Abstract
BACKGROUND & AIMS:
Defects in the epithelial tight junction (TJ) barrier contribute to development of intestinal inflammation associated with diseases. Interleukin 1 beta (IL1B) increases intestinal permeability in mice. We investigated microRNAs that are regulated by IL1B and their effects on expression of TJ proteins and intestinal permeability.
METHODS:
We used Targetscan to identify microRNAs that would bind the 3ʹ untranslated region (3ʹUTR) of occluding mRNA; regions that interacted with microRNAs were predicted using the V-fold server and Assemble2, and 3-dimensional models were created using UCSF Chimera linked with Assemble2. Caco-2 cells were transfected with vectors that express microRNAs, analyzed by immunoblots and real-time polymerase chain reaction (PCR), and grown as monolayers; permeability in response to IL1B was assessed with the marker inulin. Male C57BL/6 mice were given intraperitoneal injections of IL1B and intestinal recycling perfusion was measured; some mice were given dextran sodium sulfate to induce colitis and/or gavage with an antagonist to MIR200C-3p (antagomiR-200C) or the nonspecific antagomiR (control). Intestinal tissues were collected from mice and analyzed by histologyand real-time PCR; enterocytes were isolated by laser capture microdissection. We also analyzed colon tissues and organoids from patients with and without ulcerative colitis.
RESULTS:
Incubation of Caco-2 monolayers with IL1B increased TJ permeability and reduced levels of occludin protein and mRNA without affecting the expression of other transmembrane TJ proteins. Targetscan identified MIR122, MIR200B-3p, and MIR200C-3p, as miRNAs that might bind to the occludin 3ʹUTR. MIR200C-3p was rapidly increased in Caco-2 cells incubated with IL1B; the antagomiR-200c prevented the IL1B-induced decrease in occludin mRNA and protein and reduced TJ permeability. Administration of IL1B to mice increased small intestinal TJ permeability, compared with mice given vehicle; enterocytes isolated from mice given IL1B had increased expression of MIR200C-3p and decreased levels of occludin messenger RNA (mRNA) and protein. Intestinal tissues from mice with colitis had increased levels of IL1B mRNA and MIR200C-3p and decreased levels of occludin mRNA; gavage of mice with antagomiR-200C reduced levels of MIR200C-3p and prevented the decrease in occludin mRNA and the increase in colonic permeability. Colon tissues and organoids from patients with ulcerative colitis had increased levels of IL1B mRNA and MIR200C-3p compared with healthy controls. Using 3- dimensional molecular modeling and mutational analyses, we identified the nucleotide bases in the occluding mRNA 3ʹUTR that interact with MIR200C-3p.
CONCLUSIONS:
Intestine tissues from patients with ulcerative colitis and mice with colitis have increased levels of IL1B mRNA and MIR200C-3p, which reduces expression of occludin by enterocytes and thereby increases TJ permeability. Three-dimensional modeling of the interaction between MIR200C-3p and the occludin mRNA 3ʹUTR identified sites of interaction. The antagomiR-200C prevents the decrease in occludin in enterocytes and intestine tissues of mice with colitis, maintaining the TJ barrier.
Keywords: MicroRNA 200C, RNA Degradation, Epigenetic Modification, Translation
Introduction
Intestinal epithelial tight junctions (TJ) are the apical-most junctions and function as intercellular barrier against paracellular permeation of bacterial antigens and other noxious agents in the intestinal lumen.1–3 Defective intestinal TJ barrier is an important pathogenic factor contributing to the development of various inflammatory conditions of the gut, including inflammatory bowel disease (IBD) and necrotizing enterocolitis.4–8 Despite the recognized importance of defective intestinal TJ barrier or “leaky gut” in the pathogenesis of intestinal inflammation associated with IBD and other inflammatory diseases, there are no available therapeutic agents that target the TJ barrier.
Interleukin-1β (IL-1β) is ubiquitous and pluripotent proinflammatory cytokine that plays an essential role in the development and prolongation of intestinal inflammation in IBD and other inflammatory conditions.9–21 Patients with IBD have elevated levels of IL-1β and IL-1β has been shown to play a crucial role in animal models of intestinal inflammation.9, 22 Previous studies in IBD patients have shown a correlation between elevated level of IL-1β and decreased level of its natural antagonist IL-1ra and increased intensity of intestinal inflammation.10–12, 23, 24 In addition, there is a direct correlation between defective intestinal TJ barrier, characterized by an increase in intestinal permeability and a persistence of intestinal inflammation in patients with IBD.1, 2, 6 Previous studies from our laboratory and others have shown that IL-1β at physiologically relevant concentrations causes an increase in intestinal TJ permeability.9–12, 23, 25–28 The IL-1β induced increase in intestinal TJ permeability appeared to be mediated in part by an increase in myosin light chain kinase (MLCK) activity.13–15, 29–34 In previous studies, we showed that IL-1β causes a decrease in occludin expression in Caco-2 monolayers.10 However, the functional relevance of occludin depletion or the molecular mechanisms responsible for the occludin down-regulation remains unclear.
The major aims of this study were to determine the role of occludin depletion in IL-1β induced increase in intestinal TJ permeability and to delineate the molecular mechanisms that mediate the IL-1β induced decrease in occludin level and increase in TJ permeability, using both in vitro model system (consisting of filter grown Caco-2 monolayers) and in vivo model system (consisting of recycling intestinal perfusion of small intestine in live mice). Using in silico approach, we predicted microRNA (miR)-200c-3p to play a regulatory role in IL-1β modulation of intestinal TJ permeability by targeting occludin mRNA degradation. We also identified the 3-dimensional (3D) molecular docking interaction with the occluding messenger RNA (mRNA). Our experimental data show that IL-1β induces a rapid transcription of miR-200c-3p in enterocytes in vitro and in vivo; miR-200c-3p binds to the 3’UTR noncoding region of occludin mRNA and induces occludin mRNA degradation and occludin depletion, resulting in an increase in TJ permeability; and antagomiR-200c inhibits the dextran sulfate sodium (DSS)-induced increase in intestinal tissue miR-200c-3p, increase in intestinal permeability, and development of colitis.
Materials and Methods
Cell cultures
Caco-2 cells were maintained in a culture medium as previously described.10, 35 The cells were grown at 37˚C in a 5% CO2 environment.
Bioinformatics Analysis and In Silico 3-Dimensional Predictions
TargetScan analysis was used for initial predictions of miRNA target.36 A modified protocol described in Gan and Gunsalus37 was followed to determine the initial molecular docking interactions by secondary molecular structures of both target and miRNA. The single stranded fragments of both occludin 3’ untranslated region (3’UTR) interacting region (139–145) and human miRNA-200c-3p were individually predicted for their tertiary structure by V-fold server38 and Assemble2; Java application was used to manipulate and create energy minimized models.39 Further the structures of mature miRNA-200c-3p and fragment picked from occludin 3’UTR region (124–146; UUGCUUUAACAUCAUCAGUAUUG) were subjected to tertiary structure predication and simulations by programs RASSIE and RASCAL.40, 41 The 3D models were visualized in 3D viewer UCSF Chimera linked with Assemble2.42 The 2D structures from 3D structures were predicated for simplification by RNApdbee 2.0.43, 44 The target complex with initial interaction predicted by RASCAL software was subjected to MD simulation and trajectory analysis conducted using the Desmond software.45 The full system was prepared by using Maestro’s Preparation Wizard. The system was solvated in TIP3P water models with Sodium cations to neutralize the charges on the molecules and 0.15 M NaCl. A simulation box covering the whole system was placed with a 10 Å buffer space. The simulation was run for 2 ns at 300 K temperature and, standard pressure (1.01325 bar), OPLS-AA 2005 force field parameters were elected for utilization for both this preparation and all later simulations.45
Cloning
Construction of occludin promoter reporter was carried out using the pGL-3 basic luciferase reporter vector. The primers used and PCR condition were as previously described.35, 46 Construction of 570 bp long occludin 3′UTR reporter was carried out using the pMIR-REPORT reporter vector.35 Deletion construct of miR-200c-3p binding site were generated as previously described.35
Transfection of DNA Constructs
Caco-2 cells were transfected using lipofectamine 2000 as previously described by us.35, 47 Luciferase activity was determined using the DUAL Luciferase assay kit (Promega).35, 47
Western blot and [ 35S]Methionine Pulse-Chase Experiment
Occludin protein expression from Caco-2 cells and mouse tissue was assessed by western blot as previously described.9, 10 Caco-2 cells were pulse-labeled with [35S] methionine and the time course of 35S-labelled occludin (pulse-chase) decomposition was determined.46, 48
Determination of Caco-2 Paracellular Permeability
Caco-2 paracellular permeability was determined using an established paracellular marker inulin.10, 49 Known concentrations of permeability marker inulin (2 μM) and its radioactive tracer were added to the apical solution.35
RNA Isolation and Quantification of Gene Expression Using Real-Time PCR
Total RNA was isolated using the miRNeasy kit (Qiagen, Valencia, CA) and reverse transcription (RT) was performed using the QuantiTect reverse transcription kit (Qiagen, Valencia, CA) according to manufacturer’s protocol. The real-time PCRs were performed using PikoReal 96 Real-Time PCR system (Thermo Scientific, Waltham, MA).
