gels, and quantified with a PhosphorImager (Molecular Dynamics). Reaction rates were determined by a nonlinear least-squares fit: fraction precursor RNA = fe−kt + (1 − f), where k is the observed rate constant, and f is the fraction reactive RNA (typically 65–70%). For high-sensitivity splicing reactions (35), bI3 RNA concentrations were 50 nM, proteins were added in excess, and splicing was initiated by addition of [32P]-GTP (3,000 Ci/mmol) to 0.3 μM.
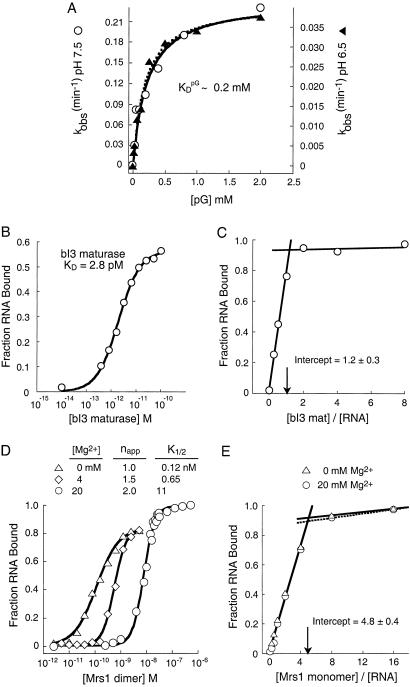
Figure 3.
Guanosine cofactor binding and protein assembly of the bI3 ribonucleoprotein. (A) Kinetic determination of pG binding. KM for pG was measured at pH 7.5 (open circles, left axis) and pH 6.5 (filled triangles, right axis). Curves were fit to kobs = kcat/(1 + KM/[pG]). (B) Equilibrium RNA binding by the bI3 maturase. (C) Stoichiometric binding by the bI3 RNA–bI3 maturase complex is consistent with formation of a 1:1 maturase–RNA complex at 20 mM Mg2+, [bI3 RNA] = 100 nM (≫KD). (D) Magnesium ion-dependent, cooperative assembly of Mrs1 with the bI3 group I intron RNA. Mrs1 dimer binding to the bI3 intron is strongly cooperative under optimal splicing conditions (circles, 20 mM Mg2+). In the absence of Mg2+, binding is noncooperative (triangles) and, at 4 mM Mg2+, transitional behavior is observed (diamonds). (E) Stoichiometric binding assay for the formation of the Mrs1–bI3 RNA complex at 0 mM (triangles) and 20 mM (circles) Mg2+; [bI3 RNA] = 100 nM (≫K1/2).
RNA-binding assays were performed by filter partitioning (34) and preincubating membranes in 40 mM Hepes (pH 7.5) and 80 mM KOAc (pH 7.4) for 30 min. Maturase and Mrs1 binding measurements contained 0.2 pM and 10 pM bI3-ΔL8 RNA, respectively, with the [BSA] supplemented to 30 μg/ml. Membranes were washed with reaction buffer before and after filtering, dried, and quantified. Maturase-binding experiments were analyzed by using a one-site binding model: fraction RNA bound = A ([P]/[P] + KD), where A is the total fraction bound RNA, [P] is the free protein concentration, and KD is the equilibrium dissociation constant. Mrs1 binding curves were analyzed by using fraction RNA bound = A ([P]n/[P]n + K1/2n), where n is the apparent Hill coefficient, and K1/2 is the [Mrs1] where one-half of the RNA is bound.
Sedimentation Equilibrium Analysis.
