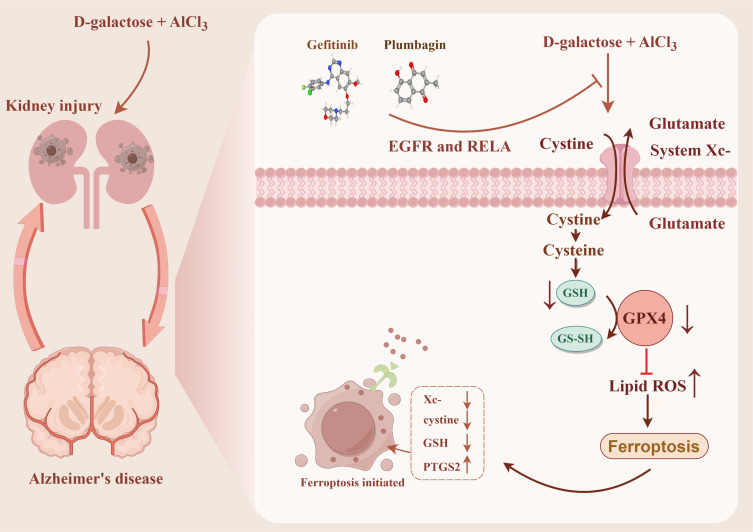
Abstract
Background
The phenomenon of “kidney-brain crosstalk” has stimulated scholarly inquiry into the correlations between kidney injury (KI) and Alzheimer’s disease (AD). Nonetheless, the precise interactions and shared mechanisms between KI and AD have yet to be fully investigated. The primary goal of this study was to investigate the link between KI and AD, with a specific focus on identifying diagnostic biomarkers for KI-related AD.
Methods
The first step of the present study was to use Mendelian randomization (MR) analysis to investigate the link between KI and AD, followed by verification of in vivo and in vitro experiments. Subsequently, bioinformatics and machine learning techniques were used to identify biomarkers for KI-associated ferroptosis-related genes (FRGs) in AD, which were validated in following experiments. Moreover, the relationship between hub biomarkers and immune infiltration was assessed using CIBERSORT, and the potential drugs or small molecules associated with the core biomarkers were identified via the DGIdb database.
Results
MR analysis showed that KI may be a risk factor for AD. Experiments showed that the combination of D-galactose and aluminum chloride was found to induce both KI and AD, with ferroptosis emerging as a bridge to facilitate crosstalk between KI and AD. Besides, we identified EGFR and RELA have significant diagnostic value. These biomarkers are associated with NK_cells_resting and B_cells_memory and could be targeted for intervention in KI-related AD by treating gefitinib and plumbagin.
Conclusion
Our study elucidates that ferroptosis may be an important pathway for kidney-brain crosstalk. Notably, gefitinib and plumbagin may be therapeutic candidates for intervening in KI-associated AD by targeting EGFR and RELA.
Keywords: kidney injury, Alzheimer’s disease, kidney-brain crosstalk, ferroptosis, bioinformatics, machine learning
Graphical Abstract
Introduction
The phenomenon of “kidney-brain crosstalk” has stimulated scholarly inquiry into the correlations between kidney injury (KI) and Alzheimer’s disease (AD). An acute kidney injury (AKI) or chronic kidney disease (CKD) may be the cause of kidney injury (KI).1 AKI is characterized by acute and transient renal dysfunction, which develops into chronic kidney disease (CKD) over time.2 Chronic kidney disease (CKD), which represents a progressive and irreversible loss of kidney function, is a long-term condition.3 Notably, the prevalence of AKI and CKD are both high worldwide.4
Alzheimer’s disease, also called AD, is the primary cause of dementia on a global scale and is a significant issue in public health.5 AD is responsible for half to three-quarters of the 50 million individuals globally who have dementia.6 de Oliveira et al7 found that gender, education, changes in coronary heart disease risk, creatinine clearance, body mass index, and APOE haplotypes all affected the rate of cognitive and functional decline in AD. Creatinine is the primary marker for kidney damage.8 Based on the concept of kidney-brain crosstalk proposed by several scholars, we explored the interconnection between kidney injury and AD. Initial research has observed that “kidney-brain crosstalk” in CKD frequently accompanies a variety of neurological disorders, including cerebrovascular disease, cognitive deficits, and neuropathy.9 Additionally, subsequent studies have shown that AKI significantly affects the extrarenal organs, especially the brain’s function.10 This crosstalk may be caused by cytokine-induced injury, sodium dysregulation, oxidative stress, and leukocyte extravasation.9 Notably, the pathophysiology of functional crosstalk between the kidney and brain is characterized by oxidative stress and inflammation. In the natural course of CKD, the above process can lead to neuropsychiatric disorders, cognitive impairments, and dementias.11 Meanwhile, in vitro and in vivo studies have proposed that the kidney plays an important role in the peripheral clearance of Aβ and tau.12,13 Stacking of Aβ and tau leads to aggravating AD pathology when the kidneys are damaged, which indicates that KI is tightly connected to the development of AD and that CKD patients are more likely to develop cognitive impairment and AD.14,15 Due to its insidious onset, long latency, and various ambiguous mechanisms, AD is difficult to detect at its early stages. A biomarker facilitates diagnosis, fosters the assessment of a disease’s severity, and allows clinicians to assess a treatment’s effectiveness using surrogates.16 KI (AKI and CKD) is often accompanied by varying degrees of neurological dysfunction, such as AD. Nonetheless, the precise interactions and shared mechanisms between KI and AD have yet to be fully investigated. Exploring the possibility of including biomarkers in diagnosing KI-related AD is proposed to identify AD patients as early as possible and help determine therapeutic interventions.
Although the underlying molecular mechanisms KI contributes to AD are complex and ambiguous, the oxidative stress in kidney-brain crosstalk points out a new direction. Oxidative stress is the central mechanism of ferroptosis.17 The process of ferroptosis, a new form of programmed cell death, is iron-dependent, oxidative, and characterized by elevated reactive oxygen species (ROS) and reduced glutathione levels.18 It is possible to induce ferroptosis by accumulating ROS, increasing reactive iron, and decreasing GPX4 activity, thereby contributing to the progression of AKI and AD.19,20 Regrettably, there is a lack of studies on biomarkers involved in the pathogenesis of AD in KI-related ferroptosis-related genes (FRGs). We took “ferroptosis” as a point cut to identify biomarkers to explore the effective treatment of KI-related AD, which might have been a good choice.
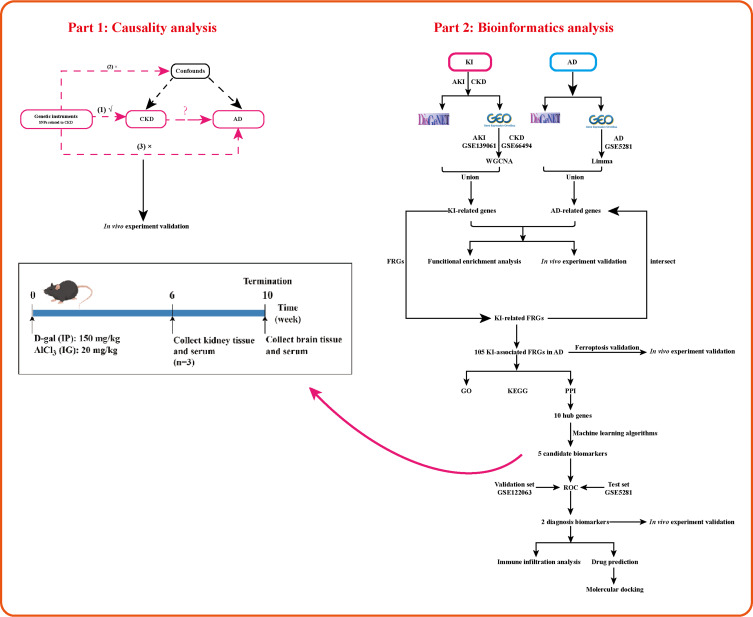
Our study used Mendelian randomization (MR) to examine the causal association between CKD and AD. We also applied multiple integrated bioinformatics tools to elucidate kidney-brain crosstalk in KI and AD, investigate the common mechanisms, and identify diagnostic biomarkers of AD in KI-associated FRGs. Subsequently, we utilized in vivo experiments to further confirm the above analysis. Finally, we explored the immune cellular signatures of diagnostic biomarkers in AD to reveal associations between pivotal genes and the immunological landscape. Generally, we have set up common biomarkers and mechanisms for KI and AD for the first time through this study, which provides clues for exploring therapeutic strategies for KI-related AD. The study flow chart is shown in Figure 1.
Figure 1.
The study flow diagram was created by Figdraw.
Materials and Methods
MR Analysis
Study Design
The instrumental variables employed in this bidirectional MR study consisted of multiple single-nucleotide polymorphisms (SNPs) that capture the overall genetic diversity within the human population. The MR approach relies on three fundamental assumptions to produce unbiased estimates: (1) the genetic instrumental variable must be associated with the exposure of interest; (2) confounding factors must not directly influence the instrumental variables.
Data Sources
The IEU Open GWAS project (https://gwas.mrcieu.ac.uk) provided us with datasets relevant to CKD and AD. Since the study included publicly available data, ethics approval was not required. The CKD data includes 12385 CKD patients and 104780 controls. The AD data contains 17008 patients with AD and 37154 controls (Table 1).
Table 1.