Determination of Mouse Intestinal Permeability
The laboratory animal care and use committee at the University of New Mexico and Penn State University approved all experimental protocols. Male C57BL/6 male mice (9–10 weeks) were purchased from Jackson Laboratory (Bar Harbor, ME). IL-1β (5 μg) effect on intestinal permeability in an in vivo mouse model system was determined using recycling intestinal perfusion method.9, 35, 50, 51
In Vivo Transfection of MIR-200C-3P Precursor
The in vivo miRNA and antisense transfection of intestinal epithelial cells were carried out as previously described.35 The transfection solution (0.5 ml) consisting of pre-miR-200c-3p (25 nM) or antisense of miR-200c-3p (25 nM) or scramble control and lipofectamine (50 μl) was injected through a 30-gauge needle into the lumen of isolated intestinal segment and the mucosal surface was exposed to transfection solution for 1 h.35 The mouse was allowed to recover for 3 days before the permeability studies were performed.35
DSS-Colitis and AntagomiR-220c Treatment
Mice received 3% DSS (36–50 KDa; MP Biomedicals, Santa Ana, CA) in autoclaved drinking water for 7 days.52 For anti-sense therapeutic studies, antagomiR-200c and nonspecific antagomiR were designed by Creative biogene, (Shirley, NY) and were administrated daily by oral gavage (800 mg/d) starting 2 days prior to DSS treatments and continued throughout DSS administration.
Laser Capture Microdissection
Frozen mouse tissue sections were fixed and processed for microdissection as previously described.35 On average, approximately 1000 cells were obtained per microdissection cap and total RNA was isolated.35
Patient Samples and Human Colon Organoids
The samples used were collected with patient consent as part of the biorepository within the Department of Surgery, Division of Colon and Rectal Surgery at the Pennsylvania State University College of Medicine. Colon organoids (colonoids) are established from donor biopsies and maintained in 3D culture as previously described.53, 54
Statistical Analysis
The experimental data was analyzed using paired t test (Graph Pad (La Jolla, CA) Prizm 6.00). All experiments were repeated at least three times to ensure reproducibility.
Results
IL-1β modulation of TJ proteins
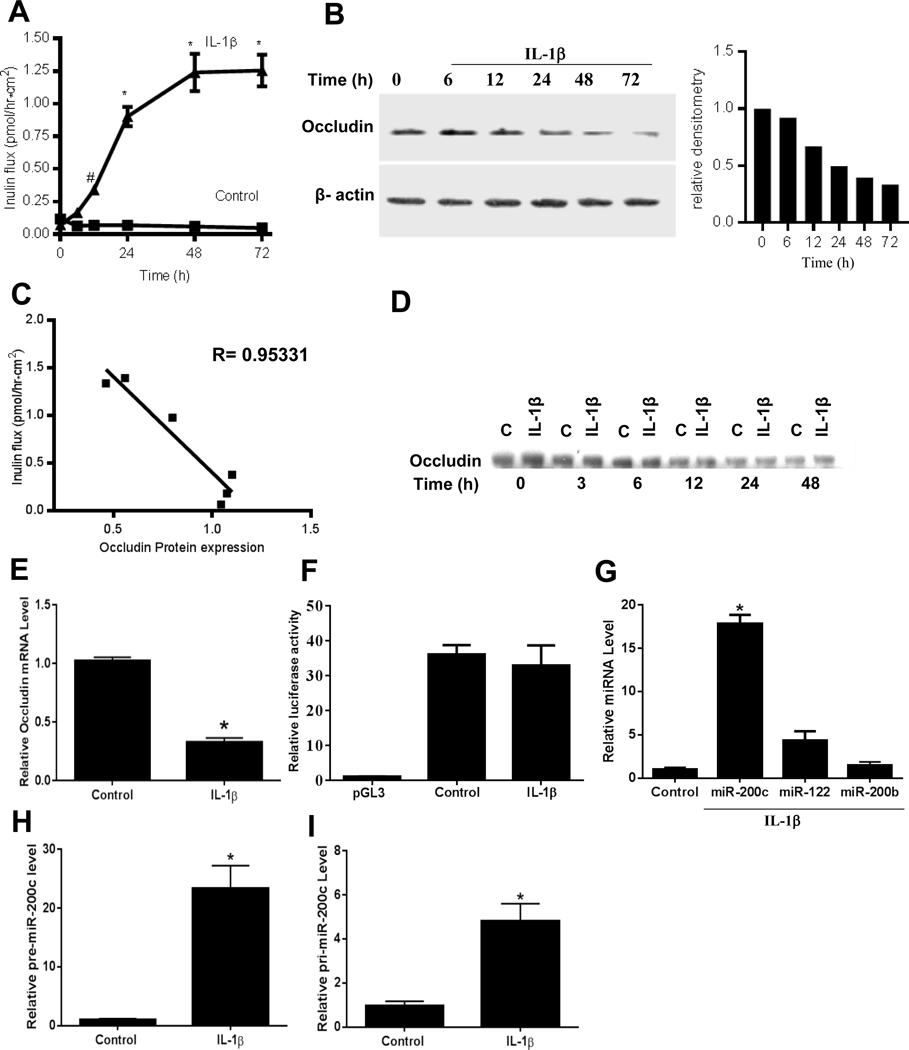
The involvement of the TJ proteins that actually form the TJ complex in mediating the IL-1β effect on TJ barrier remains unclear. In the following studies, we examined the possibility that the IL-1β induced increase in intestinal epithelial TJ permeability was mediated in part by an alteration in TJ protein expression. IL-1β at physiological concentration (10 ng/ml) caused a time-dependent increase in Caco-2 TJ permeability as assessed by mucosal-to-serosal flux of paracellular marker inulin, starting at about 24 hours (Figure 1A). The IL-1β treatment of filter grown Caco-2 monolayers also caused a time-dependent decrease in occludin expression starting at about 24 hours (Figure 1B), without affecting the expression of ZO-1 or other transmembrane TJ proteins known to affect the TJ barrier, including claudin-1, claudin-2, claudin-3, claudin-4, or claudin-5. These results suggested that the IL-1β increase in Caco-2 TJ permeability was associated with a decrease in occludin level but not other transmembrane TJ proteins. To assess the possible cause-and-effect relationship, the time course effect of IL-1β on Caco-2 TJ permeability was correlated with Caco-2 occludin expression. There was a direct linear relationship between the level of IL-1β induced decrease in occludin expression and the increase in TJ permeability, with a correlation coefficient of 0.95331, suggesting a cause-and-effect relationship (Figure 1C).
Figure 1.
IL-1β effect on occludin paracellular permeability, protein expression, occludin mRNA transcription, promoter activity and miRNA expression in filter-grown Caco-2 monolayers. (A,B) Time course effect of IL-1β on mucosal-to-serosal flux of inulin and occludin expression (C) Occludin expression vs inulin flux, the correlation coefficient of occludin expression and inulin flux was 0.95. (D) Filter-grown Caco-2 monolayers were treated with IL-1β and the time course of 35S-labelled occludin (pulse-chase) decomposition was determined. (E,F) Effect of IL-1β (10 ng/ml) over 2-hours on occludin mRNA expression and occludin promoter activity. (G) Effect of IL-1β (10 ng/ml) over 2-hour on in silico predicted miRNA expression as determined by real-time PCR. (H,I) Effect of IL-1β on pre-miRNA-200c and pri-miRNA-200c expression. *P < .001 vs control, # P < .005 vs control. pre-miR-200c: precursor microRNA of miR-200c-3p; pri-miR-200c: primary microRNA of miR-200c-3p.
To determine the intracellular mechanisms responsible for the decrease in occludin, we examined the possibility that the decrease in occludin expression was due to an accelerated degradation of occludin. By 35S-methionine labelling of occludin, the time course of 35S-labelled occludin (pulse-chase) decomposition was determined. IL-1β did not affect the degradation rate of 35S-methionine labeled occludin (Figure 1D). Next, the possibility that IL-1β causes an accelerated degradation of occludin mRNA was examined by real-time PCR. IL-1β treatment resulted in a rapid decrease in occludin mRNA level (Figure 1E). The rapid time course (within 1–2h) of occludin mRNA decrease suggested that the IL-1β effect was due to rapid degradation and not decrease in gene activity or gene transcription. To rule out the possibility that IL-1β effect was due to a decrease in occludin gene activity, IL-1β effect on occludin promoter activity was also examined. IL-1β did not affect the occludin promoter activity (Figure 1F).
