Abstract
Assembly of functional Ig and T cell receptor genes by V(D)J recombination depends on site-specific cleavage of chromosomal DNA by the RAG1/2 recombinase. As RAG1/2 action has mechanistic similarities to DNA transposases and integrases such as HIV-1 integrase, we sought to determine how integrase inhibitors of the diketo acid type would affect the various activities of RAG1/2. Both of the inhibitors we tested interfered with DNA cleavage and disintegration activities of RAG1/2, apparently by disrupting interaction with the DNA motifs bound specifically by the recombinase. The inhibitors did not ablate RAG1/2's transposition activity or capture of nonspecific transpositional target DNA, suggesting this DNA occupies a site on the recombinase different from that used for specific binding. These results further underscore the similarities between RAG1/2 and integrase and suggest that certain integrase inhibitors may have the potential to interfere with aspects of B and T cell development.
Keywords: V(D)J recombination‖transposition‖immune development
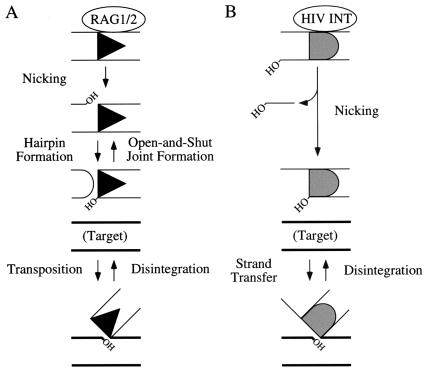
V(D)J recombination is an essential step in the development of the specific immune system. This process involves a rearrangement that joins gene segments present in multiple copies at the Ig and T cell receptor loci and ultimately leads to assembly of the functional coding regions (reviewed in ref. 1). Rearrangement is initiated by the lymphoid-specific recombinase composed of the RAG1 and RAG2 proteins (2). The RAG1/2 complex specifically recognizes the recombination signal sequences (RSSs) flanking the coding segments and cuts the DNA at these sites (2). RSSs are composed of conserved heptamer and nonamer sequence motifs separated by spacers of either 12 or 23 bases, and recombination most often results in the fusion of coding sequences flanked by RSSs with different spacer lengths. RAG1/2-mediated cleavage occurs in two steps (Fig. 1A). RAG1/2 first introduces a single-strand nick between the RSS and coding DNA. The 3′ hydroxyl at the end of the coding DNA then attacks the opposite strand in a direct transesterification reaction to produce a hairpin on the coding DNA (2–4). Transesterification takes place within a synaptic complex of the two RSSs held together by RAG1/2 (not shown). After cleavage, the recombinase continues to hold the cleaved RSS ends in this complex (5–7) for an extended period before the RSSs are joined to one another (8), whereas joining of coding ends is more rapid (8). Both types of ends are joined by a set of nonhomologous end-joining DNA repair factors (9, 10).
Figure 1.
Parallels between the biochemical activities of the RAG1/2 recombinase and HIV-1 integrase. (A) RAG1/2 binds to RSS sequences (black triangle) and catalyzes hydrolysis to introduce a nick between the RSS and flanking coding DNA (Nicking). The 3′ hydroxyl (OH) left on coding DNA by this reaction then attacks the opposite strand, leaving a hairpin at the end of coding DNA (Hairpin Formation) and a free 3′ OH at the end of the blunt RSS. The blunt RSS can attack nonspecific target DNA (thick lines; Transposition). Both hairpin formation and transposition are apparently reversible (Open-and-Shut Joint Formation and Disintegration). (B) HIV-1 integrase binds to the long terminal repeats (LTR; gray semicircle) near the ends of viral cDNA. Nicking leaves a free 3′ OH at the end of the LTR (Nicking), which can then attack target DNA (Strand Transfer). Strand transfer is reversible (Disintegration).
After cleavage, RAG1/2 can perform additional reactions that would perturb the normal joining process (Fig. 1A). RAG1/2 can carry out transposition in vitro by capturing target DNA of nonspecific sequence in the RAG1/2–RSS complex and then using the RSS ends to attack it (11, 12). This reaction also proceeds by direct transesterification but in this case results in strand transfer instead of hairpin formation. RAG1/2 has also been shown in vitro to efficiently detach the RSS from the strand transfer product created by transposition (13), in a reaction known as disintegration. Alternatively, RSS ends can reattack coding ends to form so-called “open-and-shut” or “hybrid” joints (14). Such joints are found in cells that undergo V(D)J recombination and represent a specialized form of RAG1/2 strand transfer related to those leading to transposition.