Features of GWAS Consortiums are Applied for Each Variable
| Traits | Sample Size (Cases/Controls) | Population | Consortium |
|---|---|---|---|
| Chronic kidney disease (CKD) | 12385/104780 | European | IEU |
| Alzheimer’s disease (AD) | 17008/37154 | European | IEU |
Disease-Related Microarray Data Collection and Processing
The DisGeNET database (http://www.disgenet.org/web/DisGeNET/) was applied to collect kidney injury (including acute kidney injury and CKD) and AD-related genes. Meanwhile, raw expression profiling datasets for kidney injury and AD, including GSE139061, GSE66494, GSE5281, and GSE122063, were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). As shown in Table 2, the datasets have detailed descriptive information.
Table 2.
The Information on the GSE Datasets from the GEO Database
Differentially Expressed Genes (DEGs) and Ferroptosis-Related Genes (FRGs) Identification
Our analysis of AKI, CKD, and AD datasets used the “Limma” package in R software to identify differentially expressed genes (DEGs). The filtering condition was set as |Log (fold change, FC)| > 1, p < 0.05. Among these, logFC > 1 corresponded to up-regulated genes and logFC < −1 to down-regulated genes. Finally, with the help of the “ggplot2” and “pheatmap” packages, expression patterns of DEGs were visualized as volcano maps and heatmaps. Ferroptosis-related genes (FRGs) were downloaded from the FerrDB database (http://www.zhounan.org/ferrdb/current/).
An Analysis of Weighted Gene Co-Expression Networks (WGCNA)
WGCNA is commonly applied to calculate disease-related gene modules, as described previously.21 Briefly, in order to carry out the WGCNA analysis, the package “WGCNA” was utilized. In addition, to ensure a co-expression network with signed scale-free parameters, the corresponding β and R2 were chosen as soft threshold parameters.
Functional Enrichment and Protein-Protein Interaction (PPI) Network Analysis
The biological function and concrete mechanism of FRGs were explored via Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis using the Online platform of Sanger-Box (http://vip.sangerbox.com/home.html).22 The screen criteria are FDR < 0.1, P value < 0.05. Meanwhile, the GeneMANIA tool was also adapted to investigate the function of common genes.
The interactions between KI and AD were investigated via the PPI network, constructed by the STRING database and visualized through Cytoscape.
Machine Learning Algorithms
The Least Absolute Shrinkage and Selection Operator (LASSO), Random Forest (RF), Support Vector Machine Recursive Feature Extraction (SVM-RFE), and Gradient Boosting Machine (GBM) were combined to recognize the characteristic genes.23 Finally, the overlapping genes in the three machine learning algorithms described above were defined as signature genes.
The Construction of a Receiver Operating Characteristic (ROC) Curve and a Nomogram
The diagnostic value of biomarkers was evaluated using the ROC curve, which was constructed using the survival ROC package. Besides, the nomogram was created to validate the accuracy of ROC and further verified in the calibration curve.
Immune Cell Infiltrations Analysis
Based on CIBERSORT, an immune-related algorithm that analyzes 22 types of immunocytes, we analyzed the immune landscape of CKD and AD samples.24 The Pearson correlation coefficient was used to determine whether the hub genes correlate with immune infiltrating cells. Finally, with the “vioplot” and “pheatmap” R packages, it was visualized.
Establishment of the KI and AD Mice Models
The male C57BL/6J mice (18–22 g) were supplied by the Nanjing Qinglongshan Animal Cultivation Farm (Nanjing, China). All mice were housed in a dark/light (12 h/12 h) environment at 25 ± 2 °C and 60 ± 10% relative humidity. The animals were fed standard feed and allowed to drink at their leisure. According to the animal welfare guidelines, all animal care procedures and experiments followed the 3R principle (reduction, replacement, refinement). China Pharmaceutical University’s Ethics Committee approved all animal experiments.
We randomly assigned the experimental animals into two groups (n = 10): (1) Control and (2) Model (D-gal, 150 mg/kg+AlCl3, 20 mg/kg) group. The D-gal (intraperitoneal injection) and AlCl3 (intragastric administration) were given to the Model group mice for 10 weeks.
Analysis of Kidney Coefficient
Mice were weighed once a week. Then, 50 mg/kg of sodium pentobarbital was used for anaesthesia to collect blood samples. After that, mice were sacrificed by cervical dislocation to isolate kidney tissues (both left and right). Accordingly, kidney coefficients were calculated as follows: Kidney coefficient = kidney weight (mg) × 100/ body weight (g).
Detected the Levels of Serum Creatinine (CRE), Blood Urea Nitrogen (BUN), Malondialdehyde (MDA), and Glutathione Peroxidase (GPx) in Mice
According to the manufacturer’s instructions, the serum creatinine (CRE) and urea nitrogen levels were determined using CRE assays (C011-2-1) and blood urea nitrogen (BUN, C013-2-1) kits manufactured by Jiancheng Bio (Nanjing, China). The levels of Malondialdehyde (MDA) and Glutathione peroxidase (GPx) in the kidney tissue were assayed using the MDA Assay Kit (S0131S, Beyotime, China) and the GPx Assay Kit (S0056, Beyotime, China), respectively.
Nissl Staining and Hematoxylin-Eosin (HE) Staining
As described in the previous study, Nissl and HE staining were performed.25,26 We preserved brain and kidney tissue in 4% paraformaldehyde, embedded it in paraffin, sectioned it, and mounted it on slides. After deparaffinizing and rehydrating, the tissue sections were stained with toluidine blue for Nissl bodies and hematoxylin and eosin for kidney tissue.
Establishment of the Cell Model
Rat pheochromocytoma PC12 cells were purchased by the Chinese Academy Cell Resource Center (Shanghai, China). As described previously,27 we utilized Aβ25-35 (20 μM) to induce injury in PC12 cells, establishing an in vitro model of Alzheimer’s disease (AD). The experiment was divided into three parts. In Part 1, we divided the cells into two groups: Control and Model. In Part 2, we divided the cells into three groups: Control, Model, and SC75741 (RELA inhibitor, 5 μM). In Part 3, we divided the cells into three groups: Control, Model, and Gefitinib (EGFR inhibitor, 200 nM).
Western Blot (WB) Analysis
The kidney and brain tissues were lysed with RIPA protein lysate (P0013B, Beyotime, China) with PMSF (ST506, Beyotime, China), and the protein concentration was determined with a BCA kit (P0010, Beyotime, China). Then, the detailed WB procedure was conducted according to the methods outlined in a previous study.28 Here are the antibodies used in this study: anti-Aβ1-42 (1:1000, 25524-1-AP, Proteintech), anti-PTGS2 (1:500, sc-19999, Santa Cruz,), anti-GPX4 (1:500, 67763-1-Ig, Proteintech), anti-EGFR (1:800, 51071-2-AP, Proteintech,), RELA (1:1000, 8242T, CST), and β-actin (1:20,000, 66009-1-lg, Proteintech). Finally, a kit for ECL chemiluminescence was used to enhance color development (Tanon, Shanghai).
Prediction of the Biomarker-Associated Candidate Drug
Through a search of the Drug Gene Interaction Database (DGIdb) (https://www.dgidb.org), potential drugs or molecules that interact with the hub genes have been predicted.29 Our analysis of the candidate genes led us to select the interacting drugs as potential therapeutic agents for KI-associated FRGs involved in AD progression using Cytoscape.
Analysis of Molecular Docking and Intermolecular Interactions
The molecular docking method is a powerful tool for analyzing intermolecular interactions. Here, we collected the candidate drugs (gefitinib and plumbagin) from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and downloaded the targets (EGFR and RELA) from the PDB database (http://www.rcsb.org/pdb/) to conduct molecular docking via Autodock Vina. We also analyze the intermolecular interactions between drugs (gefitinib and plumbagin) and proteins (EGFR and RELA) through LigPlot+ software.
Statistical Analysis
For MR analysis, the inverse-variance weighted (IVW) method, which assumes no unbalanced horizontal pleiotropy, was used as the main analysis method.30 We conducted complementary analyses using MR-Egger regression, MR-Weighted-median, MR-Simple-mode, and MR-Weighted-mode. Statistical analyses were conducted using the MendelianRandomization, TwoSampleMR, and MR-PRESSO packages in R software.
In the present study, all results were expressed as mean ± SEM. The experimental data were statistics via t-tests in GraphPad Prism 9.5 software (San Diego, California, United States), and P < 0.05 was considered significant.