Bioinformatics analysis and in-silico prediction
It has been previously shown that the genes regulated by miRNAs have significantly longer 3’UTR compared with the genes not-targeted by miRNAs.55 Occludin mRNA has a long 3’UTR, having 1241–1276 base pairs depending on the mRNA variant.56 Using RNAfold Web server, we calculated the mRNA fold energy of occludin 3’UTR region to be −152.96 kcal/mol (−0.241/base), which suggested a highly complex secondary 3’UTR structure. The long length and high energy state of occludin mRNA 3’UTR strongly predicted the susceptibility of this region for RNA interference mediated regulation. To identify the potential miRNAs that regulate occludin mRNA, a candidate-based approach was used with the Web server Targetscan.57
The Targetscan analysis identified 3 microRNAs, miR-122, miR-200b-3p, and miR-200c-3p, as potential candidates to bind to the occludin 3’UTR. The computer algorithm probability analysis based on miRNA binding interactions predicted miR-122, miR-200b-3p or miR-200c-3p to have over 90% probability of regulating occludin mRNA 3’UTR. The expression of these miRNAs (miR-122, miR-200b-3p, and mR-200c-3p) and the effect of IL-1β on miRNA expression were determined in filter-grown Caco-2 monolayers. IL-1β (10 ng/ml) caused a 15–20 fold increase in miR-200c-3p levels, 3–5 fold increase in miR-122, and no change in miR-200b-3p (Figure 1G). IL-1β also caused a 20-fold increase in precursor microRNA of miR-200c-3p, and a 4–5 fold increase in primary microRNA of miR-200c-3p (Figure 1H and 1I). As miR-200c-3p was rapidly and markedly up-regulated by IL-1β, miR-200c-3p was targeted as a likely candidate for post-transcriptional regulation of occludin mRNA.
Antisense Inhibition of MIR-200C-3P
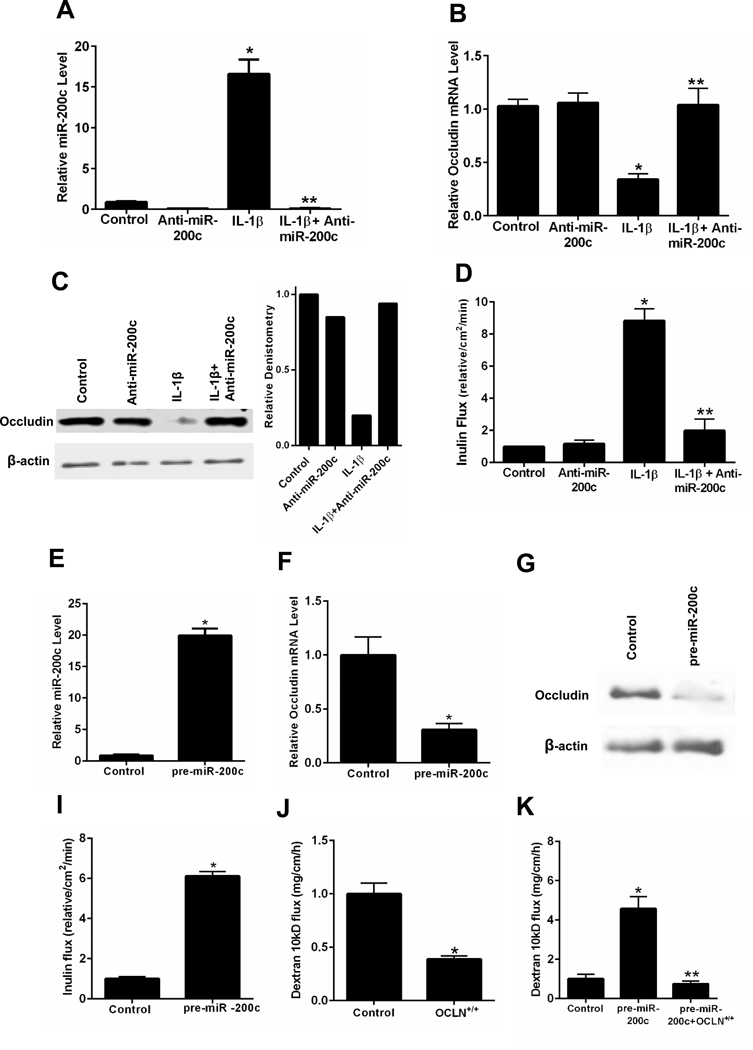
To determine the involvement of miR-200c-3p in IL-1β-induced degradation of occludin mRNA and the subsequent increase in Caco-2 TJ permeability, the effect of antisense oligoribonucleotide inhibition of miR-200c-3p was examined. For these studies, anti-miR-200c-3p was used to silence miR-200c-3p. As shown in Figure 1G, IL-1β caused a rapid increase in miR-200c-3p levels. This increase was inhibited by pre-treatment with anti-miR-200c-3p, which binds to the complimentary binding motif on miR-200c-3p (Figure 2A). The anti-miR-200c-3p prevented the IL-1β-induced decrease in occludin mRNA, occludin protein depletion and increase in Caco-2 TJ permeability (Figure 2B, 2C and 2D).
Figure 2.
Effect of antisense ribonucleotide (ASO) inhibition of miR-200c-3p and miR-200c-3p overexpression on occludin mRNA, protein expression and paracellular permeability in filter-grown Caco-2 monolayers. Effect of anti-miR-200c-3p transfection on IL-1β induced increase on miR-200c-3p expression (A), decrease in occludin mRNA expression (B), decrease in occludin protein expression and relative occludin densitometry (C), and increase in inulin flux (D). Effect of pre-miR-200c-3p transfection on miR-200c-3p expression (E), occludin mRNA expression (F), occludin protein expression (G), and Caco-2 paracellular permeability to inulin (H). Effects of occludin gene transfection on Caco-2 paracellular permeability to dextran 10kD (I), and pre-miR-200c induced increase in dextran 10kD flux (J). * P < 0.01 vs control; * * P < 0.001 vs IL-1β treatment and pre-miR-200c; OCLN+/+: plasmid pCMV3-OCLN-OFPSpark
MIR-200C-3p, Occludin mRNA Degradation and Caco-2 TJ Permeability
To validate the functional role of miR-200c-3p in post-transcriptional degradation of occludin mRNA, miR-200c-3p was overexpressed in filter-grown Caco-2 monolayers by pre-miR-200c-3p transfection. The transfection of pre-miR-200c-3p resulted in an about a 15- to 20-fold increase in miR-200c-3p (Figure 2E), a level similar to that of IL-1β effect on miR-200c-3p (Figure 1G). The pre-miR-200c-3p transfection resulted in a rapid degradation of occludin mRNA and a decrease in occludin protein level (Figure 2F and 2G). The pre-miR-200c-3p transfection also caused an increase in Caco-2 TJ permeability (Figure 2I). On the other hand, the transfection of pre-miR-122 leading to a 5-fold increase in miR-122 did not have significant effect on inulin flux (data not shown). Next, we examined the effects of occludin overexpression by full-length occludin gene transfection of Caco-2 cells using pCMV3-C-OFPSpark expression vector.58 The occludin overexpression by occludin gene transfection resulted in a 60% to 70% decrease in mucosal-to-serosal flux of dextran-10kd (Figure 2J) and also completely inhibited the pre-miR-200c induced increase in dextran flux (Figure 2K).58
Il-1β Effect on Mouse Intestinal Permeability and MIR-200C-3p
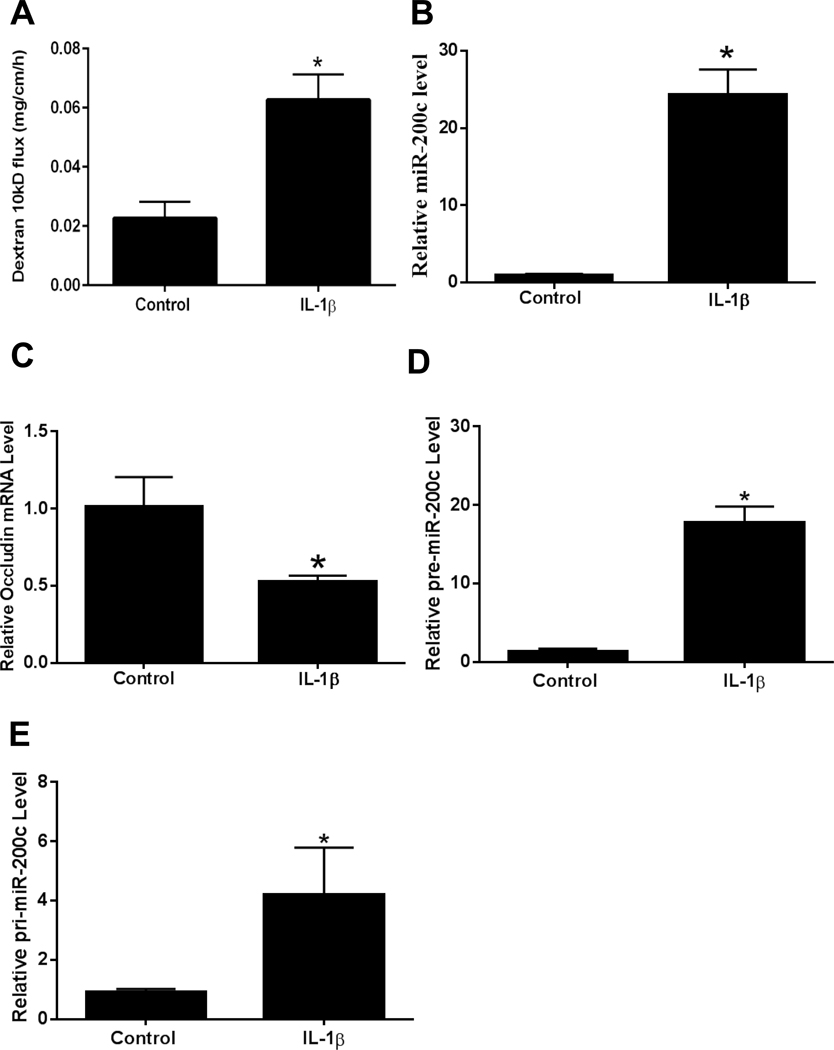
The preceding studies indicated that the IL-1β induced increase in Caco-2 intestinal epithelial TJ permeability was regulated by miR-200c-3p induced degradation of occludin mRNA. In the following studies, we examined the involvement of miR-200c-3p in IL-1β induced increase in mouse intestinal permeability by in vivo recycling small intestinal perfusion.35, 50 As previously shown by us, intraperitoneal administration of IL-1β (5 μg) caused an increase in intestinal permeability to FITC-dextran (10kd) (Figure 3A). Because intestinal tissue consists of various cell types, a pure population of intestinal epithelial cells following IL-1β administration was obtained from the intestinal mucosal surface by laser capture microdissection of surface epithelial cells as previously described.35, 59 The real time PCR analysis of mouse enterocytes isolated from the mouse intestinal tissue confirmed that IL-1β administration (5 μg intraperitoneal injection) causes a marked increase in enterocyte miR-200c-3p, a decrease in occludin mRNA level, an increase in precursor miR-200c, and an increase in primary miR-200c in vivo (Figure 3B- 3E).