Experiments (XLA analytical ultracentrifuge; Beckman), were performed at 4°C until equilibrium was established (typically 16 h). Samples were analyzed in reaction buffer at 10 mM Mg2+ and substituting 5 mM 2-mercaptoethanol for DTT to achieve low absorbance offsets. Protein molecular weights were determined at 2, 4, and 8 μM at 7,000 and 10,000 rpm (detection at 280 nm); observed molecular masses were identical, within error. For RNA–protein complexes, 70 nM bI3-ΔL8 RNA was bound with (i) 420 nM Mrs1, (ii) 580 nM Mrs1, (iii) 100 nM bI3 maturase, or (iv) 360 nM Mrs1 and 100 nM bI3 maturase and spun at 5,000 and 6,500 rpm (detection at 260 nm). Meniscus depletion runs (45,000 rpm, 5 h) established sample background. Offsets were 0.05 or less and were corrected accordingly. Sedimentation profiles (absorbance as a function of radius) were fit (Origin XLA software, Beckman) to a version of the Lamm equation (equation 2 in ref. 36), assuming a single ideal species, a good assumption given the observed tight binding constants (Fig. 3), and calculating partial specific volumes from the weighted average of the RNA (0.537 g/cm3) and protein (0.730 g/cm3) components; solution density (ρ) was 1.008.
Maturase Endonuclease and MRS1 Junction-Resolving Assays.
DNA endonuclease assays (16, 23) contained 50 mM Hepes (pH 7.5), 80 mM KOAc, 1 mM DTT, 5 μg/ml of BSA, and 0, 10, or 50 mM MgCl2, or 10 or 50 mM MnCl2 at 37°C using 20 nM–2 μM maturase. Double-stranded DNA substrates were made from 63-nt complementary strands containing 45 nt of exon–exon junction sequence at their center; sense strand = 5′-ccgga attct tgtat cttga ttatg aggtg ggtt/c tcagt atcta accct ctaag gatcc gcg-3′ (exon junction indicated with a/). Control duplexes contained 19 altered base pairs at the exon–exon junction. Strands were individually 5′-32P-end-labeled, purified by denaturing electrophoresis, and annealed with a 1.5-fold excess of unlabeled complementary strand. Native gel electrophoresis confirmed that the radiolabeled strands annealed quantitatively to form duplexes. Synthetic 10-, 32-, and 51-mer oligodeoxynucleotides were used as markers for cleavage products. DNA junction-resolving assays with Mrs1 were performed exactly as described (ref. 27; by using the branch migrating junction jmb3 and the fixed junction 1).
Results
bI3 Intron Splicing Requires Both Mrs1 and bI3 Maturase Proteins.
The first step in group I intron splicing involves attack of the 3′-hydroxyl of a guanosine nucleophile at the 5′ end of the intron to cleave the exon–intron junction. In the second step, the new free 3′-OH from the 5′-exon attacks the 3′ splice site to yield ligated exons and the G-intron product (5). We monitored splicing of the bI3 group I intron by using two approaches. Reaction kinetics are most accurately followed using a 5′-32P-end-labeled RNA precursor and a pG nucleophile. To observe potentially inefficient self-splicing reactions, we also followed splicing by using the sensitive approach (35) of detecting addition of [32P]-GTP to unlabeled RNA.
In the absence of protein cofactors, splicing by both the native and bI3-ΔL8 introns is undetectable. Addition of either Mrs1 or bI3 maturase proteins alone at concentrations from 5 to 500 nM and in the presence of up to 25 mM MgCl2 also yielded no detectable splicing products, as judged by both radiolabeled intron (Fig. 2A) or [32P]-GTP addition assays. However, assembly of the quaternary complex consisting of the bI3 intron RNA, Mrs1, the bI3 maturase, and pG results in efficient splicing and near quantitative conversion of all RNAs that undergo 5′ splice site cleavage into the ligated exon product (Fig. 2A). Formation of the ligated exon product occurred at the same rate as reaction of the RNA precursor, indicating that guanosine attack at the 5′ splice site is limiting for the overall two-step splicing reaction.
Using excess protein and saturating pG, we compared splicing reactions for the native intron with that of a simplified intron in which the 1,325-nt loop in L8 is deleted (Figs. 1A, 2 B and C). The bI3-ΔL8 RNA spliced at a rate of 0.29 min−1 and, significantly, over 65% of the RNA reacts over 30 min. The native bI3 intron splices at a rate of 0.16 min−1 and less than 20% of the RNA reacts over this period (Fig. 2B). The difference in reactive fractions for the native and simplified RNAs probably reflects a greater extent of misfolding in the much larger native RNA.
Splicing by the RNA alone at [Mg2+] up to 100 mM was undetectable by the sensitive guanosine addition assay (data not shown); we estimate that these reactions must be slower than 0.00002 min−1 to avoid detection. Thus, addition of both Mrs1 and the bI3 maturase yields a 104-fold, or greater, rate enhancement over self-splicing or splicing by incompletely formed complexes.