Results
Analysis of the Link Between KI and AD
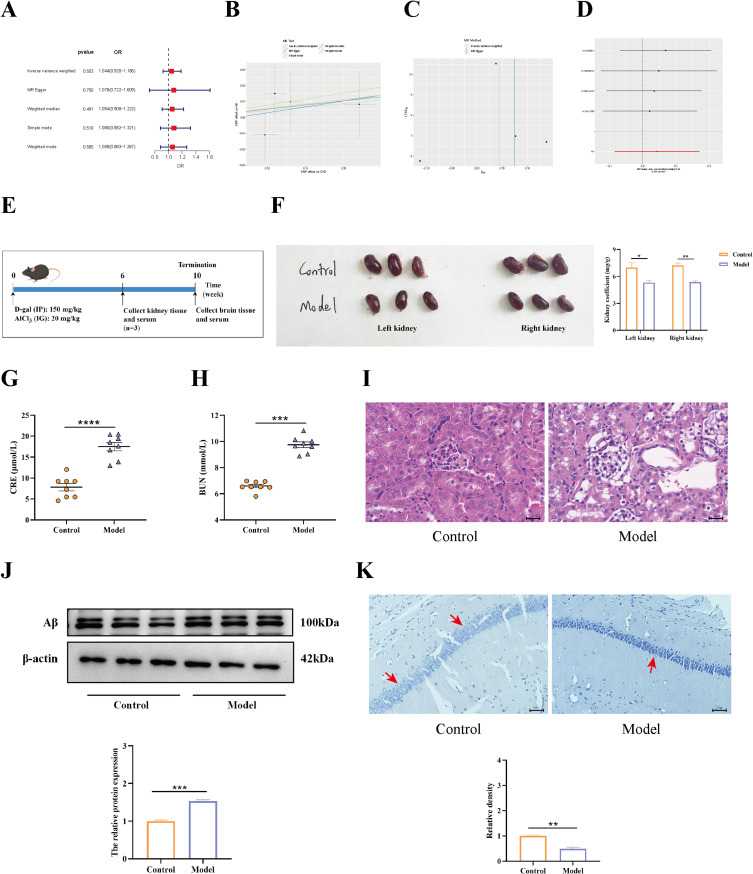
We conducted an MR analysis to determine the causal link between CKD and AD. In the study, The IVW results exhibited that, although CKD has no truly causal relationship with AD (OR = 1.044, 95% CI = 0.920–1.186, P = 0.503) (Figure 2A), there was an increased risk of AD associated with CKD prevalence (Figure 2B). Considering the Cochrane Q test statistics revealed heterogeneity (P = 0.738), a fixed-effect model was used. Also, no notable pleiotropy was observed in MR-Egger (P = 0.887) (Figure 2C). Moreover, no outliers were detected by the MR-PRESSO test (global test P = 0.806). A leave-one-out analysis was used to verify how each SNP affected the overall causal estimates (Figure 2D). Collectively, the results mentioned above suggested that the incidence of AD was significantly linked with increased risk for CKD.
Figure 2.
The results of the link between KI and AD. (A) Various MR methods (IVW, MR egger, Weighted median, Simple mode, and Weighted-mode) were used to estimate causal effects. (B) Scatter plot for the MR analysis. (C) Additionally, the funnel plots showed no notable pleiotropy (P = 0.887). (D) The leave-one-out plot of SNPs associated with CKD and their risk on AD. (E) The animal experiment flow chart was created by applying Figdraw. (F) The kidney coefficient in the control and model groups. D-gal+AlCl3-induced KI was examined by measuring serum BUN (G) levels and serum CRE (H) levels via ELISA kits. (I) The histopathological changes in the kidneys of mice treated with d-gal+AlCl3 (n = 3). The original magnification is 10×40. (J) The AD marker protein Aβ was detected via Western blot, which was normalized by β-actin (n = 3). (K) Nissl staining (magnification 10 × 40, n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.001.
A previous study confirmed that D-gal or AlCl3 can trigger KI and AD, respectively. Thus, we made the bold speculation that the occurrence of KI and AD has some linkage. To test this conjecture, we established a D-gal+AlCl3-induced mouse model (Figure 3E). At six weeks, in comparison to the control group, mice treated with D-gal+AlCl3 had significantly lower kidney coefficients (Figure 3F). At the same time, the serum CRE and BUN levels were significantly elevated in the model group (Figure 3G and H). Besides, HE staining showed that kidney tissue damage pathology was obvious in the model mice, which showed that the appearance was tubular cell vacuolization, tubular epithelial cell necrosis, and glomerular atrophy (Figure 3I). All of this demonstrated that D-gal+AlCl3 induced KI. Subsequently, on week 10 after modelling, Aβ protein expression was examined in the hippocampal tissues of the mice, and it was found that Aβ accumulated in the model mice brains (Figure 3J). Moreover, Nissl staining showed that Nissl bodies were reduced in the hippocampal tissue of the model group (Figure 3K). This suggests that D-gal+AlCl3 also induces AD pathology. The above data illustrate that D-gal+AlCl3 can induce KI and AD, in which KI onset precedes AD, demonstrating that KI is closely related to the onset of AD.
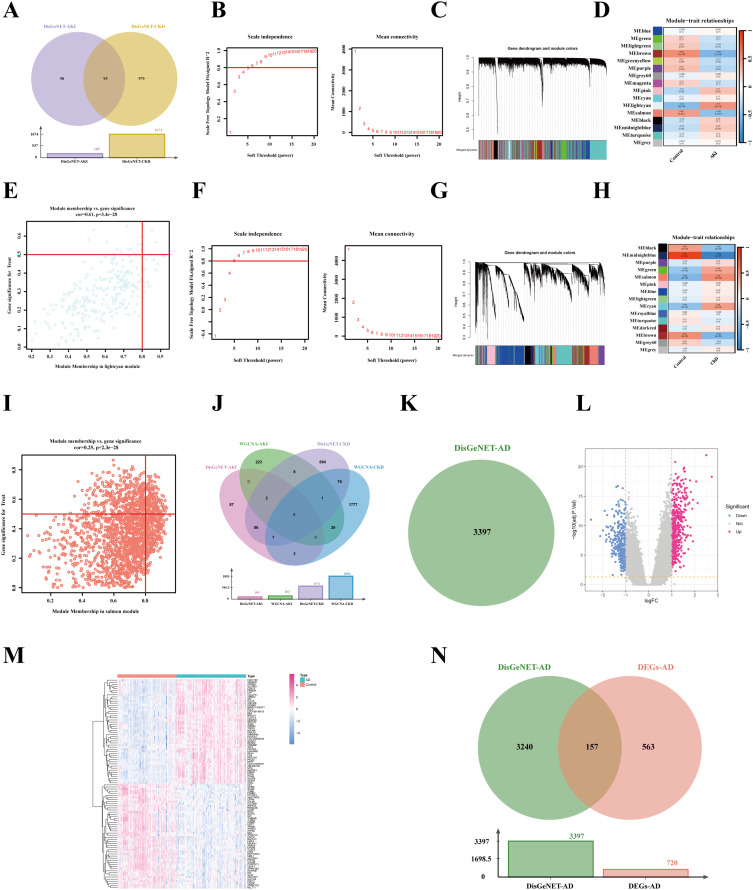
Figure 3.
Identification of KI-related genes and AD-related genes. (A) AKI and CKD diseases-related genes from the DisGeNET database. (B-E) WGCNA analysis in AKI dataset. (B) An analysis of network topology for various soft-threshold powers. (C) Cluster dendrogram of co-expression genes in AKI. (D) A relationship between a module and a trait in control and AD. (E) The correlation between genes and AD in the lightcyan module. (F-I) WGCNA analysis in CKD dataset. (F) According to the average connectivity and scale Independence, the soft threshold was selected at β = 6, based on the scale-free topology model. (G) The cluster dendrogram of co-expression genes in CKD. (H) Heatmap displaying module eigengenes in relation to CKD status. (I) Correlation plot between salmon module membership and gene significance. (J) All the KI-related genes from disease and GEO databases. (K) AD-related genes from the DisGeNET diseases database. (L–N) Differentially expressed genes (DEGs) analysis of AD datasets from the GEO database. (L) Volcano plot of DEGs between control and AD. (M) The heatmap shows the differences in partial gene expression between the control and AD groups. (N) All the AD-related genes from disease and GEO databases.
Identification of Disease-Related Genes and Gene Modules Associated with KI
As a means of collecting enough KI-related genes, we searched the DisGeNET and GEO databases under the keywords “chronic kidney disease” and “acute kidney injury”. The disease database included AKI (185) and CKD (1074) (Figure 3A). Meanwhile, in GEO datasets, we performed WGCNA analysis to identify the AKI-related module genes. Scale-free networking was ensured by choosing the power of 8 as a soft threshold (Figure 3B), which was chosen to generate hierarchical clustering trees (Figure 3C). In addition, a total of 16 modules were obtained, with the lightcyan module being the most relevant to the AKI trait (Figure 3D). Besides, we found that the lightcyan module was positively correlated with AKI (cor = 0.61, p = 3.4e-28) (Figure 3E).
Meanwhile, in the CKI dataset, we set a soft threshold of β = 6 and R2 = 0.9 to ensure a scale-free network (Figure 4F). Subsequently, 15 modules were generated, of which the salmon module was most relevant to CKD (Figure 4G and H). Furthermore, the salmon module (cor = 0.25, p = 2.3e-28) showed high genetic significance and module membership (Figure 4I).
Figure 4.
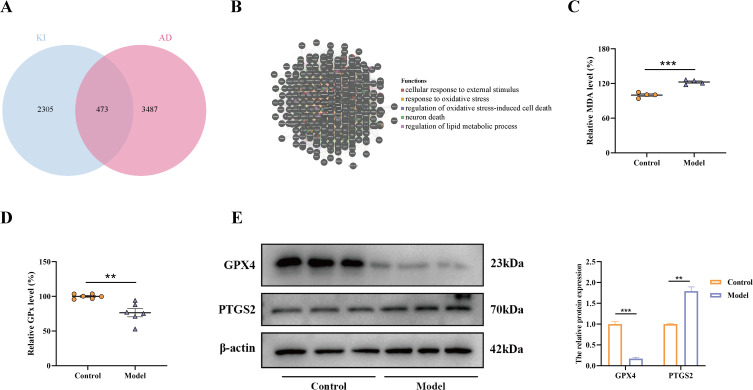
Predictive and validating the co-mechanisms of KI and AD. (A) Venn diagram indicating the intersection between the KI- and AD-related genes. (B) Functional analysis of shared genes in KI and AD via GeneMANIA tool. The serum levels of MDA (C) and GPx (D) were detected using an ELISA assay. (E) The protein levels of ferroptosis-related proteins GPX4 and PTGS2 were determined by Western blot, and β-actin was employed as internal references (n = 3). **, p < 0.01; ***, p < 0.001.