Figure 3.
Effect of in vivo intraperitoneal administration of IL-1β (5μg) on mouse intestinal epithelial cell expression of miR-200c-3p and occludin mRNA and mouse intestinal permeability after 24 h treatment period. Effect of intraperitoneal IL-1β on luminal-to-serosal FITC-dextran (MW = 10,000 g/mol) flux (A), mouse enterocyte miR-200c-3p expression (B), mouse enterocyte occludin mRNA expression (C), mouse enterocyte pre-miR-200c expression (D), and mouse enterocyte pri-miR-200c expression (E). Mouse enterocytes were isolated using LCM. * P < 0.01 vs control. pre-miR-200c: precursor microRNA of miR-200c-3p; pri-miR-200c: primary microRNA of miR-200c-3p.
Antisense Inhibition of MIR-200C-3p and Mouse Intestinal Permeability
We hypothesized that the IL-1β induced increase in mouse intestinal permeability was also mediated by miR-200c-3p degradation of occludin mRNA and occludin depletion in mouse enterocytes. The effect of in vivo mouse enterocyte anti-miR-200c-3p transfection on IL-1β- induced increase in mouse intestinal permeability was determined. The mouse small intestinal epithelial cells were selectively transfected in vivo with anti-miR-200c-3p. 35 The in vivo anti-miR-200c-3p transfection of mouse intestinal epithelial cells inhibited the IL-1β induced increase in miR-200c-3p expression, decrease in occludin mRNA level and depletion in occludin protein (Figure 4A, 4B, 4F). Anti-miR-200c-3p transfection also inhibited the IL-1β induced increase in mouse intestinal permeability (Figure 4C).
Figure 4.
The effect of anti-miR-200c-3p transfection and miR-200c-3p overexpression on mouse small intestinal permeability in vivo. Effect of anti-miR-200c-3p transfection on IL-1β induced increase in mouse enterocyte miR-200c-3p expression (A). Mouse intestinal mucosal surface was transfected anti-miR-200c-3p in vivo. Effect of anti-miR-200c-3p transfection on IL-1β induced decrease in mouse enterocyte occludin mRNA expression (B), and increase in mouse intestinal permeability(C). Mouse small intestinal mucosal surface were transfected with pre-miR-200c-3p in vivo. Enterocytes were isolated from the mucosal surface by LCM. Effect of pre-miR-200c-3p transfection on mouse enterocyte miR-200c-3p (D), and occludin mRNA (E) expression. Effect of pre-miR-200c-3p and anti-miR-200c-3p transfection on intestinal tissue occludin protein expression (F), and pre-miR-200c-3p transfection on mouse intestinal permeability in vivo (G). * P < .01 vs control, * * P < .01 vs IL-1β treatment.
MiR-200c-3p overexpression and mouse intestinal permeability
To further validate the regulatory role of miR-200c-3p in the modulation of mouse intestinal permeability, we next examined whether miR-200c-3p overexpression was sufficient to cause an increase in mouse intestinal permeability in vivo. In these studies, the effect of pre-miR-200c-3p transfection on mouse intestinal permeability was examined.35 The in vivo pre-miR-200c-3p transfection resulted in a rapid up-regulation of miR-200c-3p in mouse enterocytes (Figure 4D), similar to the level induced by intraperitoneal IL-1β administration.35 The pre-miR-200c-3p transfection also caused a decrease in mouse enterocyte occludin mRNA level, depletion of intestinal tissue occludin protein and increase in mouse intestinal permeability (Figure 4E–4G).
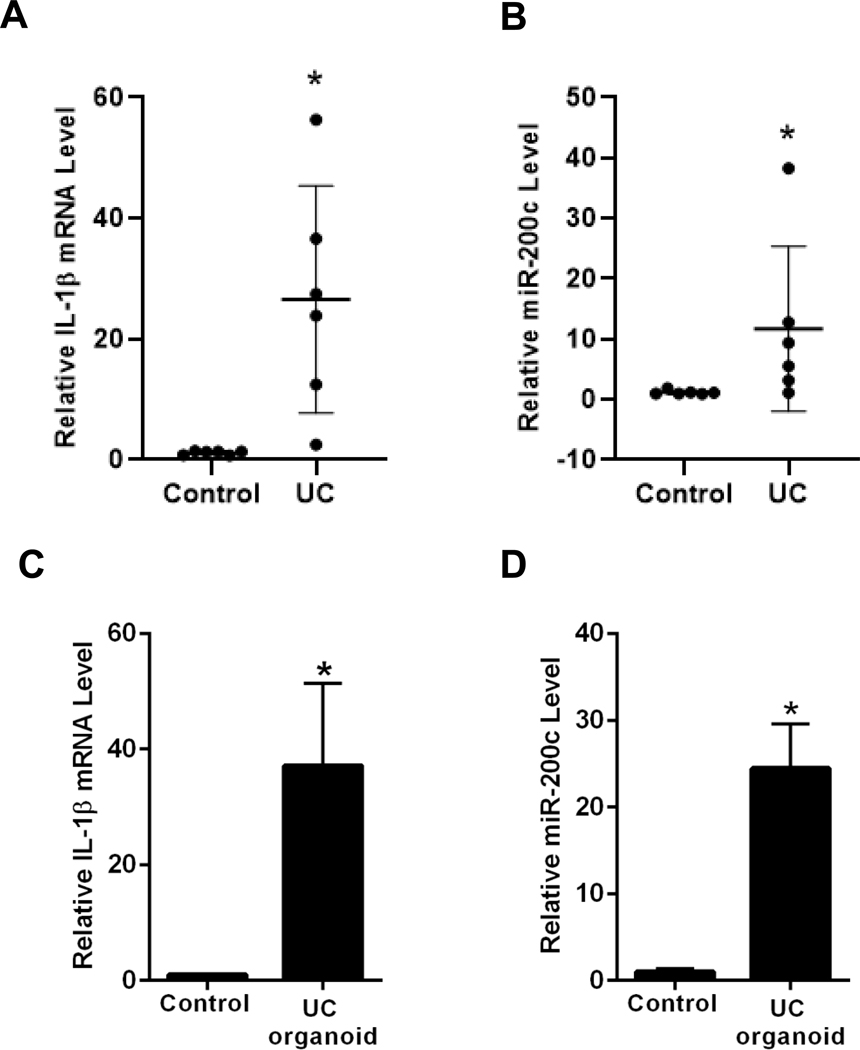
MIR200C-3p Expression in Murine DSS-induced Colitis and Human Ulcerative Colitis Tissue
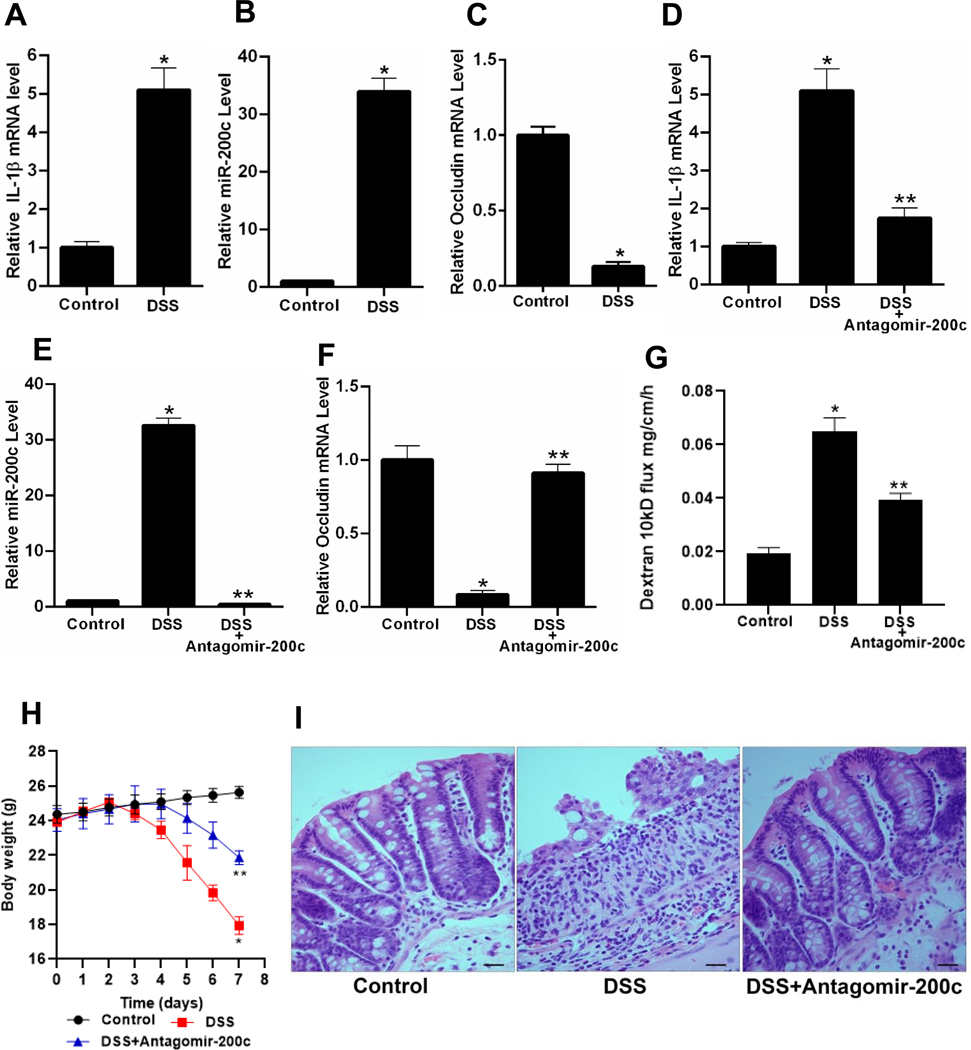