We determined the Michaelis–Menten constant
(KM) for the interaction of the pG
substrate with the bI3 ribonucleoprotein ternary complex by following
the rate of splicing as a function of [pG]. The
K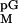 for the bI3 intron was 180 ± 20
μM (Fig. 3A, open circles).
KM is given by
(k
for the bI3 intron was 180 ± 20
μM (Fig. 3A, open circles).
KM is given by
(k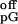 +
kcat/k
+
kcat/k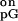 ),
making the reasonable assumption that there are no hidden intermediates
for association of this substrate with the preformed ternary complex.
If kcat is slow relative to pG
dissociation (k
),
making the reasonable assumption that there are no hidden intermediates
for association of this substrate with the preformed ternary complex.
If kcat is slow relative to pG
dissociation (k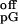 ), then
KM would reflect equilibrium binding
by pG (34, 37).
), then
KM would reflect equilibrium binding
by pG (34, 37).
We determined K at pH 6.5, which
decreased the observed kcat by 6- to
10-fold (Fig. 3A and data not shown), and found the
KM to be equal to 210 ± 60 μM,
identical, within error, to the value at pH 7.5 (compare closed and
open symbols in Fig. 3A). Thus, slowing the chemical step
after binding has no effect on KM,
supporting the assignment that the pG substrate binds the bI3
holoribonucleoprotein with a KD of
≈0.2 mM.
at pH 6.5, which
decreased the observed kcat by 6- to
10-fold (Fig. 3A and data not shown), and found the
KM to be equal to 210 ± 60 μM,
identical, within error, to the value at pH 7.5 (compare closed and
open symbols in Fig. 3A). Thus, slowing the chemical step
after binding has no effect on KM,
supporting the assignment that the pG substrate binds the bI3
holoribonucleoprotein with a KD of
≈0.2 mM.
The bI3 Maturase Assembles as a Monomer with the bI3 Group I Intron.
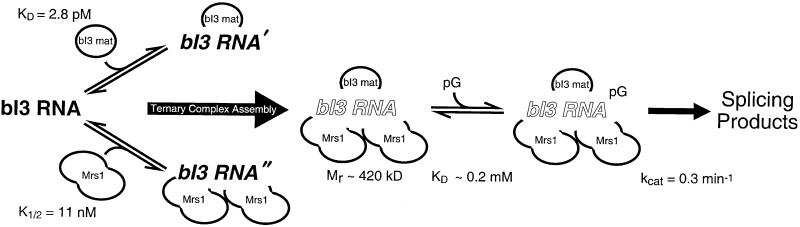
Sedimentation equilibrium analysis demonstrated that the bI3 maturase is a monomer in solution (Table 1). We determined the equilibrium-binding constant for the maturase–RNA complex by using a filter partitioning assay; the bI3 maturase binds very tightly to the bI3 intron with a KD of 2.8 pM (Fig. 3B). We established the ratio of maturase to RNA in the complex using a stoichiometric binding assay in which the [RNA] was 100 nM, or much greater than the KD (Fig. 3C). The inflection point occurs at a protein/RNA ratio of 1.2 ± 0.3, consistent with formation of a bimolecular RNA–bI3 maturase complex.
Table 1.
The bI3 holoribonucleoprotein and its component masses determined by sedimentation equilibrium
| RNA | bI3 maturase | Mrs1 | Observed Mr, kDa* | Calculated Mr, kDa |
|---|---|---|---|---|
| + | 191 ± 12 | 198 | ||
| + | 35 ± 3 | 35 | ||
| + | 92 ± 4 | 90† | ||
| + | + | 220 ± 12 | 233 | |
| + | + | 372 ± 10 | 378‡ | |
| + | + | + | 420 ± 10 | 413‡ |
Error estimates are reported as standard deviations from three or more independent data sets.
Assuming Mrs1 (Mr = 45 kDa) is a dimer.
Assuming two Mrs1 dimers bind the bI3 RNA.
Two Mrs1 Dimers Assemble Cooperatively with the bI3 Group I Intron.