Finally, the disease and GEO databases were merged to include AKI and CKD-related genes. After removing duplicates, 3193 KI-related genes were gained, including AKI (446) and CKD (2883) (Figure 4J).
Collection of Disease-Related Genes and Identified DEGs in AD
In order to obtain AD-related genes, we searched DisGeNET and GEO databases using the keyword “Alzheimer’s disease”. The results showed that DisGeNET collected 3397 AD-related genes (Figure 4K). In addition, a Limma differential expression gene analysis of the AD dataset found 720 DEGs, including 415 up-regulated genes and 305 down-regulated genes (Figure 4L and M). Next, 3960 genes were defined as AD-associated genes after removing duplicates by combining the genes obtained from the disease database with the GEO database (Figure 4N).
Predictive and Experimental Results About the Mechanism by Which KI and AD
As a result of the intersection between KI-related genes (AKI and CKD) and AD-related genes obtained above, we obtained 473 genes with common characteristics (Figure 4A). This implies a relationship between KI and AD. Then, functional enrichment was analyzed to explore the potential common pathogenic mechanisms in KI and AD. The result found that these genes were mainly associated with the biological processes of response to oxidative stress, regulation of oxidative stress-induced cell death, neuron death, and regulation of lipid metabolic process (Figure 4B). The term ferroptosis describes a new type of cell death characterized by irregular iron and lipid metabolism, oxidative stress, and low glutathione levels.31 This suggests that ferroptosis may be a common mechanism of KI and AD.
To further confirm the reliability of the prediction results, we detected the activities of MDA and GPx in kidney tissues. The results revealed that in the model group, MDA activity was significantly elevated (Figure 5C), marking the occurrence of oxidative stress. Meanwhile, in the model group, the GPx activity was reduced (Figure 5D), the GPX4 protein expression level was significantly down-regulated, and PTGS2 was significantly up-regulated (Figure 5E), indicating the occurrence of ferroptosis. Overall, the predicted results, together with the experimental results, elucidated that the common pathogenic mechanism of KI and AD may be associated with ferroptosis.
Figure 5.
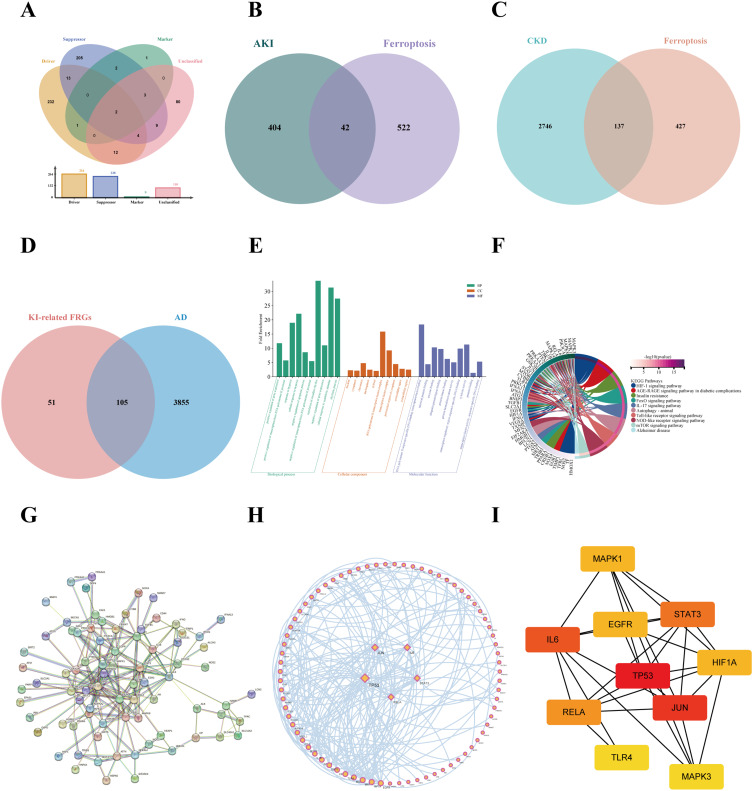
Functional enrichment analysis of KI-related FRGs in AD and protein-protein interaction (PPI) network construction. (A) Venn plot of ferroptosis-related genes (FRGs) from FerrDB database. (B) Identified the intersection of AKI-related genes with FRGs via the Venn diagrams. (C) Identified the intersection of CKD-related genes with FRGs via the Venn diagrams (D) Screened the KI-related FRGs in AD-related genes via the venn diagram. (E) An analysis of the top ten GO-BP, GO-CC, and GO-MF enrichments. (F) A circos plot representing the KEGG analysis results of shared FRGs. The PPI network was created by the STRING database (G) and further visualized through Cytoscape (H). (I) The top ten hub genes were selected via Cytohubba analysis.
Identification of KI-Related FRGs in AD
In order to determine which KI-related FRGs contribute to AD progression, we first gathered FRGs through the Ferrdb database. The 564 FRGs were obtained in the Ferrdb database, which contained driver (264), suppressor (238), marker (9), and unclassified (110) (Figure 5A). Next, a total of 156 KI-associated FRGs were identified after being duplicated, among which 42 were in AKI and 137 were related to CKD, respectively (Figure 5B and C). Eventually, we further identified 105 KI-associated FRGs associated with AD (Figure 5D).
The Functional Enrichment and PPI Network Analysis Results
Functional enrichment analysis (GO and KEGG) was performed to gain insight into the biological functions and pathways involved in these 105 common FRGs. According to GO enrichment analysis, these genes participate in several biological processes (BP), including positive regulation of gene expression, positive regulation of transcription from RNA polymerase II promoter, and hypoxia response. Their major cellular components include the nucleus, cytoplasm, and chromatin. Most of these genes have also been linked to RNA polymerase II sequence-specific DNA binding transcription factor binding, identical protein binding, and enzyme binding processes within molecular function (MF) categories (Figure 6E).
Figure 6.
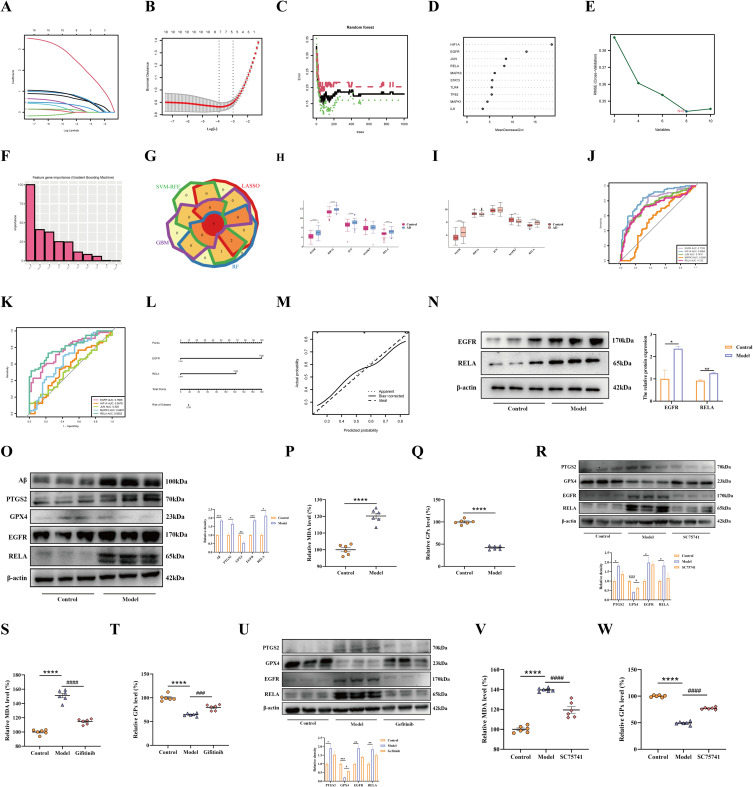
Screened candidate diagnostic biomarkers of AD progression and infiltration analysis of biomarkers. (A) Candidate gene LASSO coefficient profiles. (B) A cross-validation study was conducted to determine which tuning parameter log (Lambda) is most effective. (C) Candidate gene profiles according to RF coefficients. (D) Random forest algorithm MeanDecreaseGini for hub genes. (E) Biomarker screening using the SVM-RFE algorithm. (F) Biomarker screening using the GBM algorithm. (G) Four machine learning algorithm screening results are shown in a Venn diagram, in which the overlap genes were defined as candidate biomarkers. (H) The expression of 5 candidate biomarkers (EGFR, HIF1A, JUN, MAPK3, and RELA) between control and AD groups in the GSE5281 test dataset. (I) The expression of 5 candidate biomarkers (EGFR, HIF1A, JUN, MAPK3, and RELA) between control and AD groups in the GSE122063 validation dataset. (J) The ROC curves of the 5 candidate biomarkers (EGFR, HIF1A, JUN, MAPK3, and RELA) are based on the GSE5281 test dataset. (K) The ROC curves of the 5 candidate biomarkers (EGFR, HIF1A, JUN, MAPK3, and RELA) in the GSE122063 validation dataset. (L) Nomograms of AD. (M) Calibration curves of the nomogram. (N) Western blot detection of EGFR and RELA (n = 3). (O) The protein levels were determined by Western blot, and β-actin was employed as internal references (n = 3). The serum levels of MDA (P) and GPx (Q) were detected using an ELISA assay (n = 6). (R) The protein levels were determined by Western blot, and β-actin was employed as internal references (n = 3). The serum levels of MDA (S) and GPx (T) were detected using an ELISA assay (n = 6). (U) The protein levels were determined by Western blot, and β-actin was employed as internal references (n = 3). The serum levels of MDA (V) and GPx (W) were detected using an ELISA assay (n = 6). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; #, p < 0.05; ###, p < 0.001; ####, p < 0.0001.