The effect of murine DSS-induced colitis on intestinal tissue IL-1β and miR-200c-3p expression was examined. The daily administration of 3% DSS for 7 days resulted in an increase in colonic tissue IL-1β mRNA, an increase in miR-200c-3p and a decrease in occludin mRNA (Figure 5A–5C). The oral-gastric gavage of antagomiR-200c daily starting 2 days before and continuing throughout the DSS administration resulted in an inhibition of IL-1β mRNA increase, miR-200c-3p increase, and decrease in occludin mRNA (Figure 5D–5F). The antagomiR-200c treatment also inhibited the DSS-induced increase in colonic permeability, drop in body weight, and colitis (Figure 5G-I). The examination of human colonic tissue expression of IL-1β and miR-200c-3p levels in inflamed colonic tissues obtained from patients with active ulcerative colitis and from patients with normal-appearing colonic tissue undergoing colonic resection for noninflammatory conditions revealed a significant increase in IL-1β mRNA and miR-200c-3p levels in the inflamed tissues compared to the normal colonic tissues (Figure 6A and B). In Addition, organoids generated from the patients with ulcerative colitis also had a marked increased expression of IL-1β mRNA and miR-200c-3p compared to the organoids generated from the normal colonic tissues (Figure 6C and D). These results showed that the colonic tissue IL-1β and miR-200c-3p levels are elevated in an animal model of colitis and in patients with ulcerative colitis, and that treatment with antagomiR-200c inhibits the DSS-induced decrease in occludin mRNA and the development of colitis.
Figure 5.
The effect of murine DSS-induced colitis on cooperation between IL-1β, miR-200c-3p, and mouse intestinal permeability. Effect of DSS oral administration for 7 days on IL-1β mRNA expression (A), miR-200c-3p expression (B) and on occludin mRNA expression (C). The colonic perfusion of DSS mice to analyze the effect of antagomiR-200c on DSS induced increase in IL-1β mRNA expression (D), increase on miR-200c-3p expression (E), decrease in occludin mRNA expression (F), and increase in dextran 10kD flux (G). Effect of antagomiR-200c gavage on body weights of DSS mice (H), Histology of mice colon to see the effect of antagomiR-200c, (I). *P < .01 vs control; * *P < .01 vs DSS; Histology scale-black bar: 20μm.
Figure 6.
Expression of IL-1β mRNA and miR-200c-3p in human ulcerative colitis (UC) tissue and in human colon organoids. IL-1β mRNA expression (A), and miR-200c-3p expression (B) in human ulcerative colitis (UC) tissue. Control tissues from the six non-IBD/UC individuals and inflamed tissues from gut of the six ulcerative colitis (UC) patients were used. IL-1β mRNA expression (C), and miR-200c-3p expression (D) in ulcerative colitis (UC) vs non-IBD/UC control colon organoids. *P < .05 vs control.
In Silico Analysis of MIR200C-3p/occludin mRNA 3’UTR Molecular Interactions
Based on the in silico analysis, the occludin 3’UTR region was found to have 7mer pairing site and m8 seed match with miR-200c-3p. Targetscan indicated a high 3’ pairing score (−0.016)60, a positive seed-pairing stability score (0.062), and the positive target-site abundance score of 0.013 suggested high specificity of miR-200c-3p and occludin 3’UTR region.61 Pct (probability of conserved targeting) score of 0.72 indicated phylogenetically highly conserved interaction between miR-200c-3p and occludin 3’UTR among other mammalian species.62
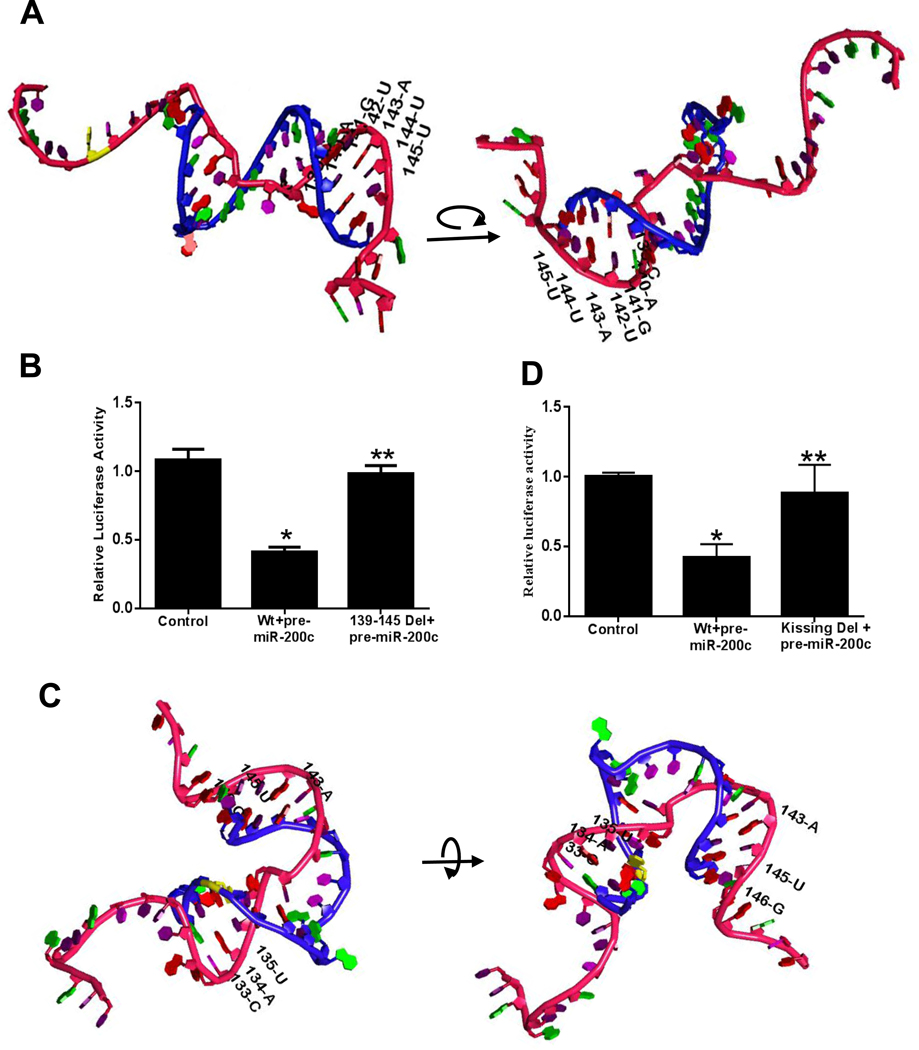
The molecular modeling studies using RASSIE and RASCAL programs and Metropolis Monte Carlo simulation 41 40, shows the primary event of dimerization or the initial “kissing” interaction between the hairpin loop of the miR-200c-3p (dimerization initiation site) and occludin 3’UTR (Figure 7). Figure 7C shows the 3D molecular modeling of the critical nucleotide bases on occludin 3’UTR responsible for the initial contact or the ‘kissing’ interaction (labeled “kissing loop”). This 3D simulation is the closest prediction of the real interaction in vivo in which miR-200c-3p and occludin 3’UTR initially interact with each other to form the final docking complex by recruiting argonaut proteins. The molecular simulation shows the progressive dimer maturation following the initial “kissing” interaction. The pivotal importance of initially interacting bases for the formation of the dimer can be clearly observed in the video showing the initial kissing and the final docking interactions of the dimer in 3-dimension (Supplementary Video 1).