The molecular mass of free Mrs1, determined by sedimentation equilibrium, is 92 ± 4 kDa, exactly twice the calculated mass of a Mrs1 monomer (Table 1). Thus, we infer free Mrs1 is a dimer in solution. Under conditions optimal for splicing (20 mM Mg2+), The Mrs1 dimer binds the bI3 intron in a strongly cooperative manner. The Mrs1 dimer concentration required to bind half of the RNA at equilibrium, the K1/2, is 11 nM, and the apparent Hill coefficient is 2.0 (Fig. 3D; circles).
We observed marked differences in the mode of Mrs1 dimer assembly as a function of [Mg2+]. In the absence of Mg2+, the K1/2 is tighter at 0.12 nM and binding is distinctly noncooperative; the apparent Hill coefficient is 1.0 (Fig. 3D; triangles). At the intermediate Mg2+ concentration of 4 mM, we observe transitional behavior such that the K1/2 is 0.65 nM, and the apparent Hill coefficient is 1.5 (Fig. 3D, diamonds). Cooperativity in Mrs1 dimer-binding plateaus by 8 mM Mg2+, where the apparent Hill coefficient is also 2.0 (data not shown). We determined the ratio of Mrs1 to RNA in the complex using a stoichiometric binding assay, where all added protein should bind quantitatively to the RNA. The inflection point at 4.8 ± 0.4 is approximately consistent with binding by two Mrs1 dimers (four Mrs1 monomers in total) at both 0 mM and 20 mM Mg2+ (Fig. 3E; triangles and circles, respectively).
Thus, there exist two Mrs1-binding sites on the bI3 RNA, each of which binds one Mrs1 dimer. In the absence of Mg2+, two Mrs1 dimers bind independently; in contrast, under optimal splicing conditions (20 mM Mg2+), two Mrs1 dimers assemble on the bI3 intron RNA through a strongly linked binding event.
The Functional bI3 Group I Intron Assembles to a 420-kDa Ribonucleoprotein.
We monitored stepwise assembly of the bI3 ribonucleoprotein by using analytical ultracentrifugation. In sedimentation equilibrium experiments, the position of a molecule in the centrifuge or complex is sensitive to the overall molecular mass such that larger complexes achieve equilibrium at larger cell radii. For tight binding stoichiometric complexes (Fig. 3), absolute molecular masses can be obtained directly by global fitting of the observed absorbance as a function of radial position (36).
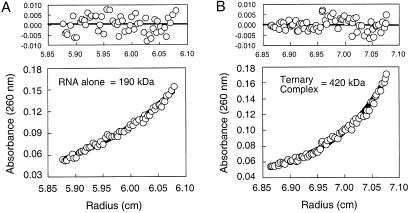
The bI3-ΔL8 RNA sediments with a molecular mass of 191 ± 12 kDa (Fig. 4A; Table 1). This value is identical, within error, to the calculated mass for this RNA, confirming that the RNA is a well behaved monomer in these experiments. The bI3 maturase also sediments as a monomer (see line 2 of Table 1). In contrast, Mrs1 sediments with a molecular mass of 92 ± 4 kDa, exactly twice the calculated mass (45 kDa) for the protein monomer. Thus, Mrs1, like its homolog Cce1 (27, 38), is a solution dimer.
Figure 4.
Macromolecular masses of (A) the bI3-ΔL8 RNA and (B) the bI3-ΔL8 RNA–Mrs1–bI3 maturase ternary complex determined by sedimentation equilibrium. Sedimentation profiles were fit to the Lamm equation (36), assuming a single species (A and B Lower). Partial specific volumes for the RNA alone and ternary complex were calculated to be 0.537 and 0.642 g/cm3 and samples were spun at 6,500 and 5,000 rpm, respectively. A and B Upper show the (small, random) fit residuals.
We could follow assembly of the bI3 holoribonucleoprotein complex from its RNA, maturase, and Mrs1 components in a straightforward way because protein absorbance is negligible compared with RNA absorbance at 260 nm and by assuming that partial specific volumes for the protein–RNA complexes are a weighted average of their protein and RNA components.
