Abstract
Objectives
Deidentification of personally identifiable information in free-text clinical data is fundamental to making these data broadly available for research. However, there exist gaps in the deidentification landscape with regard to the functionality and flexibility of extant tools, as well as suboptimal tradeoffs between deidentification accuracy and speed. To address these gaps and tradeoffs, we develop a new Python-based deidentification software, pyDeid.
Materials and Methods
pyDeid uses a combination of regular expression-based rules, fixed exclusion lists and inclusion lists to deidentify free-text data. Additional configurations of pyDeid include optional named entity recognition and custom name lists. We measure its deidentification performance and speed on 700 admission notes from a Canadian hospital, the publicly available n2c2 benchmark dataset of American discharge notes, as well as a synthetic dataset of artificial intelligence (AI) generated admission notes. We also compare its performance with the Physionet De-identification Software and the popular open-source Philter tool.
Results
Different configurations of pyDeid outperformed other tools on various metrics, with a “best” accuracy value of 0.988, best precision of 0.889, best recall of 0.950, and best F1 score of 0.904. All configurations of pyDeid were significantly faster than Philter and Physionet De-identification Software, with the fastest deidentification speed of 0.48 s per note.
Discussion and Conclusions
pyDeid allows the flexibility to prioritize between performance and speed, as well as precision and recall, while addressing some of the gaps in functionality left by other tools. pyDeid is also generalizable to domains outside of clinical data and can be further customized for specific contexts or for particular workflows.
Keywords: deidentification, personal health information, privacy, software validation
Background and significance
Electronic health records (EHRs) are a valuable resource for medical research and quality improvement.1–5 For many important research applications, personally identifiable information (PII) contained in EHRs must be deidentified in compliance with privacy laws, data sharing agreements, and research ethics. Deidentification involves removing PII and/or replacing it with surrogate information. In the United States and Canada, the Health Insurance Portability and Accountability Act (HIPAA) and the Personal Information Protection and Electronic Documents Act, respectively, outline what information constitutes PII. In general, names, addresses, and contact information such as phone numbers and email addresses are considered PII.
Manual deidentification is laborious and time-consuming, as it can take approximately 1.5 min to deidentify a single patient record in an EHR.6 Thus, tools have been developed to automate the deidentification process. Rule-based tools use regular expressions to pattern-match information in EHRs to information in data dictionaries.7–11 Tools that use machine-learning techniques can generalize better, but are slower relative to rule-based methods. Hybrid tools that use machine-learning capabilities in conjunction with rules have yielded considerable success in PII deidentification.12–15 By and large, however, existing tools lack broad generalization (eg, US vs Canadian health care contexts), are time-consuming to run, or costly to use.
In this paper, we introduce pyDeid, a Python-based refactor of the widely used Perl-based Physionet De-identification Software v1.1.10 pyDeid is a modular, open-source tool that uses both regular expressions and data dictionaries—as well as a named entity recognition (NER) option—for fast and accurate PII deidentification. pyDeid is faster than comparable tools, flexible in its deidentification approach, customizable, and has some desirable properties missing from comparable tools such as surrogate replacement, custom regular expressions, and in-memory wordlists. pyDeid fills the existing gap in the Canadian health care system, with a potential for application outside of the Canadian health care context. We present a comparison of both the speed and performance of pyDeid with 2 other rule-based tools, Physionet deid10 and Philter,14 on a set of unstructured Canadian admission notes and the popular n2c2 benchmark dataset of American discharge notes.16,17
Materials and methods
pyDeid
Overview
pyDeid is a Python-based tool that primarily uses regular expressions and lookup dictionaries (henceforth referred to as “wordlists”) to identify and replace PII with realistic surrogates.
It is a refactor of the Perl-based Physionet De-identification Software v1.1,10 borrowing many of the same regular expression rulesets as the original software while adding additional rules for the Canadian context (eg, rules to identify Ontario Health Insurance Plan [OHIP] numbers, postal codes, etc.). Additional rules were also added for edge cases that were not captured by the Physionet deidentification software, such as compound names and name prefixes (eg, Van der Meer, see “Step 1. Find PII”). The implementation is faster to use and requires no pre- or post-processing of data into a software-specific format and operates directly on raw comma separated value (CSV) data. It accepts lists of patient and doctor names as arguments to accommodate workflows where these data must be read from remote server locations. The package is modular and easily modified (see Appendix S2), and it allows for optional NER that can find less common or region-specific names.