The similarities among RAG1/2, bacterial transposases, and retroviral integrases are striking. During HIV-1 integration, HIV-1 integrase first processes the viral cDNA by introducing a nick near each of the 3′ ends (Fig. 1B; one end is shown); it then catalyzes a direct transesterification reaction by which the viral cDNA becomes fused to chromosomal DNA (reviewed in ref. 15). Like RAG1/2, integrase can catalyze the reversal of this reaction, disintegration (16). Both RAG1/2 and integrase can mediate alcoholysis during the nicking step of their respective reactions (4), and all proteins in this class are thought to share a common active site motif, including critical acidic amino acid residues that form the metal-binding site (refs. 17 and 18 and refs. therein). These observations suggest the possibility that inhibitors targeted against integrase may also inhibit RAG1/2 and may possibly interfere with its role in immune development.
Several groups have tried to develop integrase inhibitors as drugs for treatment of HIV-1 infection, and many examples of compounds that inhibit integrase in vitro have been reported (19). Although some of the compounds possess antiviral activity in cell-based assays, the majority of the earlier reported compounds are cytotoxic because of the lack of selectivity for integrase in vivo. However, the recently described diketo acids represent the first inhibitors of HIV-1 integrase that demonstrate selectivity for the strand transfer step of the integration reaction (20, 21). Diketo acids exhibit antiviral activity in cell culture, and the resistant viruses generated in these experiments are found to have mutations only in the integrase gene, providing evidence that the inhibitors are selective for integrase. One of these compounds was also recently cocrystallized with the HIV-1 integrase core domain, confirming that it binds to important residues in the integrase active site (20).
We have tested two diketo acid integrase inhibitors on RAG1/2 activity in vitro. Both compounds inhibited cleavage by RAG1/2, although one was far more effective than the other. As with HIV-1 integrase, these compounds primarily inhibit DNA binding. However, whereas with integrase it is the binding of nonspecific target DNA that is most sensitive, with RAG1/2 the binding to RSS sequences is disrupted.
Materials and Methods
Proteins.
Murine RAG1 (amino acids 384-1008) and RAG2 (amino acids 1–387) fused at their amino termini to maltose-binding protein and at their carboxy termini to polyhistidine tails and myc epitopes were coexpressed in Sf9 cells and purified as previously described (2). Recombinant high-mobility group 1 (HMG) protein (amino acids 1–163) was expressed and purified as previously described (7). Tn10 transposase was a gift from Murray Junop of this laboratory. Deoxynucleotidyl transferase was purchased from GIBCO/Life Technologies (Rockville, MD), and T4 polynucleotide kinase and all restriction enzymes were purchased from New England Biolabs.
DNA Substrates.
Oligonucleotides were purchased from Oligos Etc. (Guilford, CT) and were gel purified before use. The oligonucleotides were either labeled on the 3′ terminus by using terminal deoxynucleotidyl transferase and α-32P-cordycepin (NEN Life Sciences) or on the 5′ terminus by using T4 polynucleotide kinase and γ-[32P]ATP (NEN Life Sciences). The structure and preparation of the cleavage substrate composed of oligonucleotide pairs DAR39/DAR40 (12 RSS) and DG61/DG62 (23 RSS), the precleaved RSS transposition donors composed of oligonucleotide pairs DG9/DG10 (12 RSS) and DG2/DG4 (23 RSS), and the disintegration substrate have been described previously (2, 12, 13). Labeled substrates were purified free of unincorporated radioisotope by using QUANTPROBE G-50 microcolumns. For RAG1/2-mediated transposition reactions, either cesium chloride-purified pBR322 DNA or oligonucleotide MM565 (5′-AATCCATAGGCAGTACTGTCGACAGGCCAATGCGAGCGCA) annealed to its complement, MM566, was used as a target.
Biochemical Assays.
Buffer components and procedures for RAG1/2-mediated cleavage, transposition, target capture, and disintegration were previously described (12, 13) with modification as described in the appropriate figure legends. RSS binding by RAG1/2 was detected by mobility-shift assay as previously described (7). DMSO (10%) was included in all reactions unless otherwise indicated. Reactions were carried out in 10-μl volumes with 450 ng of RAG1/2 (present in a ratio of ≈2:1) and 50 ng of HMG1. FPLC-purified integrase inhibitors designated here as p8 [5CITEP (20)] and p10 [L-708,906 (21)] were included at the concentrations indicated. These compounds were synthesized at the National Institutes of Health (Bethesda, MD).