Figure 7.
The potential molecular interactions of miRNA-200c-3p and occludin mRNA 3’UTR predicted by in silico bioinformatics analysis. Occludin 3’UTR region was cloned into pMIR-REPORT vector in 5’to 3’ direction, and the effect of pre-miR-200c-3p transfection on occludin 3’UTR was determined by luciferase assay. (A) Schematic representation of the predicted 3D final heterodimer formation of miRNA-200c-3p and occludin mRNA 3’UTR as predicted by bioinformatics analyses (rotated for full coverage), Red backbone represents the mature miR-200c-3p, blue backbone represents the occludin 3’UTR fragment (120–151). (B) MiR-200c-3p binding site (139-C, 140-A, 141-G, 142-U, 143-A, 144-U and 145-U) was deleted by site-directed mutagenesis, and effect of pre–miR-200c-3p transfection on deletion (Del) construct was determined. (C) Schematic representation of the initial 3D molecular interactions between miRNA-200c-3p and occludin mRNA 3’UTR (rotated for full coverage), Red backbone represents the mature miR-200c-3p, blue backbone represents the occludin 3’UTR fragment (120–151). (D) Targeted mutation of bases involved in initial kissing interactions (133-C, 134-A, 135-U, 143-A, 145-U and 146-G) prevented pre-miR-200c-3p induced inhibition of occludin mRNA 3’ UTR activity. *P < 0.01 vs control, **P <0.05 vs wild type (Wt) transfected with pre–miR-200c-3p.
To validate the preceding 3D in silico predictions, the requirement of the predicted initial and final contact sites of miRNA-200c-3p and occludin 3’UTR interactions were interrogated in filter grown Caco-2 monolayers. In these studies, occludin 3’UTR was cloned into pMIR-REPORT Expression Reporter Vector System which also encodes reporter gene firefly luciferase and co-transfected into Caco-2 cells with pre-miR-200c-3p. Pre-miR-200c-3p co-transfection caused a marked increase in miR-200c-3p expression and a decrease in luciferase transcription (Figure 3A and 7B). The deletion of the predicted miR-200c-3p final binding sequence (139-C, 140-A, 141-G, 142-U, 143-A, 144-U and 145-U) on occludin 3’UTR inhibited the pre-miR-200c-3p induced decrease in transcriptional activity (Figure 7A and B). These results suggested that miR-200c-3p inhibits occludin 3’UTR (or transcription activity) by binding to its complementary sequence on the occludin 3’UTR as predicted by the Targetscan analysis (Figure 7A and B).
Next, the requirement of the predicted initial “kissing” interactions which allows miR-200c-3p to dock and then migrate to the final binding site on the occludin 3’UTR was studied. The in silico RNA-RNA tertiary complex analysis predicted 133-C, 134-A, 135-U, 143-A, 145-U and 146-G as the initial docking site (Figure 7C). To investigate this possibility, the predicted nucleotide motif on occludin mRNA 3’UTR was also selectively mutated. The targeted mutation of the predicted bases involved in the kissing interaction caused a near-complete inhibition of transcriptional activity (Figure 7D), confirming that the kissing or docking interaction was necessary for the final miRNA/occludin 3’UTR binding and the subsequent mRNA degradation.
Discussion
The intestinal epithelial TJ barrier acts as an intercellular barrier against diverse range of microbes residing in the gut. This barrier regulates the passive paracellular permeation of various water soluble molecules and bacterial antigens in the intestinal lumen. The TJs, which are the apical-most intercellular junctions, form a functional and physical barrier in-between cells to prevent paracellular penetration of luminal molecules. Defective intestinal TJ barrier, characterized by an increased intestinal permeability to paracellular markers, has been shown to play a key pathogenic role in promoting and propagating intestinal inflammation by allowing increased antigenic penetration.1–8 Recent studies from our laboratory and others have shown that transmembrane TJ protein occludin plays an important regulatory role in restricting the paracellular flux of large macromolecules.51, 63–66 In contrast, claudin-2 proteins form a small channel or pore pathway that permits the flux of small sized molecules < 4 Å in molecular radius.67 In this study, we examined the role of transmembrane TJ protein occludin in IL-1β induced increase in intestinal epithelial TJ permeability. Herein, we show that miRNA-induced occludin depletion plays a crucial role in IL-1β induced increase in intestinal TJ permeability and provide a novel insight into the molecular interactions that mediate the occludin depletion in enterocytes in vitro and in vivo.
In the present study, we examined the involvement of occludin and scrutinized various leads in search of a novel, therapeutically relevant mechanism of intestinal TJ barrier regulation. Using computer algorithms and bioinformatics analysis, we identified miR-122, miR-200b-3p and miR-200c-3p as likely candidates to bind to occludin 3’UTR and to have high probability of functional activity. Our studies showed that miR-200c-3p was rapidly and markedly increased in response to IL-1β treatment and that miR-200c-3p plays a central regulatory role in IL-1β modulation of intestinal TJ permeability by targeting the posttranscriptional degradation of occludin mRNA. Our data showed that IL-1β causes a rapid increase in miR-200c-3p expression in filter-grown Caco-2 monolayers; the antisense inhibition of miR-200c-3p inhibited the IL-1β increase in TJ permeability, demonstrating the requirement of miR-200c-3p up-regulation in the increase in TJ permeability. Using live mice, we also showed that the IL-1β induced increase in mouse intestinal permeability in vivo was also regulated by an increase in miR-200c-3p. In the in vivo study, we also transfected pre-miR-200c-3p and anti-miR-200c-3p antisense oligoribonucleotide into mouse enterocytes.35 These in vivo transfection studies demonstrated that the overexpression of miR-200c-3p in mouse enterocytes was an essential process that causes a degradation of occludin mRNA and subsequent increase in intestinal permeability, and that antisense blocking of miR-200c-3p expression in mouse enterocytes inhibits the degradation of occludin mRNA and increase in mouse intestinal permeability. Thus, our results show that the IL-1β induced increase in miR-200c-3p expression was responsible for the degradation of occludin mRNA and the increase in intestinal TJ permeability. The potential clinical and translational application of therapeutic targeting of miR-200c-3p was also demonstrated by the animal and human studies showing an increase in IL-1β and miR-200c-3p expression in colonic tissue and organoids derived from inflamed colonic tissue of patients with ulcerative colitis and that antagomiR-200c treatment of mice inhibits the DSS-induced increase in colonic permeability and the development of colitis.
We also performed a detailed in silico analysis to predict the molecular docking interactions between miR-200c-3p and occludin 3’UTR, which allows miRNA to dock within the 3D mRNA-3’UTR molecular structure. Detailed computational analyses predicted both the early and the final molecular interactions. The tertiary molecular modeling studies of miR-200c-3p and occludin mRNA predicted a strong compatibility and complementary molecular interactions that initiated the initial contact and the final docking as shown in Figure 7 and the movie video of the 3D molecular interactions. The in silico approach predicted the critical bases (133-C, 134-A, 135-U, 143-A, 145-U and 146-G) involved in the initial docking interaction that lead to the miRNA-mRNA complex formation. The site-directed mutagenesis studies of the predicted docking sites confirmed the requirement of the initial 3D kissing interactions to modulate the occludin mRNA 3’UTR transcriptional activity.