The Health Insurance Portability and Accountability Act lists 18 categories of PII, including nontextual identifiers such as vehicle license plate numbers, fingerprints, photographs, and others. pyDeid currently supports the deidentification of 8 categories of PII, which cover 7 of the 18 HIPAA identifiers. The supported categories include names, dates, locations (smaller than the level of province, including cities, postal codes, and street names), identifiers such as medical reference numbers, social insurance numbers, OHIP numbers (which are not considered PII under HIPAA), email addresses, and telephone numbers. It can easily be extended to additional HIPAA categories such as IP addresses, URLs, and vehicle identification numbers.
Algorithm
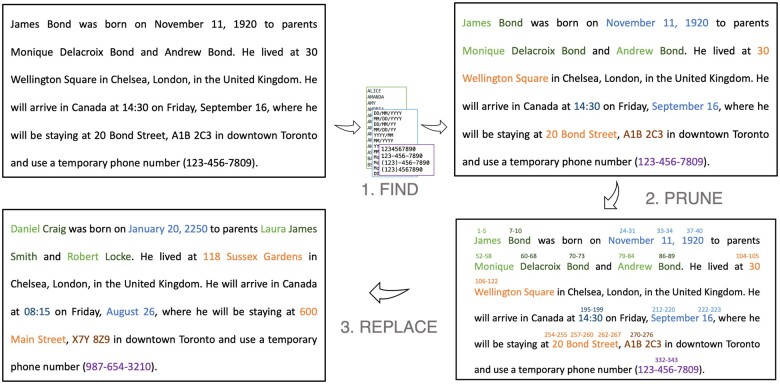
The pyDeid algorithm can be broken down into 3 major “steps”: (1) find PII in the note using the Physionet De-identification Software ruleset with some modifications, (2) correct for or “prune” overlapping instances of PII, and (3) replace the found PII with surrogates.
Step 1. Find PII
1.A. Wordlist lookup. This step begins by splitting the note text into continuous alphanumeric strings or “tokens.” The tokens are then matched against a set of wordlists and tagged with corresponding labels if a match is found. These wordlists are described below:
Name lists. Includes lists of common and popular male and female first and last names from the publicly available Ontario Data Catalogue, and ambiguous female and male first names, and ambiguous last names as published with the original Physionet De-identification Software v1.1. Custom lists of names, such as the list of patients, doctors, or nurses from a given health care facility, can also be provided as input to pyDeid, without having to be read from persistent storage.
Prefixes and titles (eg, Mr, van der, de la).
Area codes and locations. In our deployment, locations primarily include places and neighborhoods in Ontario, Canada (eg, Richmond Hill, Bathurst Manor). These lists can be modified to suit other locations and contexts as well.
Common words. Terms that tend to be non-PII.
Medical terminology. These include medical phrases which contain proper nouns that may otherwise be tagged as PII (eg, Douglas’ pouch) and other medical terminology (eg, X-ray, trach) from the SNOMED vocabulary (also published with the original Physionet De-identification Software v1.1).
1.B. Ruleset matching. The “find PII” step continues by matching the note against a set of regular expressions (henceforth referred to as “rules”) for each PII category of interest. Some tokens are tagged as “ambiguous” if they are found in a particular wordlist but do not readily correspond to a rule for a given PII category. These rules and examples of ambiguous cases within each PII category are described below:
-
1. Names.
The token is preceded by or followed by some indicator (“name is,” “Mr,” “MD,” “Professor,” “prepared by,” etc.).
The token is preceded by or followed by an unambiguous name, suggesting a “First, Last,” or “Last, First” name pattern.
Multiple names are listed in succession (eg, Drs A, B, and C), including compound or hyphenated last names.
Tokens that were not tagged in the first pass can be tagged as name PII by a set of additional rules. For example, names adjacent to certain titles, prefixes, suffixes, name indicators (eg, “Mr,” “de la,” “nurse,” “resident,” “prepared by,” “name is”), and names present in the ‘Last name, First name’ format. Additional rules check for compound and hyphenated names.