We determined the mass of the bimolecular bI3 maturase–ΔL8 RNA complex by using an excess of (weakly absorbing) maturase to be 220 ± 12 kDa (Table 1). This value is consistent with formation of a 1:1 maturase–RNA complex (calculated mass = 233 kDa) and provides independent support for the conclusion, described above, that the bI3 maturase recognizes the bI3 RNA as a monomer.
To determine rigorously the stoichiometry of the Mrs1-bound RNA species, we mixed Mrs1 with the bI3-ΔL8 RNA at 6:1 and 8:1 molar ratios. Complexes formed at both protein–RNA ratios, detected by the RNA absorbance at 260 nm, sediment to give an average molecular mass of 372 ± 10 kDa (Table 1). This value is identical to the calculated mass for a (Mrs1)4–bI3 RNA complex of 378 kDa. This observed molecular mass further supports the conclusion drawn above: Mrs1 binds as two dimers to the bI3 group I intron RNA.
To form the ternary complex, we incubated one equivalent of the bI3 maturase and five equivalents of Mrs1 with the bI3 intron RNA. The radial absorbance profile was fit to a model for a single ideal species and gave a molecular mass of 420 ± 10 kDa (Fig. 4B; Table 1). The molecular mass determined by sedimentation equilibrium is in outstanding agreement with the calculated mass for this complex (413 kDa; Table 1). The bI3 holoribonucleoprotein comprises the bI3 RNA, a maturase monomer, and two Mrs1 dimers.
Both the bI3 Maturase and Mrs1 Have Lost Functions Characteristic of Their Close Homologs.
ORFs for LAGLIDADG site-specific endonucleases, homologous with the bI3 maturase, appear to have colonized group I introns on multiple occasions (23, 24). Using oligodeoxynucleotide duplexes spanning 45 nt at the cyt b exon 3–exon 4 junction, we attempted to detect latent DNA endonuclease activity under conditions that yield efficient cleavage in other systems (16, 23). We failed to detect specific cleavage for duplexes radiolabeled at either the coding or noncoding strands, in the presence of Mg2+ or Mn2+, or at high concentrations of bI3 maturase (data not shown). After 5 h, we did observe a low (≤1%) level of cleavage, but this activity was observed for both duplex and single-stranded DNAs and was not specific for the correct junction sequence.
Mrs1 is homologous to the S. cerevisiae Cce1 protein (27, 28, 30). Under conditions where Cce1 efficiently cleaves DNA junctions (27), we were unable to detect Holliday junction cleavage with either migrating or fixed DNA junctions (data not shown). Thus, both the bI3 maturase and Mrs1 appear to have lost or are severely compromised in the DNA cleavage activity of their structural homologs.
Discussion
Assembly of the bI3 Group I Intron Ribonucleoprotein.
Our mechanism for assembly and splicing by the bI3 ribonucleoprotein is outlined in Fig. 5. Self-splicing by the bI3 intron is undetectable, suggesting that the RNA does not achieve a catalytically active conformation in the absence of protein cofactors. The bI3 maturase and Mrs1 proteins bind independently to the RNA and with distinct modes of assembly. The maturase binds as a monomer, whereas two Mrs1 dimers assemble with strong cooperativity on the bI3 RNA. The ternary complex has a molecular mass of 420 kDa and binds the guanosine nucleophile with a KD of ≈0.2 mM. The active ribonucleoprotein quaternary complex splices with a kcat of 0.3 min−1. The reaction is particularly efficient, in that nearly all of the cleaved 5′ exon is converted to ligated exon product. Guanosine attack at the 5′ exon is the rate-determining step for the overall splicing reaction.
Figure 5.
Assembly of the bI3 ribonucleoprotein catalyst comprised of the bI3 RNA, bI3 maturase, Mrs1, and the guanosine nucleotide.
Evolution in Splicing of the bI3 Group I Intron.
Detectable splicing by the bI3 intron depends on two protein facilitators in vitro, and the evolutionary origin of each protein is distinct. Moreover, both the bI3 maturase and Mrs1 appear to have lost or to be compromised in the DNA nuclease-based functions that characterize their closest homologs.