To further validate the independent regulatory function of miR-200c-3p on occludin mRNA degradation and TJ permeability, the sufficiency of miR-200c-3p overexpression on occludin mRNA and TJ barrier regulation was also demonstrated. The miR-200c-3p over-expression studies by pre-miR-200c-3p transfection demonstrated that miR-200c-3p overexpression by itself was sufficient to induce a rapid degradation of occludin mRNA and cause an increase in intestinal TJ permeability in live mice and also in Caco-2 monolayers. The overexpression of miR-200c-3p, similar to the level produced by IL-1β, had a similar functional effect on enterocyte occludin mRNA degradation and increase in TJ permeability. In addition, occludin overexpression by occludin gene transfection prevented the miR-200c-3p increase in Caco-2 TJ permeability. These data confirmed that the IL-1β effect on intestinal TJ permeability could be entirely accounted for by the increase in enterocyte miR-200c-3p expression and its effect on occludin mRNA. The alternative possibility that miR-200c-3p may also affect other transmembrane proteins was also considered, but bioinformatics algorithm did not identify any miR-200c-3p binding sequence on 3’ UTR regions of other transmembrane TJ proteins, including claudin and JAM family of proteins. As previous studies have shown nuclear factor (NF)-κB mediated increase in MLCK gene and protein expression to be important in proinflammatory mediator induced increase in intestinal TJ permeability68, 69, as an alternative possibility, we also considered the possible involvement of myosin light kinase in miR-200c-3p induced increase in TJ permeability. In our studies, pre-miR-200c-3p transfection did not cause an activation of NF-κB or increase in MLCK mRNA or protein level, ruling out this possibility (data not shown). Previous studies have shown occludin to be a component of the TJ strands, which provide sealing function at the level of TJ’s between two adjacent cells. The occludin overexpression in MDCK cells resulted in thicker TJ strands and improved barrier function70 and targeted occludin depletion correlated with a loss of TJ barrier function.71 Recent studies have shown that occludin knockdown in intestinal epithelial cells both in vitro and in vivo leads to an increased intestinal TJ permeability via a non-restrictive or “leak” pathway and that occludin depletion leads to a preferential increase in flux of macromolecules.51, 63–66 Our results described herein show for the first time that miR-200c-3p has a functional role in occludin mRNA degradation and occludin depletion and in regulating intestinal TJ permeability.
In conclusion, we used in silico modeling to predict potential miRNA targets and the 3D molecular interactions that may mediate IL-1β regulation of intestinal TJ permeability. We tested the in silico predictions using in vivo and in vitro models to determine the molecular mechanisms involved in miR-200c-3p and occludin mRNA molecular interaction and the subsequent IL-1β induced opening of the intestinal TJ barrier. We identified the nucleotide bases critical to the 3D miRNA-mRNA molecular docking process. IL-1β actuates the miR-200c-3p machinery that brings about the degradation of occludin miRNA in intestinal epithelial cells in vitro and in vivo. MIR200C-3p binds to the occludin 3’UTR, initiating its degradation process, culminating in occludin depletion and increase in TJ permeability. Our results also demonstrate the feasibility of repressing/neutralizing miR-200c-3p in an animal model of IBD to protect the intestinal TJ barrier and the development of intestinal inflammation. Thus, our data suggest that microRNA could be a potential therapeutic target to preserve the intestinal TJ barrier function.
Supplementary Material
Funding
This research project was supported by a Veterans Affairs (VA) Merit Review grant from the VA Research Service and National Institute of Diabetes and Q4 Digestive and Kidney Diseases Grant R0–1-DK-64165–01, R0–1-DK- 106072– 01, and J. Lloyd Huck endowment (to T. Y. Ma).
Abbreviations used in this paper:
- DSS
dextran sulfate sodium
- IBD
inflammatory bowel disease
- IL-1b
Interleukin-1b
- MLCK
myosin light chain kinase
- miRNA
microRNA
- mRNA
messenger RNA
- TJ
Tight junction
- UTR
Untranslated region
- PCR
polymerase chain reaction
- 3D
3 dimensional
Footnotes
Credit Authorship Contributions
Manmeet Rawat, PhD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Project administration: Lead; Software: Equal; Validation: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Equal). Meghali Nighot, PhD (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting). Rana Al-Sadi, PhD (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting). Yash Gupta, PhD (Data curation: Supporting; Software: Lead; Visualization: Supporting). Dharmaprakash Viszwapriya, PhD (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting). Gregory Yochum, PhD (Resources: Supporting). Walter Koltun, MD (Resources: Supporting). Thomas Y. Ma, M.D., Ph.D. (Conceptualization: Equal; Formal analysis: Equal; Funding acquisition: Lead; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – review & editing: Lead; approved final version of manuscript: Lead).
Conflict of interest
The authors disclose no conflicts.
References
- 1.Konig J, Wells J, Cani PD, et al. Human Intestinal Barrier Function in Health and Disease. Clin Transl Gastroenterol 2016;7:e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma TY. Intestinal epithelial barrier dysfunction in Crohn’s disease. Proc Soc Exp Biol Med 1997;214:318–27. [DOI] [PubMed] [Google Scholar]
- 3.Madara JL. Loosening tight junctions. Lessons from the intestine. J Clin Invest 1989;83:1089–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol 2009;9:799–809. [DOI] [PubMed] [Google Scholar]
- 5.Arrieta MC, Madsen K, Doyle J, et al. Reducing small intestinal permeability attenuates colitis in the IL10 gene-deficient mouse. Gut 2009;58:41–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyatt J, Vogelsang H, Hubl W, et al. Intestinal permeability and the prediction of relapse in Crohn’s disease. Lancet 1993;341:1437–9. [DOI] [PubMed] [Google Scholar]
- 7.Arnott ID, Kingstone K, Ghosh S. Abnormal intestinal permeability predicts relapse in inactive Crohn disease. Scand J Gastroenterol 2000;35:1163–9. [DOI] [PubMed] [Google Scholar]
- 8.Ma TY, Anderson JM, Turner JR. Chapter 38 - Tight Junctions and the Intestinal Barrier. In: Johnson LR, Ghishan FK, Kaunitz JD, Merchant JL, Said HM, Wood JD, eds. Physiology of the Gastrointestinal Tract (Fifth Edition). Boston: Academic Press, 2012:1043–1088. [Google Scholar]
- 9.Al-Sadi R, Guo S, Dokladny K, et al. Mechanism of interleukin-1beta induced-increase in mouse intestinal permeability in vivo. J Interferon Cytokine Res 2012;32:474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Sadi RM, Ma TY. IL-1beta causes an increase in intestinal epithelial tight junction permeability. J Immunol 2007;178:4641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinarello CA. Biologic basis for interleukin-1 in disease. Blood 1996;87:2095–147. [PubMed] [Google Scholar]
- 12.O’Neill LA, Dinarello CA. The IL-1 receptor/toll-like receptor superfamily: crucial receptors for inflammation and host defense. Immunol Today 2000;21:206–9. [DOI] [PubMed] [Google Scholar]
- 13.Al-Sadi R, Ye D, Said HM, et al. Cellular and molecular mechanism of interleukin-1beta modulation of Caco-2 intestinal epithelial tight junction barrier. J Cell Mol Med 2011;15:970–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Sadi R, Ye D, Said HM, et al. IL-1beta-induced increase in intestinal epithelial tight junction permeability is mediated by MEKK-1 activation of canonical NF-kappaB pathway. Am J Pathol 2010;177:2310–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Sadi R, Ye D, Dokladny K, et al. Mechanism of IL-1beta-induced increase in intestinal epithelial tight junction permeability. J Immunol 2008;180:5653–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinarello CA. The interleukin-1 family: 10 years of discovery. FASEB J 1994;8:1314–25. [PubMed] [Google Scholar]
- 17.Ligumsky M, Simon PL, Karmeli F, et al. Role of interleukin 1 in inflammatory bowel disease--enhanced production during active disease. Gut 1990;31:686–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura M, Saito H, Kasanuki J, et al. Cytokine production in patients with inflammatory bowel disease. Gut 1992;33:933–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunne A, O’Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci STKE 2003;2003:re3. [DOI] [PubMed] [Google Scholar]
- 20.Isaacs KL, Sartor RB, Haskill S. Cytokine messenger RNA profiles in inflammatory bowel disease mucosa detected by polymerase chain reaction amplification. Gastroenterology 1992;103:1587–95. [DOI] [PubMed] [Google Scholar]
- 21.Monteleone G, Fina D, Caruso R, et al. New mediators of immunity and inflammation in inflammatory bowel disease. Curr Opin Gastroenterol 2006;22:361–4. [DOI] [PubMed] [Google Scholar]
- 22.Reinecker HC, Steffen M, Doehn C, et al. Proinflammatory cytokines in intestinal mucosa. Immunol Res 1991;10:247–8. [DOI] [PubMed] [Google Scholar]
- 23.Cominelli F, Pizarro TT. Interleukin-1 and interleukin-1 receptor antagonist in inflammatory bowel disease. Aliment Pharmacol Ther 1996;10 Suppl 2:49–53; discussion 54. [DOI] [PubMed] [Google Scholar]
- 24.Hyams JS, Fitzgerald JE, Wyzga N, et al. Relationship of interleukin-1 receptor antagonist to mucosal inflammation in inflammatory bowel disease. J Pediatr Gastroenterol Nutr 1995;21:419–25. [DOI] [PubMed] [Google Scholar]