Based on KEGG pathway enrichment analysis, the above genes were mainly enriched for HIF-1 signalling, AGE-RAGE signalling, IL-17 signalling, and Toll-like receptor signalling (Figure 6F).
Next, to further elucidate the relationship between the above 105 common FRGs, we created a PPI network using STRING and further visualized it in Cytoscape (Figure 6G and H). The results indicated that these genes were interrelated. Meanwhile, we employed the MCC algorithm in Cytohubba to evaluate the hub genes in the PPI network, and high scores indicated that the corresponding proteins were the core of the network (Figure 6I). Thus, we selected the top 10 genes (TP53, JUN, IL6, STAT3, RELA, MAPK1, HIF1A, EGFR, MAPK3, and TLR4) as the hub genes in the PPI network.
Machine Learning-Based Selection of Diagnostic Biomarkers of KI-Related AD Progression
To further screen the signature genes, we integrated four machine learning algorithms. First, we utilized LASSO regression analysis to calculate the lowest binomial deviance of seven feature genes (EGFR, HIF1A, JUN, MAPK3, RELA, TLR4, and TP53) among the top 10 hub FRGs screened by PPI (Figure 6A and B). Next, the RF algorithms identified the characterized genes among the 10 hub genes, whose MeanDecreaseGini > 5 (HIF1A, EGFR, JUN, RELA, MAPK3, STAT3, TLR4, TP53) were screened as characterized genes (Figure 6C and D). Then, SVM-RFE was then applied to screen the eight genes with the lowest error and highest accuracy, including HIF1A, EGFR, JUN, RELA, MAPK3, STAT3, TP53, TLR4 (Figure 6E). Following this, five genes with importance scores of importance ≥ 25 were screened using the GBM algorithm (HIF1A, MAPK3, EGFR, JUN, and RELA) (Figure 6F). Lastly, the characterized genes identified by the above four algorithms were merged, where the five genes (EGFR, HIF1A, JUN, MAPK3, and RELA) in the overlapping portion were identified as pivotal genes (Figure 6G).
Biomarker Diagnostic Value Assessment and Validation
We first assessed the expression levels of the above five candidate signature genes. In terms of the dataset GSE5281, we compared the expression of five candidate genes between Control and AD groups. Other than MAPK3, all four genes (EGFR, HIF1A, JUN, and RELA) showed statistically significant differences in expression between the two groups (Figure 7H). Meanwhile, in the validation set GSE122063, except for HIF1A, JUN, EGFR, MAPK3, and RELA were differentially expressed between the two groups (P<0.05) (Figure 7I).
Figure 7.
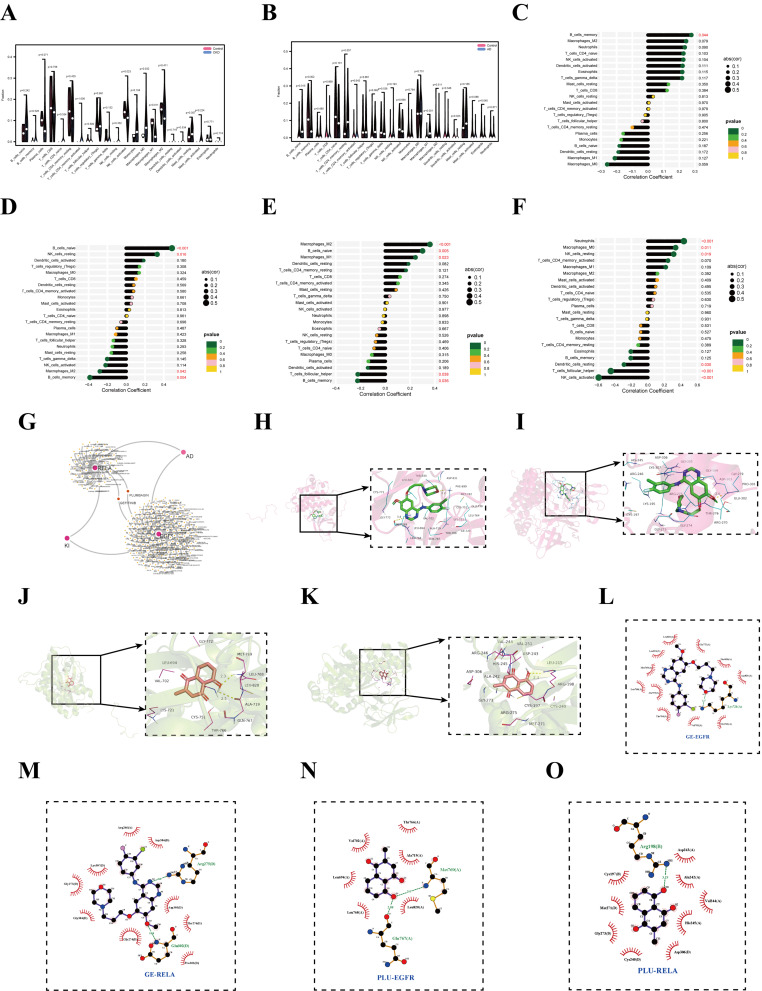
Predicting candidate drugs and validating drug-biomarker interactions. (A) Violin plot of immune-infiltrating cells between the CKD and control groups. (B) Violin plot of immune infiltration in the AD and control groups. Lollipop diagram of a correlation between EGFR (C) and RELA (D) in the CKD dataset. The lollipop diagram of a correlation between EGFR (E) and RELA (F) in the AD dataset. (G) The DGIdb database has explored 238 drugs and small molecules, of which 185 target EGFR and 53 target RELA, which was visualized by Cytoscape. (H) GE and EGFR molecular docking. (I) GE and RELA molecular docking. (J) PB and EGFR molecular docking. (K) PB and RELA molecular docking. Intermolecular interaction analyses of the GE and EGFR (L), and RELA (M). Intermolecular interaction analyses of the PB and EGFR (N), and RELA (O). The spoke arcs in red represent hydrophobic contacts and the dashed green line represents hydrogen bonds.
As a further test of the diagnostic utility of the five signature genes above, we developed ROC curves. In terms of the test set GSE5281, area under the curve (AUC) values were calculated for these 5 genes, including the EGFR (0.7783), HIF1A (0.8364), JUN (0.7437), MAPK3 (0.5581), and RELA (0.722), among which EGFR, HIF1A, JUN, and RELA possessed a high diagnostic value (AUC > 0.7) (Figure 7J). Subsequently, ROC curves for the above five genes were further constructed in the validation set GSE122063, which showed AUC values of EGFR (0.7666), HIF1A (0.5578), JUN (0.525), MAPK3 (0.6607), and RELA (0.8052), respectively (Figure 7K). Consistent with the results of the test set, both EGFR and RELA had an AUC > 0.8, suggesting a better diagnostic value. Combining these results, we constructed a nomogram for EGFR and RELA, and the score of each gene corresponded to the incidence of AD (Figure 7L). The nomogram showed that EGFR and RELA performed well in diagnosing AD progression, which was further confirmed by the calibration curve (Figure 7M). The above results suggest the excellent diagnostic value of EGFR and RELA.
The expression of EGFR and RELA was detected in AD mice as a further verification of the reliability of the aforementioned results. As predicted, the Western blot result matched the above results. The EGFR and RELA all increased in the brains of AD mice (Figure 7N).
To further verify the accuracy of the above results, we performed additional analysis in in vitro experiments. First, in Experiment 1, we induced PC12 cell injury using Aβ25-35 and found that Aβ expression was elevated in the PC12 cells of the model group (Figure 7O). Subsequently, we further found that both EGFR and RELA expression was upregulated in PC12 cells of the model group (Figure 7O). At the same time, the ferroptosis-related index GPX4 protein expression was significantly down-regulated, while PTGS2 was significantly up-regulated (Figure 7O). Additionally, MDA activity was elevated (Figure 7P), and GPx activity was decreased (Figure 7Q), signaling the occurrence of ferroptosis. The RELA inhibitor SC75741 was added in experiment II, and further analysis revealed that RELA was inhibited and that the ferroptosis-related indexes mentioned above were partially improved (Figure 7R–T). It was demonstrated that RELA was a key target of ferroptosis. In addition, in Experiment 3, we added the EGFR inhibitor Gefitinib, which inhibited the expression of EGFR. Consequently, the ferroptosis-related indices were partially improved (Figure 7U–W), demonstrating that EGFR is a key target in ferroptosis.
Results of Immune Cell Infiltration Analysis
Enrichment analysis also discovered that KI-associated FRGs in AD were also involved in immunomodulation. Thus, we investigated the immune infiltration patterns of 22 immune cell types and elucidated the correlation between candidate biomarkers (EGFR and RELA) and immune cells in the CKD (GSE66494) dataset and AD dataset (GSE5281), respectively. The violin plots show that the CKD samples showed significantly different NK_cells_activated, Macrophages_M0, and Macrophages_M1 cells than the control samples (Figure 7A). Moreover, Macrophages_M1 cell was significantly reduced in CKD samples compared to normal samples, while NK_cells_activated and Macrophages_M0 cells were significantly increased. Furthermore, we analyzed AD datasets for immune cell infiltration. There was a decrease in Dendritic cells_activated and Eosinophils cells, while there was an increase in B cells_naive, B cells_memory, and Macrophages_M1 cells in AD patients (Figure 7B). Interestingly, Macrophages_M1 cells were decreased in CKD samples while increasing in AD samples, suggesting that modulation of Macrophages_M1 cells is critical for interfering with AD progression in CKD patients.