Other.
All quantification of gels was by phosphor-storage autoradiography by using Molecular Dynamics TYPHOON 8600 and Molecular Dynamics IMAGEQUANT 5.1 software.
Results
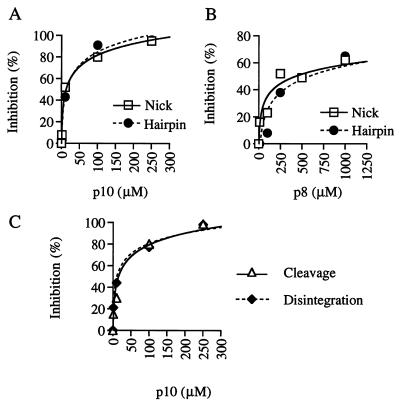
Two related diketo acid compounds previously found to inhibit HIV-1 integrase, designated here as p8 [5CITEP (20)] and p10 [L-708,906 (21)], were examined for their ability to inhibit RAG1/2-mediated activities in vitro. In these reactions, RAG1/2, HMG1, labeled 12 RSS substrate, and unlabeled 23 RSS were first combined, Ca2+ was added to allow stable RAG1/2–RSS complex formation, and then Mg2+ was added to initiate cleavage. When added before Ca2+, both compounds inhibited the nicking and transesterification steps of cleavage (Fig. 2 A and B). The compound designated p10, with an IC50 of 20 μM, was at least 10 times more effective than p8. Although the remaining experiments presented herein focus on p10, similar results with regard to the various RAG1/2 activities were observed by using ≈10-fold higher concentrations of p8. As with cleavage, p10 inhibited RAG1/2-mediated disintegration of RSS from target DNA when it was added to the reaction before Ca2+ (Fig. 2C; transposition is discussed below). Under these conditions, the apparent IC50s for cleavage and disintegration were indistinguishable.
Figure 2.
Effect of HIV integrase inhibitors on RAG1/2 biochemical activities. The effect of p10 (A) and p8 (B) on nicking and transesterification (hairpin formation) was measured in oligonucleotide cleavage assays performed as described in Materials and Methods. Reaction mixtures were incubated for 30 min after the addition of Mg2+. (C) The effect of p10 on cleavage and disintegration was determined as described in Materials and Methods. All reactions included 10% DMSO. In all cases, inhibitor was added before divalent metal cation. Percent inhibition relative to solvent control is shown.
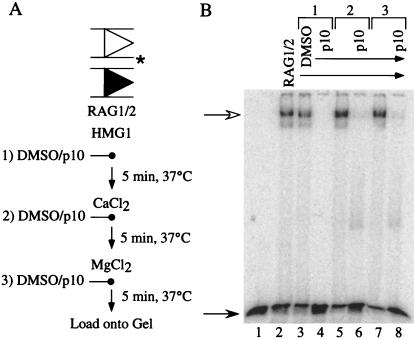
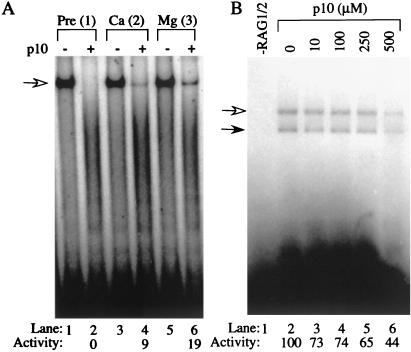
The parallel effects of p10 on these activities suggested it might be acting at a common step such as DNA binding. Indeed, p10 disrupted interactions between RAG1/2 and RSS-containing DNA. We performed mobility-shift assays to examine the effect of p10 on RSS binding. Under these conditions in which both 12 and 23 RSSs were present, the primary complex formed in the presence of RAG1/2, HMG1, and divalent cation was the synaptic complex with RAG1/2 bound to both RSSs (Fig. 3B, lane 2, and data not shown). When p10 was added to the reaction mixture before addition of Ca2+, RAG1/2 was prevented from binding to the labeled substrate (Fig. 3B, lane 4). p10 was also able to disrupt RAG1/2–RSS complexes formed in the presence of Ca2+ or Ca2+ plus Mg2+, resulting in faster-migrating smeared bands (Fig. 3B, lanes 6 and 8; these bands are very faint). In these reactions, p10 or solvent was added immediately after Ca2+ (lanes 5 and 6) or Mg2+ (lanes 7 and 8). These data indicated that p10 could bind to and alter a complex of RAG1/2 bound to RSS containing DNA so that it was no longer stable during the mobility-shift assay.