Some checks are run on the names identified by these scenarios to reduce false positives, such as for whether the tokens are title cased, whether the token and surrounding tokens correspond to a medical phrase contained in the Medical Terminology wordlist, and for compound names, whether the token preceding or following it was an unambiguous name.
2. IDs. Tokens corresponding to medical record numbers and health insurance numbers are tagged using regular expressions that match the format of those identification numbers, preceded by some indicator that provides context to the identifier in the case of an ambiguous string of digits.
3. Contacts. Phone numbers (with or without extensions) are tagged as such using regular expressions corresponding to many popular formats. For potential phone numbers with missing or additional digits, line breaks, and spaces, they are more likely to be tagged as PII if the first 3 digits correspond to a Canadian area code (eg, 416) to improve deidentification precision. To reduce false positives, if numerical tokens are preceded by words indicating medical measurements (eg, BP, HR), then the following numbers are not tagged as PII. Note that pyDeid does not currently identify incomplete numbers (eg, 7-digit pager numbers without the area code). Regular expressions are also used to identify email addresses.
4. Dates. Regular expressions for many popular date formats are used to identify potential date PII. This includes patterns such as “12-Apr-05,” “2005/04,” “April 12th,” “12 of April,” and “Apr. of 2005.” In ambiguous cases, such as 12/04, false positives are reduced by checking that the day, month, and year components can form a valid date. That is, the day must fall between 1 and 31, month between 1 and 12, or “January/Jan” and “December/Dec,” and the year must be between some minimum and maximum valid year that can be specified by the user. Moreover, the date must not appear after a medical term (eg, cc, BP, dose). Although not considered PII under HIPAA, years are found using the context surrounding the token to limit false positives. This is to limit the risk that a missed year would provide information about which dates in the text were replaced with surrogate dates. The note is additionally searched for holidays using regular expressions that match Canadian holiday names, as well as seasons and date ranges.
5. Locations. Locations are identified using regular expressions for address formats (eg, “1 Main Street”), along with prefixes and suffixes that are indicative of an address or place (eg, “Avenue,” “Apt”). Postal codes are tagged as such using regular expressions for Canadian postal codes (ie, A1B 2C3), though this can be modified to suit other formats as well (eg, American zip codes, 12345—see Appendix S2). Wordlists are used to identify cities and local place names.
The “find PII” step results in a list of tokens tagged with their start and end positions, as well as tags corresponding to the rules or wordlists to which the token was matched (Figure 1).
Figure 1.
Illustrative example of each processing step in the pyDeid algorithm.
Note that given the text is whitespace tokenized, names such as “JohnDoe” would not be tagged as PII. However, PII split across new lines is handled appropriately.
Step 2. Prune PII
The list is first sorted by start position. As tokens in this list may overlap partially or fully with each other, this additional step is used to generate a list of nonoverlapping PII tokens to be replaced in the original string. First, tokens with only “ambiguous”-type tags are removed from the list. This means that the token found no “supporting” rule during the find PII step that would address its ambiguity and “strengthen” its case as PII. Next, 2 scenarios are checked to resolve potentially overlapping tokens:
The next PII in the list is fully contained within the current PII. In this case, the “containing” PII takes precedence and the next PII is dropped. For example, in the string “20 Bond Street,” “Bond” may be tagged as a name, and would be fully contained within “20 Bond Street,” tagged as an address. The final PII would be “20 Bond Street” and is replaced only once, with a surrogate address.
The start position of the next PII occurs after the start position of the current PII, but before the end position of the current PII, and the end position of the next PII is after the end position of the current PII. In this case, we combine the PII, taking the string starting at the start position of the current PII, and ending at the end position of the next PII. For example, consider the PII “20th of March,” followed by the PII “March, 2020.” In this case, the “prune PII” step would collapse these PII into “20th of March, 2020” and perform 1 replacement in the string. If there is a type mismatch between the 2 overlapping strings, the type is flagged as a generic <PII> type.
The “prune PII” step results in a list of nonoverlapping PII sorted by start position.