Proteins containing LAGLIDADG motifs are derived from a family of DNA endonucleases, whose ORFs appear to have invaded group I introns on multiple occasions (22–24). A subset of LAGLIDADG proteins additionally function as RNA maturases; this RNA binding and splicing facilitation activity is thought to have evolved from the existing DNA-binding function (Fig. 6A) (16, 22, 39). The maturase domain (Fig. 1B), which functions as a splicing cofactor, is translated entirely from the intron RNA itself (Figs. 1 and 2A), similar to the I-AniI (16) and bI4 (40) maturases. Thus, the functional portion of the bI3 maturase has had the opportunity to coevolve with the bI3 intron.
Figure 6.
Evolutionary series for the bI3 maturase (A) and Mrs1 (B). Activities associated with each protein or class of proteins are emphasized at the left. Branched arrows indicate there exist other outcomes (not shown) at each step of diversification.
Several lines of evidence suggest that the maturase has lost its original DNA endonuclease activity. First, DNA endonuclease activity is a prerequisite for intron mobility, and the bI3 intron does not home into intronless genomes (41). Second, despite significant effort, we do not detect cleavage of cyt b exon 3–exon 4 DNA junctions by purified bI3 maturase. Finally, structural analysis of related DNA binding proteins indicates that two LAGLIDADG motifs form an interface consisting of parallel α-helices and bind Mg2+ at their carboxyl termini (42–44). The second aspartyl residue (underlined) in the motif is essential for catalysis (45) and coordinates directly a catalytic Mg2+ ion (43) in homologous endonucleases. This residue is a structurally nonconservative lysyl residue in the C-terminal motif (P2) in the bI3 maturase (Fig. 1B, strain 777–3A). A plausible evolutionary series for the bI3 maturase is shown in Fig. 6A.
The closest homolog to Mrs1 in the current databases is the S. cerevisiae DNA junction-resolving enzyme Cce1. Cce1 is a member of the RNase H fold superfamily of dimeric resolving enzymes, which share a set of four conserved sequence motifs despite having low sequence similarity overall (Fig. 6B) (29, 30). These four motifs correspond to residues at the active site as visualized in the structure of one member of this family, RuvC (29, 30).
Although Mrs1 shares a much higher level of sequence identity with Cce1 than with any of the resolving enzymes, regions conserved between Cce1 and Mrs1 do not correspond to the regions most highly conserved among the DNA resolving enzymes. Motif 1 (29, 30) is missing altogether, and the other motifs are less strongly conserved than in the highly divergent resolving enzymes that make up the RNase H fold superfamily. Conversely, sequences strongly conserved between Cce1 and Mrs1 correspond to regions not conserved among the resolving enzymes. The likely explanation for these observations is that the resolving enzymes have retained essential catalytic residues over a very long period of evolutionary divergence because of their conserved function. In contrast, Cce1 and Mrs1 have diverged relatively recently, consistent with the phylogenetic restriction of Mrs1 to the genus Saccharomyces and thus have a higher overall sequence identity. The poor conservation of important catalytic residues in Mrs1, the lack of detectable junction-resolving activity (see above), and the observation that Mrs1 functions as a dimer of dimers (Fig. 3D; Table 1) all emphasize that Mrs1 has evolved to fulfill a new role in the cell (Fig. 6B).
Group I Introns as Tools for Understanding the Pervasiveness of Ribonucleoproteins.
The evolution and present day dispersal of group I introns is complex and likely reflects both the retention of some ancient introns and also significant horizontal transfer of introns (reviewed in refs. 9, 21, and 22). However, no matter what their evolutionary origin, analysis of intron encoded maturase activities and the co-opting of other cellular proteins to function as group I intron splicing cofactors provides an authentic opportunity to evaluate the specific mechanistic and structural advantages and opportunism in evolving from an RNA catalyst to an obligatory ribonucleoprotein. There are diverse evolutionary series (Fig. 6) by which once RNA-based reactions have become protein dependent, a frequent occurrence in the transition from any RNA world to modern metabolism. Principles derived from the assembly and splicing of the bI3 group I intron (Fig. 5) and from studies in comparative ribonucleoprotein assembly and catalysis (46) are likely to inform evolution of other ribonucleoprotein complexes, especially cases where RNA-centered functions have been enhanced or obscured by the accumulation of multiple protein cofactors.