However, the ratio of immune cells in KI and AD is only one aspect of their common pathogenesis. We used Pearson correlation analysis to confirm the relationship between these two shared candidate genes and immune cells. According to the CKD dataset, there was a significant positive correlation between B_cells_memory cell and EGFR (Figure 7C). The RELA gene was positively correlated with B_cells_naive and NK_cells_resting cells, while it was negatively correlated with Macrophages_M2 and B_cells_memory cells (Figure 7D). In the AD dataset, we found some relationships between the pivotal genes EGFR and RELA and immune cells, in which EGFR was significantly positively correlated with Macrophages_M2, B_cells_naive, and Macrophages_M1 cells, while significantly negatively correlated with T_cells_follicular_helper and B_ cells_memory cells (Figure 7E). Besides, RELA was positively correlated with Neutrophils, Macrophages_M0, and NK_cells_resting cells (p < 0.05), whereas it was negatively correlated with Dendritic_cells_resting, T_cells_follicular_helper, and NK_cells_activated cells (p < 0.05) (Figure 7F). T Accordingly, immune-related cells may also play a role in AD pathogenesis. Overall, there was a consistent correlation between candidate biomarkers (EGFR and RELA) and NK_cells_resting cell, while there was an opposite correlation with B_cells_memory cell in CKD and AD.
Prediction of Targeted Drugs with Candidate Genes
In the current study, we predicted the target drug with the candidate genes (EGFR and RELA) via the DGIdb database. The drug-gene interaction results revealed 238 potential target drugs or compounds for KI-related AD therapy. Among them, 185 drugs targeted EGFR, and 53 drugs targeted RELA. Then, further analysis of the drug-gene networks was conducted using Cytoscape software, which found that gefitinib (GE) and plumbagin (PB) co-targeted EGFR and RELA, respectively (Figure 7G). Due to this, GE and PB have been identified as potential therapeutic agents for KI-associated AD.
To validate the above results further, we carried out molecular docking of GE and PB with EGFR and RELA, respectively. The results, as expected, showed that GE improved AD by interacting with EGFR (Figure 7H) and RELA (Figure 7I) at an optimal docking binding of −7.4/-7.5 kcal/mol (Table 3). Meanwhile, PB alleviated AD by interacting with EGFR (Figure 7J) and RELA (Figure 7K) at an optimal docking binding of −6.5/-6.8 (kcal/mol) (Table 3). Moreover, the intermolecular interactions of small molecules (GE and PB) with proteins (EGFR and RELA) were mainly through hydrogen bond and hydrophobic interactions (Figure 7L–O).
Table 3.
The Energy Docking Results of Molecular Docking
| Molecule Name | Binding Affinity (kcal/mol) | |
|---|---|---|
| EGFR (1m14) | RELA (1K3Z) | |
| GEFITINIB | −7.4 | −7.5 |
| PLUMBAGIN | −6.5 | −6.8 |
Discussion
Considering the immense societal burden of AD, identifying patients with early-stage AD (prodromal AD) is critical.32 However, in its early stages, AD can be difficult to detect due to its insidious and irreversible onset.33 Encouragingly, introducing the “kidney-brain crosstalk” concept has pointed us in a new direction. Past studies have proposed that oxidative stress is a crucial pathological process in kidney-brain crosstalk, and mediated ferroptosis is also a common pathogenic mechanism in KI and AD. Thus, ferroptosis may largely act as a trigger, which contributes to KI-induced AD. Most previous studies have focused on only identifying diagnostic biomarkers for KI or AD. Thus, disregarding the possibility of kidney-brain crosstalk contributes to the progression of KI to AD, leading to a paucity of studies on diagnostic-related markers of KI-related AD. Based on this, searching for core diagnostic biomarkers involved in the pathogenesis of AD in the KI-associated FRGs is a major step toward deciphering kidney-brain crosstalk and exploring an effective therapeutic route.
Here, through MR analysis, we first determined that an increased risk of CKD development induces an increased risk of AD development. These tentatively confirm the possibility of kidney-brain crosstalk. Following this, based on previous studies, it has been suggested that the excess D-gal will lead to an increase in reactive oxygen species (ROS) and late glycosylation end products. The above phenomena will further cause oxidative stress, cellular damage, and inflammation, which in turn promote organ damage, including brain, liver, and kidney damage.34 Meanwhile, Al can cross the blood-brain barrier and deposit in multiple brain regions, leading to neurodegenerative diseases (AD) and also inducing multiple organ diseases (such as kidney and hepatic).35 We integrated D-gal and AlCl3 to explore the relationship between KI and AD. As expected, mice were treated with D-gal + AlCl3, which can cause KI and AD, and the occurrence of KI preceded AD. Therefore, we ventured to speculate that KI occurrences would further promote AD. At the same time, bioinformatics-based analysis predicted that the common pathogenic mechanism of KI and AD may be related to ferroptosis. Hence, the kidney tissues were analyzed in a D-gal+AlCl3-induced mice model for MDA, GPx, GPX4, and PTGS2 expression, and MDA and PTGS2 were significantly upregulated, while GPx and GPX4 were significantly downregulated. Meanwhile, the results of in vitro AD model were consistent with those of in vivo experiments. The MDA content indirectly reflects the level of lipid peroxidation in the body, and an increase implies a significant decrease in the antioxidant capacity.36 GPX4, the sensor of oxidative stress and cell death signalling, and its decreased levels lead to a significant elevation of reactive oxygen species in vivo.37 Thus, it is considered an important target for triggering ferroptosis. Our study from the experimental level confirms that KI and AD pathogenesis are associated with ferroptosis, which is consistent with previous findings.38,39
Subsequently, we identified five biomarkers of KI-associated FRGs in AD by combining four machine-learning algorithms (LASSO, RF, SVN-RFE, and GBM). Among these, EGFR and RELA can be used for the early diagnosis of AD progression in KI patients. Meanwhile, our study also explored new therapeutic avenues using immune infiltration analysis, which may help find new targets for KI-associated AD. The study revealed a positive correlation between EGFR/RELA and NK_cells_resting. In contrast, there was an opposite correlation with B_cells_memory. Previous studies have shown that the depletion of NK cells enhances neurogenesis, reduces neuroinflammation, and improves cognitive function in AD mice.40 Additionally, therapeutic B-cell depletion reverses the progression of Alzheimer’s disease.41 Meanwhile, the depletion of NK cells or B cells also attenuates KI.42,43 These findings suggest that targeting NK cells or B cells may be a novel strategy to combat AD and KI. Notably, ferroptosis is closely associated with NK cell- and B cell-mediated immune responses.31,44 Based on this, we speculate that EGFR and RELA might ameliorate KI-associated AD by modulating immune processes during ferroptosis. Regrettably, the specific pathways of their actions remain to be further investigated.
An epidermal growth factor receptor (EGFR) is a type of growth factor receptor found on the surface of cells that plays a crucial role in the proliferation, differentiation, and survival of cells.45 Some studies indicated that EGFR can mediate the ferroptosis process, which involves the pathogenesis of lupus nephritis, cardiomyocyte ischemia-reperfusion injury, and osteoarthritis,46–48 and that the effect of EGFR in disease is mainly attributed to regulating ferroptosis. EGFR has previously been identified in the kidney, and its expression is more common in diseased kidneys than normal ones.49 In addition, studies have found that EGFR contributes to AD development. EGFR inhibitors may improve cognitive function in AD by attenuating Aβ pathology,50 suggesting that EGFR plays an important role in AD. Our findings suggest that EGFR is upregulated in AD, and its underlying mechanism may be related to immune processes during ferroptosis. These suggest that higher EGFR levels reflect a more severe disease. RELA is a transcription factor for nuclear factor-κB p65 (NF-κB p65), which is closely related to many cellular signalling processes as well as the pathology of diseases. Activating the NF-κB signalling pathway contributes to neurological diseases by regulating ferroptosis.51 Consequently, RELA has the ability to control ferroptosis, which plays a crucial role in disease. Ke et al52 proposed that elevated expression of RELA increases the level of inflammatory factors in serum and kidney tissues, damages kidney podocytes and tethered cells, and ultimately leads to kidney injury. Moreover, Qian et al53 supported that RELA is an immunologically relevant biomarker associated with AD and mild cognitive impairment. ICAM-1 can protect neurons against Aβ and improve cognitive behaviors in 5xFAD mice by inhibiting NF-κB.54 Our results showed an upregulation of RELA expression in KI-associated AD, and the underlying mechanism may be associated with immune processes during ferroptosis. In summary, EGFR and RELA may be potential therapeutic targets for KI-associated AD, and their specific action pathways need to be evaluated in further studies.