Step 3. Replace PII
The sorted list of PII is looped through sequentially. For each PII found and its corresponding PII type, the substring defined by the start and end positions of the PII is cut from the note and in its place, a randomly generated surrogate for that PII type is inserted. In the special case of dates, to preserve the medical timeline of events, surrogate dates are shifted by the same amount of time within a particular note (a single CSV cell of free text). For instance, if a patient was admitted on November 11 and had an X-ray done on November 17, 2016, then the surrogate dates for those events would maintain the difference of 6 days in between the 2 dates (eg, April 5 and April 11, 2022). Other PII are randomly replaced by a surrogate PII of the same type without specific constraints. PII for which the type cannot be determined are replaced with a generic surrogate such as <PII>. Additionally, if multiple instances of the same name are identified as PII, the same surrogate will be used for both.
Input. pyDeid expects tabular data in CSV format as input.
Output. pyDeid generates 2 output files: (1) a deidentified file with the same structure as the original in CSV format and (2) a PII replacement file in CSV or JSON format. The PII replacement file contains the list of PII generated during the “prune PII” step with their start and end positions in the original note, as well as their surrogates generated during the “replace PII” step, with their start and end positions in the new deidentified note.
Customization. pyDeid can easily be modified or extended to generalize to a variety of contexts with minimal effort. Within a particular context, such as Canadian health care, pyDeid also allows the ability to capture custom regular expressions by specifying them during runtime. Thus, users can add their own rules or wordlists to suit their context or use case (Appendix S2).
Named entity recognition . Named entity recognition is an “information retrieval” task to identify structured information such as people and locations in unstructured text.18 pyDeid allows for an additional NER step following step 1.B. using the spaCy Python library from Explosion. Personally identifiable information identified by NER is treated the same as PII found using the regular expression rules, and receive the same pruning and replacement defined in the basic algorithm. By default, pyDeid uses the pretrained small, CPU optimized, English NER pipeline.
Data description
Our primary analysis used 700 admission notes from patients admitted to General Internal Medicine at St Michael’s Hospital in Toronto, Ontario, Canada between May 2014 and October 2017.
Each admission note was manually annotated by a subject matter expert for PII and subsequently reannotated by a second rater. The following categories of PII were specified based on prior work and HIPAA guidelines: names and initials, identifying numbers, contact information, locations, and dates (see Table S1 in Appendix S1 for more details).
There were 797 072 tokens in the dataset, of which 48 491 represented instances of PII. On average, there were 1139 tokens in each note, of which approximately 96 tokens represented PII (6.08%). A complete breakdown of the number and percentage of PII tokens in each category is provided in Appendix S1.
In order to evaluate the generalizability of pyDeid to other regions and contexts, deidentification performance was evaluated on medical notes from the test split of the n2c2 dataset, which is a widely used corpus of medical discharge summaries obtained from American health care institutions,17,18 with ground truth labels relevant for comparison (see Appendix S2).
Evaluation metrics
First, the following descriptive measures of overall performance at the token level are provided by comparing deidentification results to the manual token annotations.
- Overall accuracy. Defined by identifying whether a token represents PII or not:
- Category accuracy. Of those tokens identified as PII, the proportion assigned to the correct PII category (ie, categorizing a name as a name instead of a date):
Precision.
Recall.
- F1: The harmonic mean of precision and recall.
Speed. Speed was assessed as the total time taken to deidentify the set of 700 admission notes as well as the average time taken to process each note. Each tool was run 10 times to obtain an average estimate of the time it took to process all 700 notes. All runs were performed on a Windows 10 system with 16GB of RAM, and an AMD Ryzen 5 PRO 4650U CPU, using Python v3.8.8 and Perl v5.32.1. A one-way analysis of variance was conducted to test the difference in speed across the 3 tools, and between different configurations of pyDeid.
Comparison of tools
We compare the performance of pyDeid against 2 popular, regular expression-based tools for deidentification of clinical text:
deid
deid is the open-source, Perl-based automated deidentification software on which pyDeid is based. It uses a combination of regular expressions and lexical matching to identify PII in clinical text and was developed on a corpus of nursing notes from the MIMIC II database.10 The 1.1 version of the software was used for this comparison, without any modification for the Canadian context.