Acknowledgments
We thank Richard Waring (Temple University) for generous gifts of bI3 intron clones; D. M. J. Lilley for support of early work on Mrs1; A. Tripathy (University of North Carolina Macromolecular Interactions Facility) for assistance with equilibrium sedimentation experiments; J. Lazowska, A. Longo, and K. Bjornson for discussion; and L. Blackwell for comments on the manuscript. K.M.W. is a Searle Scholar in the Biomedical Sciences. M.F.W. is a Royal Society University Research Fellow. D.M.O. received a fellowship from the Fundação de Amparo à Pesquisa do Estado de São Paulo. This work was supported by the National Institutes of Health (K.M.W.) and by a Human Frontiers Science Program Long Term Fellowship (G.S.B.).
Abbreviation
- pG
guanosine 5′-monophosphate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.McKay D B, Wedekind J E. In: The RNA World. 2nd Ed. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Press; 1999. pp. 265–286. [Google Scholar]
- 2.Joyce G F. In: The RNA World. 2nd Ed. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Press; 1999. pp. 687–690. [Google Scholar]
- 3.Gutell R R, Power A, Hertz G Z, Putz E J, Stormo G D. Nucleic Acids Res. 1992;20:5785–5795. doi: 10.1093/nar/20.21.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michel F, Westhof E. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 5.Cech T R. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 6.Kurz J C, Fierke C A. Curr Opin Chem Biol. 2000;4:553–558. doi: 10.1016/s1367-5931(00)00131-9. [DOI] [PubMed] [Google Scholar]
- 7.Yusupov M M, Yusupova G Z, Baucom A, Lieberman K, Earnest T N, Cate J H, Noller H F. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 8.Weeks K M. Curr Opin Struct Biol. 1997;7:336–342. doi: 10.1016/s0959-440x(97)80048-6. [DOI] [PubMed] [Google Scholar]
- 9.Lambowitz A M, Caprara M G, Zimmerly S, Perlman P S. In: The RNA World. 2nd Ed. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Press; 1999. pp. 451–486. [Google Scholar]
- 10.Golden B L, Gooding A R, Podell E R, Cech T R. Science. 1998;282:259–264. doi: 10.1126/science.282.5387.259. [DOI] [PubMed] [Google Scholar]
- 11.Green R, Szostak J W. Science. 1992;258:1910–1915. doi: 10.1126/science.1470913. [DOI] [PubMed] [Google Scholar]
- 12.Jaeger L, Michel F, Westhof E. Nucleic Acids Mol Biol. 1996;10:33–51. [Google Scholar]
- 13.Lehnert V, Jaeger L, Michel F, Westhof E. Chem Biol. 1996;3:993–1009. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
- 14.Lisacek F, Diaz Y, Michel F. J Mol Biol. 1994;235:1206–1217. doi: 10.1006/jmbi.1994.1074. [DOI] [PubMed] [Google Scholar]
- 15.Hur M, Geese W J, Waring R B. Curr Genet. 1997;32:399–407. doi: 10.1007/s002940050294. [DOI] [PubMed] [Google Scholar]
- 16.Ho Y, Kim S J, Waring R B. Proc Natl Acad Sci USA. 1997;94:8994–8999. doi: 10.1073/pnas.94.17.8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costa M, Michel F. EMBO J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaeger L, Michel F, Westhof E. J Mol Biol. 1994;236:1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 19.Lazowska J, Claisse M, Gargouri A, Kotylak Z, Spyridakis A, Slonimski P P. J Mol Biol. 1989;205:275–289. doi: 10.1016/0022-2836(89)90341-0. [DOI] [PubMed] [Google Scholar]