- 25.Al-Sadi R, Guo S, Ye D, et al. Mechanism of IL-1beta modulation of intestinal epithelial barrier involves p38 kinase and activating transcription factor-2 activation. J Immunol 2013;190:6596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemetz A, Nosti-Escanilla MP, Molnar T, et al. IL1B gene polymorphisms influence the course and severity of inflammatory bowel disease. Immunogenetics 1999;49:527–31. [DOI] [PubMed] [Google Scholar]
- 27.Carvalho FA, Aitken JD, Gewirtz AT, et al. TLR5 activation induces secretory interleukin-1 receptor antagonist (sIL-1Ra) and reduces inflammasome-associated tissue damage. Mucosal Immunol 2011;4:102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hultgren OH, Berglund M, Bjursten M, et al. Serum interleukin-1 receptor antagonist is an early indicator of colitis onset in Galphai2-deficient mice. World J Gastroenterol 2006;12:621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boivin MA, Ye D, Kennedy JC, et al. Mechanism of glucocorticoid regulation of the intestinal tight junction barrier. Am J Physiol Gastrointest Liver Physiol 2007;292:G590–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen L, Black ED, Witkowski ED, et al. Myosin light chain phosphorylation regulates barrier function by remodeling tight junction structure. J Cell Sci 2006;119:2095–106. [DOI] [PubMed] [Google Scholar]
- 31.Ye D, Ma I, Ma TY. Molecular mechanism of tumor necrosis factor-alpha modulation of intestinal epithelial tight junction barrier. Am J Physiol Gastrointest Liver Physiol 2006;290:G496–504. [DOI] [PubMed] [Google Scholar]
- 32.Ma TY, Tran D, Hoa N, et al. Mechanism of extracellular calcium regulation of intestinal epithelial tight junction permeability: role of cytoskeletal involvement. Microsc Res Tech 2000;51:156–68. [DOI] [PubMed] [Google Scholar]
- 33.Clayburgh DR, Rosen S, Witkowski ED, et al. A differentiation-dependent splice variant of myosin light chain kinase, MLCK1, regulates epithelial tight junction permeability. J Biol Chem 2004;279:55506–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma TY, Hollander D, Tran LT, et al. Cytoskeletal regulation of Caco-2 intestinal monolayer paracellular permeability. J Cell Physiol 1995;164:533–45. [DOI] [PubMed] [Google Scholar]
- 35.Ye D, Guo S, Al-Sadi R, et al. MicroRNA regulation of intestinal epithelial tight junction permeability. Gastroenterology 2011;141:1323–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gan HH, Gunsalus KC. Tertiary structure-based analysis of microRNA-target interactions. RNA 2013;19:539–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y, Huang Y, Gong Z, et al. Automated and fast building of three-dimensional RNA structures. Sci Rep 2012;2:734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jossinet F. S2S-Assemble2: a Semi-Automatic Bioinformatics Framework to Study and Model RNA 3D Architectures, 2014. [Google Scholar]
- 40.Yamasaki S HT, Asai K, et al. Tertiary structure prediction of RNA-RNA complexes using a secondary structure and fragment-based method. J Chem Inf Model 2014;54:672–82. [DOI] [PubMed] [Google Scholar]
- 41.Yamasaki S, Nakamura S, Fukui K. Prospects for tertiary structure prediction of RNA based on secondary structure information. J Chem Inf Model 2012;52:557–67. [DOI] [PubMed] [Google Scholar]
- 42.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 2004;25:1605–12. [DOI] [PubMed] [Google Scholar]
- 43.Zok T, Antczak M, Zurkowski M, et al. RNApdbee 2.0: multifunctional tool for RNA structure annotation. Nucleic Acids Res 2018;46:W30–W35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Antczak TZ M, Popenda M, Lukasiak P, Adamiak RW, Blazewicz J, Szachniuk M. RNApdbee – a webserver to derive secondary structures from pdb files of knotted and unknotted RNAs. Nucleic Acids Research;42:W368–W372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kevin J. Bowers EC, Huafeng Xu, Dror Ron O., Eastwood Michael P., Gregersen Brent A., Klepeis John L., Kolossváry István, Moraes Mark A., Sacerdoti Federico D., Salmon John K., Shan Yibing, and Shaw David E.. Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. Proceedings of the ACM/IEEE Conference on Supercomputing (SC06), Tampa, Florida 2006. [Google Scholar]
- 46.Dokladny K, Ye D, Kennedy JC, et al. Cellular and molecular mechanisms of heat stress-induced up-regulation of occludin protein expression: regulatory role of heat shock factor-1. Am J Pathol 2008;172:659–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ye D, Ma TY. Cellular and molecular mechanisms that mediate basal and tumour necrosis factor-alpha-induced regulation of myosin light chain kinase gene activity. J Cell Mol Med 2008;12:1331–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wick DA, Seetharam B, Dahms NM. Biosynthesis and secretion of the mannose 6-phosphate receptor and its ligands in polarized Caco-2 cells. Am J Physiol 1999;277:G506–14. [DOI] [PubMed] [Google Scholar]
- 49.Ma TY, Iwamoto GK, Hoa NT, et al. TNF-alpha-induced increase in intestinal epithelial tight junction permeability requires NF-kappa B activation. Am J Physiol Gastrointest Liver Physiol 2004;286:G367–76. [DOI] [PubMed] [Google Scholar]
- 50.Clayburgh DR, Barrett TA, Tang Y, et al. Epithelial myosin light chain kinase-dependent barrier dysfunction mediates T cell activation-induced diarrhea in vivo. J Clin Invest 2005;115:2702–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Al-Sadi R, Khatib K, Guo S, et al. Occludin regulates macromolecule flux across the intestinal epithelial tight junction barrier. Am J Physiol Gastrointest Liver Physiol 2011;300:G1054–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wirtz S, Neufert C, Weigmann B, et al. Chemically induced mouse models of intestinal inflammation. Nat Protoc 2007;2:541–6. [DOI] [PubMed] [Google Scholar]
- 53.In J, Foulke-Abel J, Zachos NC, et al. Enterohemorrhagic Escherichia coli reduce mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell Mol Gastroenterol Hepatol 2016;2:48–62 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sato T, Stange DE, Ferrante M, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011;141:1762–72. [DOI] [PubMed] [Google Scholar]
- 55.Hu Z. Insight into microRNA regulation by analyzing the characteristics of their targets in humans. BMC Genomics 2009;10:594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kohaar I, Ploss A, Korol E, et al. Splicing diversity of the human OCLN gene and its biological significance for hepatitis C virus entry. J Virol 2010;84:6987–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lewis BP, Shih IH, Jones-Rhoades MW, et al. Prediction of mammalian microRNA targets. Cell 2003;115:787–98. [DOI] [PubMed] [Google Scholar]
- 58.Subramanian VS, Marchant JS, Ye D, et al. Tight junction targeting and intracellular trafficking of occludin in polarized epithelial cells. Am J Physiol Cell Physiol 2007;293:C1717–26. [DOI] [PubMed] [Google Scholar]
- 59.George MD, Wehkamp J, Kays RJ, et al. In vivo gene expression profiling of human intestinal epithelial cells: analysis by laser microdissection of formalin fixed tissues. BMC Genomics 2008;9:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grimson A, Farh KK, Johnston WK, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 2007;27:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia DM, Baek D, Shin C, et al. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol 2011;18:1139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Friedman RC, Burge CB. MicroRNA target finding by comparative genomics. Methods Mol Biol 2014;1097:457–76. [DOI] [PubMed] [Google Scholar]
- 63.Buschmann MM, Shen L, Rajapakse H, et al. Occludin OCEL-domain interactions are required for maintenance and regulation of the tight junction barrier to macromolecular flux. Mol Biol Cell 2013;24:3056–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Odenwald MA, Turner JR. The intestinal epithelial barrier: a therapeutic target? Nat Rev Gastroenterol Hepatol 2017;14:9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mir H, Meena AS, Chaudhry KK, et al. Occludin deficiency promotes ethanol-induced disruption of colonic epithelial junctions, gut barrier dysfunction and liver damage in mice. Biochim Biophys Acta 2016;1860:765–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buckley A, Turner JR. Cell Biology of Tight Junction Barrier Regulation and Mucosal Disease. Cold Spring Harb Perspect Biol 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Itallie CM, Holmes J, Bridges A, et al. Claudin-2-dependent changes in noncharged solute flux are mediated by the extracellular domains and require attachment to the PDZ-scaffold. Ann N Y Acad Sci 2009;1165:82–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nighot M, Rawat M, Al-Sadi R, et al. Lipopolysaccharide-Induced Increase in Intestinal Permeability Is Mediated by TAK-1 Activation of IKK and MLCK/MYLK Gene. Am J Pathol 2019;189:797–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Al-Sadi R, Guo S, Ye D, et al. TNF-alpha Modulation of Intestinal Tight Junction Permeability Is Mediated by NIK/IKK-alpha Axis Activation of the Canonical NF-kappaB Pathway. Am J Pathol 2016;186:1151–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCarthy KM, Skare IB, Stankewich MC, et al. Occludin is a functional component of the tight junction. J Cell Sci 1996;109 ( Pt 9):2287–98. [DOI] [PubMed] [Google Scholar]
- 71.Li D, Mrsny RJ. Oncogenic Raf-1 disrupts epithelial tight junctions via downregulation of occludin. J Cell Biol 2000;148:791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.