Philter
Protected Health Information filter (Philter) is an open-source Python package, which uses a combination of pattern matching, statistical modeling, exclusion lists and inclusion lists to identify PII in clinical text and was developed at the University of California San Francisco.14 Protected Health Information filter was chosen as the comparator tool because it is freely available, open source, entirely regex based, and modifiable to other regions and contexts. Although tools such as MIST and NLM-Scrubber have been shown to perform faster than either deid or Philter,19 and are freely available, neither tool is easily modified to other contexts. MIST is a machine-learning-based tool and NLM-Scrubber is not available open source, making them inappropriate as alternatives to pyDeid.
Configuration of tools
pyDeid is assessed in 4 configurations: (1) basic configuration, (2) basic configuration with NER enabled, (3) basic configuration with custom name lists of doctors and patients included, and (4) a configuration with both NER and custom name lists.
Protected Health Information filter produces an internal coordinate map, with start and end positions, and PII for all found instances of PII. This map was used to compare against the 700 manually annotated admission notes. Protected Health Information filter did not easily allow for the specification of a particular subset of PII categories to identify and so the default settings were used.
Physionet’s deid allows in its configuration file to turn on or off particular broad categories of PII. Filters for age, ethnicity, state names, country names, company names, and hospital names were turned off as they are not considered to be PII in Canada.
Pre- and postprocessing of notes for comparison
Electronic health record data are commonly extracted by hospital sites in CSVs format and many downstream extract, transform, and load pipelines accept CSV files as input. pyDeid was optimized for CSV files with this in mind. Other programs may require preprocessing to reformat the CSV prior to deidentification.
pyDeid. No additional pre- or postprocessing was required from the original CSV file.
deid. The program requires the notes to be read from and written to a single, continuous text file separated by a custom header and footer pattern.
Philter. Each note was parsed from the original CSV to be read from and written into individual text files.
Analysis of relative performance between tools was done by comparing the manual annotations with a tool-specific output file that contained information about the start and end position of each PII token found in the note, as well as the type of PII. The format of this file varied between tools. For example, Philter did not classify found PII to the same level of granularity as deid or pyDeid.
Results
Performance
The performance of each tool, including the different configurations of pyDeid, is reported in Table 1, with the results broken down by category reported in Table S2 (Appendix S1).
Table 1.
Performance of all deidentification tools on 700 Canadian admission notes.a
| pyDeid configuration |
||||||
|---|---|---|---|---|---|---|
| Basic | NER | Name lists | Name lists+NER | Philter | deid | |
| Overall accuracy | 0.987 | 0.970 | 0.988 | 0.971 | 0.972 | 0.981 |
| Category accuracy | 0.986 | 0.977 | 0.987 | 0.977 | — | 0.964 |
| Precision | 0.889 | 0.688 | 0.877 | 0.686 | 0.710 | 0.798 |
| Recall | 0.906 | 0.934 | 0.932 | 0.950 | 0.924 | 0.874 |
| F1 | 0.879 | 0.792 | 0.904 | 0.797 | 0.803 | 0.834 |
| Speed (min) mean (SD) | 5.55 (0.04) | 5.78 (0.06) | 6.08 (0.16) | 9.63 (0.63) | 74.43 (5.39) | 10.84 (0.76) |
| Speed (s/note) mean (SD) | 0.48 (0.003) | 0.50 (0.005) | 0.52 (0.013) | 0.83 (0.054) | 6.38 (0.462) | 0.93 (0.065) |
Abbreviation: NER, named entity recognition.
Performance measures (except for speed) are calculated at the token level (as opposed to spans of text). Bolded cells indicate the best-performing tool for a given performance metric.
The base configuration of pyDeid had higher precision than Philter and deid (0.889 vs 0.710 and 0.798, respectively). The recall score of the base configuration of pyDeid was lower than Philter (0.906 vs 0.924), but pyDeid configurations with NER improved recall beyond the performance of Philter (0.950 and 0.934 vs 0.924). Recall was higher for the base configuration of pyDeid than deid (0.906 vs 0.874). F1 scores were better for pyDeid than both Philter and deid (0.879 vs 0.803 and 0.834). P-values for all precision, recall, and F1 score comparisons were less than .001 based on randomization tests for differences20 on 1 050 000 permutations.
pyDeid generally performs as well or better in PII identification than the other tools measured, with dramatic increases in speed. The NER configurations achieve a small improvement in recall with a large decrease in precision. However, pyDeid with custom name lists performs better on all metrics measured than any of the compared tools.