- 20.Dalgaard J Z, Klar A J, Moser M J, Holley W R, Chatterjee A, Mian I S. Nucleic Acids Res. 1997;25:4626–4638. doi: 10.1093/nar/25.22.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dujon B. Gene. 1989;82:91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- 22.Lambowitz A M, Belfort M. Annu Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- 23.Loizos N, Tillier E R, Belfort M. Proc Natl Acad Sci USA. 1994;91:11983–11987. doi: 10.1073/pnas.91.25.11983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall P, Davis T B, Lemieux C. Eur J Biochem. 1994;220:855–859. doi: 10.1111/j.1432-1033.1994.tb18688.x. [DOI] [PubMed] [Google Scholar]
- 25.Kreike J, Schulze M, Pillar T, Korte A, Rodel G. Curr Genet. 1986;11:185–191. doi: 10.1007/BF00420605. [DOI] [PubMed] [Google Scholar]
- 26.Kreike J, Schulze M, Ahne F, Lang B F. EMBO J. 1987;6:2123–2129. doi: 10.1002/j.1460-2075.1987.tb02479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White M F, Lilley D M J. J Mol Biol. 1996;257:330–341. doi: 10.1006/jmbi.1996.0166. [DOI] [PubMed] [Google Scholar]
- 28.Wardleworth B N, Kvaratskhelia M, White M F. J Biol Chem. 2000;275:23725–23728. doi: 10.1074/jbc.M002612200. [DOI] [PubMed] [Google Scholar]
- 29.Aravind L, Makarova K S, Koonin E V. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lilley D M J, White M F. Proc Natl Acad Sci USA. 2000;97:9351–9353. doi: 10.1073/pnas.97.17.9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheeler V C, Prodromou C, Pearl L H, Williamson R, Coutelle C. Gene. 1996;169:251–255. doi: 10.1016/0378-1119(95)00812-8. [DOI] [PubMed] [Google Scholar]
- 32.Sonnhammer E L L, von Heijne G, Krogh A. In: Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. Glasgow J, Littlejohn T, Major F, Lathrop R, Sankoff D, Sensen C, editors. AAAI Press; 1998. pp. 175–182. [PubMed] [Google Scholar]
- 33.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 34.Weeks K M, Cech T R. Biochemistry. 1995;34:7728–7738. doi: 10.1021/bi00023a020. [DOI] [PubMed] [Google Scholar]
- 35.Gott J M, Shub D A, Belfort M. Cell. 1986;47:81–87. doi: 10.1016/0092-8674(86)90368-5. [DOI] [PubMed] [Google Scholar]
- 36.McRorie D K, Voelker P J. Self-Associating Systems in the Analytical Ultracentrifuge. Fullerton, CA: Beckman; 1993. [Google Scholar]
- 37.McConnell T S, Cech T R, Herschlag D. Proc Natl Acad Sci USA. 1993;90:8362–8366. doi: 10.1073/pnas.90.18.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kvaratskhelia M, George S J, Cooper A, White M F. Biochemistry. 1999;38:16613–16619. doi: 10.1021/bi9921788. [DOI] [PubMed] [Google Scholar]
- 39.Szczepanek T, Lazowska J. EMBO J. 1996;15:3758–3767. [PMC free article] [PubMed] [Google Scholar]
- 40.Banroques J, Perea J, Jacq C. EMBO J. 1987;6:1085–1091. doi: 10.1002/j.1460-2075.1987.tb04862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meunier B, Tian G L, Macadre C, Slonimski P P, Lazowska J. In: Structure, Function and Biogenesis of Energy Transfer Systems. Quagliariello E, Papa S, Palmieri F, Saccone C, editors. Amsterdam: Elsevier; 1990. pp. 169–174. [Google Scholar]
- 42.Jurica M S, Monnat R J, Jr, Stoddard B L. Mol Cell. 1998;2:469–476. doi: 10.1016/s1097-2765(00)80146-x. [DOI] [PubMed] [Google Scholar]
- 43.Chevalier B S, Monnat R J, Jr, Stoddard B L. Nat Struct Biol. 2001;8:312–316. doi: 10.1038/86181. [DOI] [PubMed] [Google Scholar]
- 44.Silva G H, Dalgaard J Z, Belfort M, Van Roey P. J Mol Biol. 1999;286:1123–1136. doi: 10.1006/jmbi.1998.2519. [DOI] [PubMed] [Google Scholar]
- 45.Gimble F S, Stephens B W. J Biol Chem. 1995;270:5849–5856. doi: 10.1074/jbc.270.11.5849. [DOI] [PubMed] [Google Scholar]
- 46.Webb A E, Rose M A, Westhof E, Weeks K M. J Mol Biol. 2001;309:1087–1100. doi: 10.1006/jmbi.2001.4714. [DOI] [PubMed] [Google Scholar]