To improve the diagnosis and treatment of AD, we must urgently explore new biomarkers to develop effective therapeutic agents. Therefore, we explored 238 potential drugs or small molecules for treating KI-related AD in the DGIdb database by taking the biomarkers (EGFR and RELA) as a diagnosis of KI-related AD as a clue. Notably, GE and PB target both EGFR and RELA genes and are considered candidates for treating KI-related AD. GE is an epidermal growth factor receptor-related tyrosine kinase inhibitor for oral administration and is mainly used for treating lung adenocarcinoma.55 Unexpectedly, GE also exhibited the ability to modulate Aβ biosynthesis and clearance in brain tissue then intervene in AD.56 The alkaloid PB has anti-inflammatory and antioxidant properties and is found in the roots of Plumbaginaceae, Droseraceae, and Ebenaceae, which also have analgesic and anti-bacterial properties.57 Previous findings consistently showed that PB can ameliorate streptozotocin-induced memory dysfunction in AD by inhibiting β-secretase.58 The above is in line with our prediction that GE and PB can target EGFR and RELA to regulate ferroptosis and intervene in the AD progression of KI patients. It was inferred that early pharmacologic intervention in patients with KI improves kidney function and inhibits the onset and progression of AD, ultimately significantly prolonging the life span of patients.
In summary, we integrated bioinformatics analysis and in vivo experimental validation to pioneeringly uncover the correlation between KI and AD, identified common mechanisms of KI and AD, and revealed biomarkers of AD in KI-associated FRGs. It should be noted, however, that our study has some limitations. First, the amount of data obtained is still limited, and further clinical trials to assess the correlation between clinical parameters and hub biomarkers still need to be improved. Second, although these data suggest that GE and PB can intervene in AD progression in patients with KI, there is still a lack of studies demonstrating the efficacy of GE and PB targeting EGFR and RELA to regulate ferroptosis for treating KI-associated AD. Third, we did not consider genetic variability in patient populations in our study. Last, the model used in the study is not a recognized model of kidney injury. Given the study’s limitations, as part of follow-up studies, identifying a diagnostic biomarker and candidate drug should be tested in vivo, in vitro, and in humans.
Conclusion
In conclusion, our study first combined bioinformatics and experimental validation approach confirmed that the ferroptosis-related genes EGFR and RELA are the hub genes of KI-related AD. In addition, these signatured genes are associated with immune cell infiltration and immunoinflammatory pathways, suggesting that they may regulate KI-associated AD by modulating immune processes during ferroptosis. In conclusion, our study reveals for the first time the value of ferroptosis in KI-related AD for potential mechanisms and therapeutic strategies to effectively address KI-related AD.
Acknowledgement
We thank the Home for Researchers editorial team (www.home-for-researchers.com) for their language editing services, and Figdraw (www.figdraw.com) for the picture materials.
Funding Statement
The National Natural Science Foundation of China (No. 82201220), the Natural Science Foundation of Jiangsu Province (No. BK20190149), the Postdoctoral Science Foundation of China (No. 2020M681669), and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX24_2248) all provided support for the work.
Abbreviations
AD, Alzheimer’s disease; AKI, Acute kidney injury; AUC, Area under the curve; BP, Biological process; BUN, Blood urea nitrogen; CC, Cellular component; CKD, Chronic kidney disease; CRE, Creatinine; DEGs, Differentially expressed genes; FRGs, Ferroptosis-related genes; GBM, Gradient Boosting Machine; GE, Gefitinib; GEO, Gene Expression Omnibus; GO, Gene Ontology; GPx, Glutathione peroxidase; HE, Hematoxylin and eosin; KEGG, Kyoto Encyclopedia of Genes and Genomes; KI, Kidney injury; LASSO, Least Absolute Shrinkage and Selection Operator; MDA, Malondialdehyde; MF, Molecular function; PB, Plumbagin; PPI, Protein–protein interaction; RF, Random Forest; ROC, Receiver operating characteristic; SVM-RFE, Support Vector Machine Recursive Feature Extraction.
Data Sharing Statement
The present study-related data are included in the article. For further inquiries, please contact the corresponding author.
Ethical Approval and Consent to Participate
Our study is exempt from approval based on national legislation guidelines, such as item 1 and 2 of Article 32 of the Measures for Ethical Review of Life Science and Medical Research Involving Human Subjects dated February 18, 2023, China. Besides, the animal studies were approved by China Pharmaceutical University’s Ethics Committee.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declared that they have no conflict of interests.
References
- 1.Burke JP, Aljishi M, Francis L. et al. Protocol and establishment of a Queensland renal biopsy registry in Australia. BMC Nephrol. 2020;21(1):320. doi: 10.1186/s12882-020-01983-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chawla LS, Kimmel PL. Acute kidney injury and chronic kidney disease: an integrated clinical syndrome. Kidney Int. 2012;82(5):516–524. doi: 10.1038/ki.2012.208 [DOI] [PubMed] [Google Scholar]
- 3.Lousa I, Reis F, Beirão I, et al. New Potential Biomarkers for Chronic Kidney Disease Management-A Review of the Literature. Int J Mol Sci. 2020;22(1):43. doi: 10.3390/ijms22010043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill NR, Fatoba ST, Oke JL, et al. Global Prevalence of Chronic Kidney Disease - A Systematic Review and Meta-Analysis. PLoS One. 2016;11(7):e0158765. doi: 10.1371/journal.pone.0158765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roussos A, Kitopoulou K, Borbolis F, et al. Caenorhabditis elegans as a Model System to Study Human Neurodegenerative Disorders. Biomolecules. 2023;13(3):478. doi: 10.3390/biom13030478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison C, Dadar M, Shafiee N, et al. Regional brain atrophy and cognitive decline depend on definition of subjective cognitive decline. Neuroimage Clin. 2022;33:102923. doi: 10.1016/j.nicl.2021.102923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Oliveira FF, Pereira FV, Pivi GAK, et al. Effects of APOE haplotypes and measures of cardiovascular risk over gender-dependent cognitive and functional changes in one year in Alzheimer’s disease. Int J Neurosci. 2018;128(5):472–476. doi: 10.1080/00207454.2017.1396986 [DOI] [PubMed] [Google Scholar]
- 8.Hou J, Shang L, Huang S, et al. Postoperative Serum Creatinine Serves as a Prognostic Predictor of Cardiac Surgery Patients. Front Cardiovasc Med. 2022;9:740425. doi: 10.3389/fcvm.2022.740425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu R, Kiernan MC, Murray A, et al. Kidney-brain crosstalk in the acute and chronic setting. Nat Rev Nephrol. 2015;11(12):707–719. doi: 10.1038/nrneph.2015.131 [DOI] [PubMed] [Google Scholar]
- 10.Grigoryev DN, Liu M, Hassoun HT, et al. The local and systemic inflammatory transcriptome after acute kidney injury. J Am Soc Nephrol. 2008;19(3):547–558. doi: 10.1681/ASN.2007040469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calabrese V, Scuto M, Salinaro. AT, et al. Hydrogen Sulfide and Carnosine: modulation of Oxidative Stress and Inflammation in Kidney and Brain Axis. Antioxidants. 2020;9(12):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian DY, Cheng Y, Zhuang ZQ, et al. Physiological clearance of amyloid-beta by the kidney and its therapeutic potential for Alzheimer’s disease. Mol Psychiatry. 2021;26(10):6074–6082. doi: 10.1038/s41380-021-01073-6 [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Jin WS, Bu XL, et al. Physiological clearance of tau in the periphery and its therapeutic potential for tauopathies. Acta Neuropathol. 2018;136(4):525–536. doi: 10.1007/s00401-018-1891-2 [DOI] [PubMed] [Google Scholar]
- 14.H. X, Garcia-Ptacek S, Trevisan M, et al. Kidney Function, Kidney Function Decline, and the Risk of Dementia in Older Adults: a Registry-Based Study. Neurology. 2021;96(24):e2956–e2965. doi: 10.1212/WNL.0000000000012113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang CY, He FF, Su H, et al. Association between chronic kidney disease and Alzheimer’s disease: an update. Metab Brain Dis. 2020;35(6):883–894. doi: 10.1007/s11011-020-00561-y [DOI] [PubMed] [Google Scholar]