We also measure the performance of the base configuration of pyDeid on the n2c2 benchmark dataset, to validate the generalizability of the software to other regions and contexts. Protected Health Information filter outperforms pyDeid on recall and F1 score, whereas pyDeid outperforms Philter on the precision measure. Conversely, deid outperforms pyDeid on precision, but not recall or F1 score (see Appendix S2 for more details) (Table 2).
Table 2.
Base pyDeid performance on the n2c2 dataset.
| Category accuracy | Overall accuracy | Precision | Recall | F1 | |
|---|---|---|---|---|---|
| Base pyDeid | 0.991 | 0.760 | 0.821 | 0.777 | 0.798 |
Speed
There was a significant main effect of deidentification tool on speed, F(5, 54)=1494, P <.001. Posthoc tests with a Tukey adjustment for multiple comparisons indicated that Philter was considerably slower than deid (P< 001) and all versions of pyDeid (Ps<.001). deid was slower than the basic, Name List, and NER versions of pyDeid (Ps<.001) but did not differ significantly in speed from the Name Lists+NER configurations of pyDeid (Ps>.200).
Across the 4 configurations of pyDeid, the NER configuration was significantly slower than the basic configuration (P=.033) and the configuration with name lists (P=.004), and the configuration with Name Lists and NER was slower than with name lists only (P<.025).
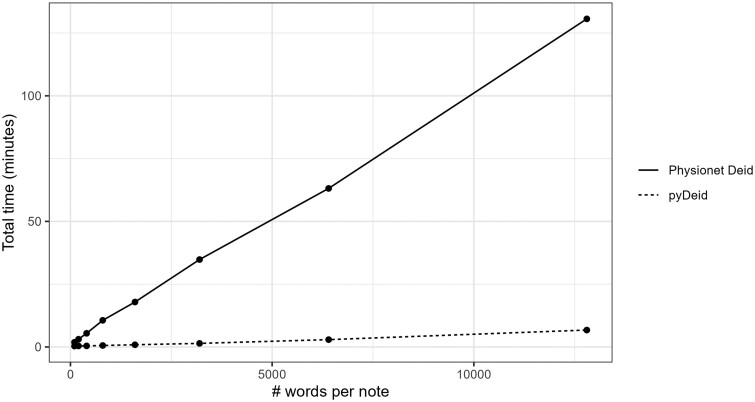
Lastly, we compared the deidentification speed of pyDeid and deid using synthetic medical notes of different sizes generated with 6% PII prevalence using ChatGPT (Appendix S3). The focus of this comparison is to assess the speed with which the tools process a given note, and does not consider the deidentification performance. Protected Health Information filter was excluded as it was the slowest deidentification tool (Table 1). As the size of the medical notes increased, deid was considerably slower than pyDeid (Figure 2).
Figure 2.
Speed comparison between the original Physionet deid tool and pyDeid.
Discussion
We present pyDeid: an updated, open-source tool for deidentification of PII in free-text medical notes. pyDeid fills a gap in the deidentification space for a tool that is fast and scalable to large notes, is customizable to other contexts, performs surrogate replacement, and is able to take advantage of in-memory name lists containing data that cannot be securely written to and read from persistent storage. Additionally, pyDeid is flexible in its approach to use regular expressions and NER to balance between precision and recall as appropriate for the use case, and is able to accept user-specified custom regular expressions while achieving a deidentification performance that is comparable to or better than existing tools.
Notably, the base configuration of pyDeid outperformed deid—the rule-based Perl software on which pyDeid is based,10 in precision, recall, and the F1 score. This is mostly attributable to the regional differences in PII in the Canadian medical notes on which these tools were tested compared to the American context for which deid was developed, which led most notably to lower false positive rates for Names. However, we note that deid has higher precision and F1 score than the NER versions of pyDeid, primarily due to the high false positive rate for names in the NER configuration. We additionally show that the non-NER configurations of pyDeid are nearly twice as fast as the deid software on the 700 admission notes containing an average of 1139 tokens per note. For high volume operational workflows, this difference in speed is crucial. Additionally, pyDeid can scale much better as the size of each individual note increases from 1000 tokens to 10 000 tokens (Figure 2).