Figure 3.
Mobility-shift assays to examine the effect of p10 on RAG1/2 interaction with RSS. (A) Scheme for binding experiments. Labeled 12 RSS (2 nM; position of label is indicated by an asterisk), 23 RSS (12.5 nM), RAG1/2 (450 ng), and HMG1 (50 ng) were combined in reaction buffer (5 min, 37°C); Ca2+ was added to 4 mM (5 min, 37°C); Mg2+ was added to 4 mM (5 min, 37°C); glycerol was added to 10%, and reactions were loaded onto shift gels. 100% DMSO (1 μl) or 5 mM p10 in 100% DMSO (1 μl) was added at one of the times indicated (1, 2, or 3). (B) Mobility-shift assays were performed as described with p10 or solvent being added as per condition 1, 2, or 3. Filled arrow, free substrate; empty arrow, complex of RAG1/2 with 12 and 23 RSSs.
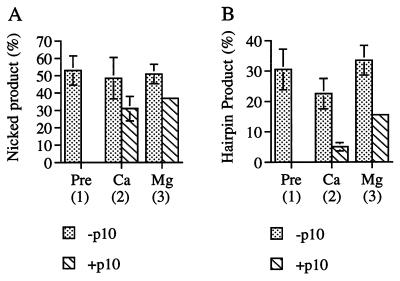
Despite the ability of p10 to disrupt preformed RAG1/2–RSS complexes in mobility-shift assays, these complexes were partially competent for cleavage even after the addition of p10. p10 completely inhibited both nicking and hairpin formation when it was added to the reaction before Ca2+ (Fig. 4 A and B, “Pre”). When p10 was added immediately after Ca2+, nicking occurred at close to normal levels, but hairpin formation was strongly inhibited (Fig. 4 A and B, “Ca”). Hairpin formation was less strongly inhibited when p10 was added immediately after Mg2+ (Fig. 4B, “Mg”). If EDTA was added immediately after either Ca2+ or Mg2+, both nicking and hairpin formation were completely blocked (data not shown), confirming that the activity observed when p10 was present was not the result of cleavage occurring before the addition of inhibitor. These results were unexpected because p10 appeared to strongly disrupt the RAG1/2–RSS complex, as judged by gel mobility. They indicated that RAG1/2 continued to bind to the RSS after p10 was added, but the complex was altered so that it was less competent for hairpin formation.
Figure 4.
Effect of p10 on cleavage competence of RAG1/2–RSS complexes. Reactions were assembled as described in Fig. 3 except that the final incubation was extended to 30 min, at which point reactions were stopped by the addition of formamide to 50%. Products were separated on denaturing polyacrylamide gels, and percent of substrate converted to nicked product (A) and hairpin product (B) was determined. “Pre,” “Ca,” and “Mg” correspond to conditions 1, 2, and 3 in Fig. 3; reactions included 10% DMSO (−p10) or 10% DMSO plus 0.5 mM p10 (+p10). Results are the average of three independent trials with standard deviation indicated by error bars.
Although the p10 inhibitor could block RSS binding if it was added before the RSS (Fig. 3B), it did not completely block capture of a nonspecific DNA that could serve as a target for transpositional strand transfer. To study target capture by the RAG1/2–RSS complex, we used a labeled oligonucleotide of nonspecific sequence as target and unlabeled 12 and 23 RSSs. Reactions were analyzed by native gel electrophoresis as for the band-shift assays. In the presence of the RSS pair, RAG1/2, HMG1 and Mg2+, a shifted band was evident (Fig. 5). This band depended on the presence of the 12 and 23 RSSs and was sensitive to proteinase K (data not shown), suggesting that it represented the complex of RAG1/2 with RSS DNA and target. As was the case with cleavage, p10 inhibited target capture when it was added to a mixture of RAG1/2, RSSs and target DNA before Ca2+ (Fig. 5A, lanes 1 and 2). However, target capture was observed if RAG1/2 was allowed to form a complex with the 12 and 23 RSSs in the presence of Ca2+ or Mg2+ before the addition of p10 (Fig. 5A, lanes 3–6). Target capture under these conditions was reduced to 10–20% of the levels seen in control reactions and was least effected when p10 was added just after Mg2+ (Fig. 5A), consistent with reductions in cleavage (Fig. 4B).