- 16.Shorr AF, Nelson DR, Wyncoll DL, et al. Protein C: a potential biomarker in severe sepsis and a possible tool for monitoring treatment with drotrecogin alfa (activated). Crit Care. 2008;12(2):R45. doi: 10.1186/cc6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun W, Yan J, Ma H, et al. Autophagy-Dependent Ferroptosis-Related Signature is Closely Associated with the Prognosis and Tumor Immune Escape of Patients with Glioma. Int J Gen Med. 2022;15:253–270. doi: 10.2147/IJGM.S343046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Zhang L, Yao C, et al. Epithelial Membrane Protein 1 Promotes Sensitivity to RSL3-Induced Ferroptosis and Intensifies Gefitinib Resistance in Head and Neck Cancer. Oxid Med Cell Longev. 2022;2022:4750671. doi: 10.1155/2022/4750671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni L, Yuan C, Wu X. Targeting ferroptosis in acute kidney injury. Cell Death Dis. 2022;13(2):182. doi: 10.1038/s41419-022-04628-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bao WD, Pang P, Zhou XT, et al. Loss of ferroportin induces memory impairment by promoting ferroptosis in Alzheimer’s disease. Cell Death Differ. 2021;28(5):1548–1562. doi: 10.1038/s41418-020-00685-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan C, Yang H, Su P, et al. OTUB1 suppresses Hippo signaling via modulating YAP protein in gastric cancer. Oncogene. 2022;41(48):5186–5198. doi: 10.1038/s41388-022-02507-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen W, Song Z, Zhong X, et al. Sangerbox: a comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta. 2022;1(3):e36. doi: 10.1002/imt2.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Mao R, Lau CT, et al. Identification of useful genes from multiple microarrays for ulcerative colitis diagnosis based on machine learning methods. Sci Rep. 2022;12(1):9962. doi: 10.1038/s41598-022-14048-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng L, Yang K, Kuang Q, et al. A Novel Risk Model for lncRNAs Associated with Oxidative Stress Predicts Prognosis of Bladder Cancer. J Oncol. 2022;2022:8408328. doi: 10.1155/2022/8408328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X, Wang H, Guo D, et al. Curcumin modulates gut microbiota and improves renal function in rats with uric acid nephropathy. Ren Fail. 2021;43(1):1063–1075. doi: 10.1080/0886022X.2021.1944875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Tong J, Liu J, et al. Down-regulation of ROCK2 alleviates ethanol-induced cerebral nerve injury partly by the suppression of the NF-κB signaling pathway. Bioengineered. 2020;11(1):779–790. doi: 10.1080/21655979.2020.1795404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai Y, Huang G, Ren M, et al. Synthesizing network pharmacology, bioinformatics, and in vitro experimental verification to screen candidate targets of Salidroside for mitigating Alzheimer’s disease. Naunyn Schmiedebergs Arch Pharmacol. 2024. doi: 10.1007/s00210-024-03555-0 [DOI] [PubMed] [Google Scholar]
- 28.Bellut M, Bieber M, Kraft P, et al. Delayed NLRP3 inflammasome inhibition ameliorates subacute stroke progression in mice. J Neuroinflammation. 2023;20(1):4. doi: 10.1186/s12974-022-02674-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan K, Zhang P, Jin J, et al. Integrative analyses of hub genes and their association with immune infiltration in adipose tissue, liver tissue and skeletal muscle of obese patients after bariatric surgery. Adipocyte. 2022;11(1):190–201. doi: 10.1080/21623945.2022.2060059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng MG, Liu F, Liang Y, et al. Association between frailty and depression: a bidirectional Mendelian randomization study. Sci Adv. 2023;9(38):eadi3902. doi: 10.1126/sciadv.adi3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Kang R, Kroemer G, et al. Ferroptosis in infection, inflammation, and immunity. J Exp Med. 2021;218(6). doi: 10.1084/jem.20210518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khurshid B, Rehman U, Muhammad S, et al. Toward the Noninvasive Diagnosis of Alzheimer’s Disease: molecular Basis for the Specificity of Curcumin for Fibrillar Amyloid-β. ACS Omega. 2022;7(25):22032–22038. doi: 10.1021/acsomega.2c02995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu N, Yuan Z, Tang Q. Improving Alzheimer’s Disease Detection for Speech Based on Feature Purification Network. Front Public Health. 2021;9:835960. doi: 10.3389/fpubh.2021.835960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng S. Protective effect of Polygonatum sibiricum Polysaccharide on D-galactose-induced aging rats model. Sci Rep. 2020;10(1):2246. doi: 10.1038/s41598-020-59055-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abu-Elfotuh K, Hussein FH, Abbas AN, et al. Melatonin and zinc supplements with physical and mental activities subside neurodegeneration and hepatorenal injury induced by aluminum chloride in rats: inclusion of GSK-3β-Wnt/β-catenin signaling pathway. Neurotoxicology. 2022;91:69–83. doi: 10.1016/j.neuro.2022.05.002 [DOI] [PubMed] [Google Scholar]
- 36.He R, Cui M, Lin H, et al. Melatonin resists oxidative stress-induced apoptosis in nucleus pulposus cells. Life Sci. 2018;199:122–130. doi: 10.1016/j.lfs.2018.03.020 [DOI] [PubMed] [Google Scholar]
- 37.Ursini F, Maiorino M. Lipid peroxidation and ferroptosis: the role of GSH and GPx4. Free Radic Biol Med. 2020;152:175–185. doi: 10.1016/j.freeradbiomed.2020.02.027 [DOI] [PubMed] [Google Scholar]
- 38.Zhao Z, Wu J, Xu H, et al. XJB-5-131 inhibited ferroptosis in tubular epithelial cells after ischemia-reperfusion injury. Cell Death Dis. 2020;11(8):629. doi: 10.1038/s41419-020-02871-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou X, Tang X, Li T, et al. Inhibition of VDAC1 Rescues Aβ (1-42)-Induced Mitochondrial Dysfunction and Ferroptosis via Activation of AMPK and Wnt/β-Catenin Pathways. Mediators Inflamm. 2023;2023:6739691. doi: 10.1155/2023/6739691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Fung ITH, Sankar P, et al. Depletion of NK Cells Improves Cognitive Function in the Alzheimer Disease Mouse Model. J Immunol. 2020;205(2):502–510. doi: 10.4049/jimmunol.2000037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim K, Wang X, Ragonnaud E, et al. Therapeutic B-cell depletion reverses progression of Alzheimer’s disease. Nat Commun. 2021;12(1):2185. doi: 10.1038/s41467-021-22479-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan AJ, Alikhan MA, Odobasic D, et al. Innate IL-17A-producing leukocytes promote acute kidney injury via inflammasome and Toll-like receptor activation. Am J Pathol. 2014;184(5):1411–1418. doi: 10.1016/j.ajpath.2014.01.023 [DOI] [PubMed] [Google Scholar]
- 43.Renner B, Strassheim D, Amura CR, et al. B cell subsets contribute to renal injury and renal protection after ischemia/reperfusion. J Immunol. 2010;185(7):4393–4400. doi: 10.4049/jimmunol.0903239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui JX, Xu XH, He T, et al. L-kynurenine induces NK cell loss in gastric cancer microenvironment via promoting ferroptosis. J Exp Clin Cancer Res. 2023;42(1):52. doi: 10.1186/s13046-023-02629-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voldborg BR, Damstrup L, Spang-Thomsen M, et al. Epidermal growth factor receptor (EGFR) and EGFR mutations, function and possible role in clinical trials. Ann Oncol. 1997;8(12):1197–1206. doi: 10.1023/A:1008209720526 [DOI] [PubMed] [Google Scholar]
- 46.Hu T, Yu WP, Zou HX, et al. Role of dysregulated ferroptosis‑related genes in cardiomyocyte ischemia‑reperfusion injury: experimental verification and bioinformatics analysis. Exp Ther Med. 2023;26(5):534. doi: 10.3892/etm.2023.12233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun H, Peng G, Chen K, et al. Identification of EGFR as an essential regulator in chondrocytes ferroptosis of osteoarthritis using bioinformatics, in vivo, and in vitro study. Heliyon. 2023;9(9):e19975. doi: 10.1016/j.heliyon.2023.e19975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu W, Chen X. Identification of hub ferroptosis-related genes and immune infiltration in lupus nephritis using bioinformatics. Sci Rep. 2022;12(1):18826. doi: 10.1038/s41598-022-23730-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakopoulou L, Stefanaki K, Boletis J, et al. Immunohistochemical study of epidermal growth factor receptor (EGFR) in various types of renal injury. Nephrol Dial Transplant. 1994;9(7):764–769. [PubMed] [Google Scholar]
- 50.Choi HJ, Jeong YJ, Kim J, et al. EGFR is a potential dual molecular target for cancer and Alzheimer’s disease. Front Pharmacol. 2023;14:1238639. doi: 10.3389/fphar.2023.1238639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao J, Ma C, Xia D, et al. Icariside II preconditioning evokes robust neuroprotection against ischaemic stroke, by targeting Nrf2 and the OXPHOS/NF-κB/ferroptosis pathway. Br J Pharmacol. 2023;180(3):308–329. doi: 10.1111/bph.15961 [DOI] [PubMed] [Google Scholar]
- 52.Ke G, Chen X, Liao R, et al. Receptor activator of NF-κB mediates podocyte injury in diabetic nephropathy. Kidney Int. 2021;100(2):377–390. doi: 10.1016/j.kint.2021.04.036 [DOI] [PubMed] [Google Scholar]
- 53.Qian XH, Xl L, Chen SD, et al. Integrating peripheral blood and brain transcriptomics to identify immunological features associated with Alzheimer’s disease in mild cognitive impairment patients. Front Immunol. 2022;13:986346. doi: 10.3389/fimmu.2022.986346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guha S, Paidi RK, Goswami S, et al. ICAM-1 protects neurons against Amyloid-β and improves cognitive behaviors in 5xFAD mice by inhibiting NF-κB. Brain Behav Immun. 2022;100:194–210. doi: 10.1016/j.bbi.2021.11.021 [DOI] [PubMed] [Google Scholar]
- 55.Gautam A, Pal K. Gefitinib conjugated PEG passivated graphene quantum dots incorporated PLA microspheres for targeted anticancer drug delivery. Heliyon. 2022;8(12):e12512. doi: 10.1016/j.heliyon.2022.e12512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dhamodharan J, Sekhar G, Muthuraman A. Epidermal Growth Factor Receptor Kinase Inhibitor Ameliorates β-Amyloid Oligomer-Induced Alzheimer Disease in Swiss Albino Mice. Molecules. 2022;27(16):5182. doi: 10.3390/molecules27165182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petrocelli G, Marrazzo P, Bonsi L, et al. Plumbagin, a Natural Compound with Several Biological Effects and Anti-Inflammatory Properties. Life. 2023;13(6). doi: 10.3390/life13061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakhate KT, Bharne AP, Verma. VS, et al. Plumbagin ameliorates memory dysfunction in streptozotocin induced Alzheimer’s disease via activation of Nrf2/ARE pathway and inhibition of β-secretase. Biomed Pharmacother. 2018;101:379–390. doi: 10.1016/j.biopha.2018.02.052 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The present study-related data are included in the article. For further inquiries, please contact the corresponding author.