Protected Health Information filter14 similarly uses a rule-based approach combined with a natural language processing pipeline to categorize tokens as PII. Recall performance was high and comparable between Philter and pyDeid, indicating that the probability of missing PII was not markedly different between the 2 tools. Precision was comparable between Philter and the NER configurations of pyDeid, whereas pyDeid outperformed Philter with its base and Name List configurations. Low precision in the deidentification process can be problematic for some downstream research use cases because valuable, non-PII information may be lost through inadvertent deidentification. Moreover, Philter was considerably slower than pyDeid, which limits its use in high volume workflows. Additionally, pyDeid offers surrogate replacement, which Philter does not. Surrogate replacement is a desirable property for the protection of patient privacy, in that missed PII is indistinguishable from found and replaced PII.
Another advantage of pyDeid is the availability of different configuration options that can be used to satisfy the needs and constraints of different deidentification use cases. When patient and doctor name lists are available, we show that there are meaningful gains to software recall, reducing missed PII for increased privacy, with very little tradeoff in precision and speed. If a use case allows for a sacrifice to speed and precision, enabling the NER option further improves recall. Another important property of the NER is that it is more effective at identifying uncommon or nontraditionally Canadian names that may not be contained in public name lists, which is valuable for the equity of patient privacy. If available, the use of custom doctor and patient name lists can also help mitigate this bias.
There are several methodological limitations of pyDeid. First, although pyDeid is conceptualized as a Python-based refactor of the Physionet deid tool, we iteratively reviewed pyDeid’s performance and added additional rules to the regex ruleset as needed, such as accounting for compound names (eg, “Van der Meer”) and name prefixes (eg, “Dr”). This is because Ontario is one of the most diverse places in the world with respect to race, language, ethnicity, and country of origin. Thus, any tool designed to detect PII must be able to handle and identify a diverse range of names without resulting in differential handling of privacy along the above axes of marginalization. Incorporating these additional rules into pyDeid ensures that equity-deserving populations have their privacy preserved. Nonetheless, we acknowledge that this process may have led to some bias in our tool comparison process, specifically, between deid and pyDeid.
Second, the largest error rate was associated with identifying locations (up to 50%, Table S3 in Appendix S1), because many locations in the medical notes were explicitly named as proper nouns (eg, the name of a nursing home) rather than street addresses. Future versions of pyDeid can enable location tagging in the NER step to identify named locations with greater accuracy. Third, when the NER option is enabled, there is a marked increase in the number of names falsely identified as PII (Table S2 in Appendix S1). This is likely due to the use of spaCy, which generally identifies named entities and may not distinguish them from medical terms, for instance. Future versions of pyDeid with NER can be improved by incorporating medspaCy that similarly identifies named entities in the clinical domain.21 We provide a short tutorial demonstrating how medspaCy components can be incorporated into pyDeid on the Github page for pyDeid.22 Finally, we report performance primarily in data from a single health care organization in Canada. Our confidence in the tool’s performance is strengthened by its strong performance in the publicly available n2c2 dataset from Partners HealthCare System in Boston, but additional validation across other health care organizations would be valuable.
Conclusion
Our results show that pyDeid performs as well as or better than existing deidentification tools across various use cases and is easily generalizable to other contexts. We also show that there are significant speed differences between existing regular expression-based implementations of deidentification tools and pyDeid, demonstrating scalability of pyDeid with increasingly large datasets. Thus, pyDeid serves as a fast, modular, and accurate tool for removing PII in free-text clinical notes and medical data.
Supplementary Material
Acknowledgments
We would like to thank Weihan Liu and Mohammad Arshad Imrit for their invaluable insights and expertise in addressing a software bug in pyDeid during the review process.
Contributor Information
Vaakesan Sundrelingam, Li Ka Shing Knowledge Institute, St Michael’s Hospital, Toronto, ON M5B 1T8, Canada.