Figure 5.
Effect of p10 on target capture and transposition. (A) Unlabeled 12 and 23 RSSs (50 nM each), RAG1/2 (450 ng), and HMG1 (50 ng) were combined in reaction buffer (5 min, 37°C); Ca2+ was added to 4 mM (5 min, 37°C); Mg2+ was added to 4 mM (30 min, 37°C); glycerol was added to 10%, and reactions were loaded onto shift gels. Reaction volume (1/10) of 100% DMSO or 5 mM p10 in 100% DMSO and labeled target oligonucleotide (10 nM) were added under conditions 1 (Pre), 2 (Ca), or 3 (Mg), as described in Fig. 3. Activity as percent of solvent control is shown. Open arrow, target capture complex. (B) Labeled 12 RSS and unlabeled 23 RSS (50 nM each), RAG1/2 (450 ng), and HMG1 (50 ng) were combined in reaction buffer (5 min, 37°C); Ca2+ was added to 4 mM (5 min, 37°C); Mg2+ was added to 4 mM (60 min, 37°C). p10 was then added to the concentration indicated, followed by the addition of 100 ng of supercoiled pBR322 target DNA (30 min, 37°C). Deproteinized products were separated on agarose gels. Open arrow, single-ended transposition product; filled arrow, double-ended transposition product. Activity as percent of solvent control is shown.
p10 had relatively little effect on transposition into supercoiled plasmid DNA provided that RAG1/2 was allowed to cleave the RSS substrates before the addition of inhibitor (Fig. 5B). In these experiments, cleavage of labeled RSS substrate was conducted for 60 min, followed by the addition of inhibitor. Supercoiled plasmid target was then added, and reactions were incubated for an additional 30 min before deproteinization and separation on agarose gels. Transposition products resulting from insertion of one or both RSSs into the target appear as labeled open circle or linear plasmids, respectively (Fig. 5B). Significant levels of transposition were still seen at concentrations of p10 (250 μM) sufficient to completely abolish cleavage (see Figs. 5B and 2A), and p10 had no apparent effect on the ratio of single- to double-ended insertion (Fig. 5B). This result was true even though the target DNA was added after p10.
Inhibition by p10 was not a general feature of all transposases and integrases. For example, transposition by Tn10 transposase was not affected unless p10 was present at very high concentrations (greater than 1 mM; data not shown). This observation was particularly interesting in light of the mechanistic relation between RAG1/2 and Tn10 transposase, both of which cleave DNA by using similar two-step processes. The drug also had no effect on DNA digestion by the restriction enzymes EcoRI, AvaI, NotI, and BsaI (data not shown).
Discussion
The RAG1/2 protein complex involved in initiating V(D)J recombination is related to transposases and retroviral integrases. This relation has provided significant functional and biochemical insights into the development of combinatorial immunity and the evolution of the specific immune system. The recent description of HIV-1 integrase inhibitors as potential therapeutic agents suggested the possibility that these inhibitors might also interfere with V(D)J recombination.
Our results show that the HIV-1 integrase inhibitor, designated herein as p10, inhibits many of the biochemical activities of RAG1/2—including nicking between the RSS and coding flank DNA, hairpin formation by transesterification, and disintegration of transposition intermediates—with an IC50 of 20 μM. This inhibition is because of interference of complex formation between RAG1/2 and RSS-containing DNA. When p10 was added before Ca2+, formation of the RAG1/2–RSS complex was blocked, and when p10 was added after Ca2+, the RAG1/2–RSS complex was altered such that it could not survive gel electrophoresis. However, these complexes were still partially competent for cleavage if Mg2+ was added, indicating that they had not been entirely disrupted. These data indicate that p10 can bind to the complex of RAG1/2 with its substrate, but that only binding of p10 to RAG1/2 before stable interaction with the RSSs results in complete abrogation of activity. In contrast to its effect on RSS binding, p10 did not block binding of the RAG1/2–RSS complex to nonspecific DNA nor did it block transpositional strand transfer. Thus RSS and target binding probably occur in distinct regions of the protein, only one of which is blocked by p10. The target DNA-binding site may well be the same as the one occupied first by the coding flank and then transiently by the coding end during RAG1/2-mediated cleavage.
Although p10 inhibited DNA cleavage by RAG1/2 as well as the activities of HIV-1 integrase, the effect of the inhibitor in the two systems was somewhat different. Overall, HIV-1 integrase was about 20 times more sensitive to p10, and the various activities of HIV-1 integrase were distinct in their sensitivities to inhibitors of this class. The strand transfer reaction was much more sensitive than either the 3′ end processing (i.e., nicking) or disintegration reactions (21). It was also observed that the presence of long terminal repeat (LTR) sequences found at the ends of viral DNA enhanced the interaction between inhibitor and integrase (21). These data were interpreted to mean that these compounds interfere with the binding of target DNA but not LTR DNA. In contrast, p10 inhibitor blocked RSS binding by RAG1/2 but not capture of nonspecific target DNA and subsequent transposition, although the increased sensitivity of hairpin formation relative to nicking paralleled the different effects of p10 on various integrase activities.
Our results are all derived from a cell-free system reconstituted with purified components. We have not been able to determine whether p10 or other similar inhibitors are able to interfere with RAG1/2 activity in intact cells because of the cellular toxicity exhibited by these compounds under the conditions used for rearrangement of V(D)J substrates in cell culture (unpublished observations). It will be important to ascertain the effects of future HIV-1 integrase inhibitor compounds on RAG1/2 activity to avoid potential deleterious side effects. The in vitro assays outlined here should be helpful in that screening process.
Acknowledgments
We thank Murray Junop for purified Tn10 reagents, Joanne Hesse and Melanie Simpson for tissue culture advice as well as our other colleagues in the Laboratory of Molecular Biology for advice and comments.
Abbreviations
- RSS
recombination signal sequence
- HMG
high-mobility group
References
- 1.Gellert M. Adv Immunol. 1997;64:39–64. doi: 10.1016/s0065-2776(08)60886-x. [DOI] [PubMed] [Google Scholar]
- 2.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 3.van Gent D C, Ramsden D A, Gellert M. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 4.van Gent D C, Mizuuchi K, Gellert M. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 5.Agrawal A, Schatz D G. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 6.Hiom K, Gellert M. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 7.Jones J M, Gellert M. Proc Natl Acad Sci USA. 2001;98:12926–12931. doi: 10.1073/pnas.221471198. . (First Published October 23, 2001; 10.1073/pnas.221471198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramsden C A, Gellert M. Genes Dev. 1995;9:2409–2420. doi: 10.1101/gad.9.19.2409. [DOI] [PubMed] [Google Scholar]
- 9.Gao Y, Sun Y, Frank K M, Dikkes P, Fujiwara Y, Seidl K J, Sekiguchi J M, Rathbun G A, Swat W, Wang J, et al. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 10.Taccioli G E, Rathbun G A, Oltz E M, Stamato T D, Jeggo P A, Alt F. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal A, Eastman Q M, Schatz D G. Nature (London) 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 12.Hiom K, Melek M, Gellert M. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 13.Melek M, Gellert M. Cell. 2000;101:625–633. doi: 10.1016/s0092-8674(00)80874-0. [DOI] [PubMed] [Google Scholar]
- 14.Melek M, Gellert M, van Gent D C. Science. 1998;280:301–303. doi: 10.1126/science.280.5361.301. [DOI] [PubMed] [Google Scholar]
- 15.Craigie R. J Biol Chem. 2001;276:23213–23216. doi: 10.1074/jbc.R100027200. [DOI] [PubMed] [Google Scholar]
- 16.Chow S A, Vincent K A, Ellison V, Brown P O. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 17.Kim D R, Dai Y, Mundy C L, Yang W, Oettinger M A. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landree M A, Wibbenmeyer J A, Roth D B. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neamati N, Marchand C, Pommier Y. Adv Pharmacol. 2000;49:147–165. doi: 10.1016/s1054-3589(00)49026-5. [DOI] [PubMed] [Google Scholar]
- 20.Goldgur Y, Craigie R, Cohen G H, Fujiwara T, Yoshinaga T, Fujishita T, Sugimoto H, Endo T, Murai H, Davies D R. Proc Natl Acad Sci USA. 1999;96:13040–13043. doi: 10.1073/pnas.96.23.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazuda D J, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler J A, Espeseth A, Gabryelski L, Schleif W, Blau C, et al. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]