Shireen Parimoo, Centre for Computational Medicine, Hospital for Sick Children, Toronto, ON M5G 0A4, Canada.
Frances Pogacar, MAP Centre for Urban Health Solutions, St Michael’s Hospital, Toronto, ON M5B 1T8, Canada.
Radha Koppula, Li Ka Shing Knowledge Institute, St Michael’s Hospital, Toronto, ON M5B 1T8, Canada.
Saeha Shin, Li Ka Shing Knowledge Institute, St Michael’s Hospital, Toronto, ON M5B 1T8, Canada.
Chloe Pou-Prom, Data Science and Advanced Analytics, Unity Health Toronto, Toronto, ON M5C 2T2, Canada.
Surain B Roberts, Li Ka Shing Knowledge Institute, St Michael’s Hospital, Toronto, ON M5B 1T8, Canada; Institute of Health Policy, Management and Evaluation, University of Toronto, ON M5T 3M6, Canada.
Amol A Verma, Li Ka Shing Knowledge Institute, St Michael’s Hospital, Toronto, ON M5B 1T8, Canada; Institute of Health Policy, Management and Evaluation, University of Toronto, ON M5T 3M6, Canada; Department of Medicine, University of Toronto, Toronto, ON M5S 3H2, Canada; Division of General Internal Medicine, Unity Health, Toronto, ON M5B 1W8, Canada.
Fahad Razak, Li Ka Shing Knowledge Institute, St Michael’s Hospital, Toronto, ON M5B 1T8, Canada; Institute of Health Policy, Management and Evaluation, University of Toronto, ON M5T 3M6, Canada; Department of Medicine, University of Toronto, Toronto, ON M5S 3H2, Canada; Division of General Internal Medicine, Unity Health, Toronto, ON M5B 1W8, Canada.
Author contributions
Vaakesan Sundrelingam, Shireen Parimoo, Surain B. Roberts, Fahad Razak, and Amol A. Verma conceptualized the study. Vaakesan Sundrelingam, Shireen Parimoo, Saeha Shin, and Radha Koppula performed data collection, annotation, and analysis. Frances Pogacar and Chloe Pou-Prom helped to develop and test the pyDeid software package. All authors critically revised the manuscript for important intellectual content.
Supplementary material
Supplementary material is available at JAMIA Open online.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. A.A.V. receives salary support as the Temerty Professor of AI Research and Education in Medicine. The development of the GEMINI data platform has been supported with funding from the Canadian Cancer Society, the Canadian Frailty Network, the Canadian Institutes of Health Research, the Canadian Medical Protective Association, Green Shield Canada Foundation, the Natural Sciences and Engineering Research Council of Canada, Ontario Health, the St Michael’s Hospital Association Innovation Fund, and the University of Toronto Department of Medicine, with in-kind support from partner hospitals and Vector Institute. Funders had no role in the design, conduct, or interpretation of this study.
Conflicts of interest
Fahad Razak and Amol A. Verma are part-time employees of Ontario Health (Provincial Clinical Leads) beyond the scope of this work. Other authors have no competing interests to declare.
Data availability
Research ethics board approval was obtained from all participating hospitals. This study qualifies for exception from informed consent as outlined in Article 3.7A & B of the Tri-Council Policy Statement: Ethical Conduct for Research Involving Humans, 2nd Edition (TCPS2). Data from this manuscript can be accessed upon request to the corresponding author, to the extent that is possible in compliance with local research ethics board requirements and data sharing agreements. The code is publicly available, and the open source is available at https://github.com/GEMINI-Medicine/pyDeid.
References
- 1. Afzal N, Sohn S, Abram S, et al. Mining peripheral arterial disease cases from narrative clinical notes using natural language processing. J Vasc Surg. 2017;65:1753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Finlayson SG, LePendu P, Shah NH. Building the graph of medicine from millions of clinical narratives. Sci Data. 2014;1:140032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iqbal E, Mallah R, Rhodes D, et al. ADEPt, a semantically-enriched pipeline for extracting adverse drug events from free-text electronic health records. PLoS One. 2017;12:e0187121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wagner T, Shweta F, Murugadoss K, et al. Augmented curation of clinical notes from a massive EHR system reveals symptoms of impending COVID-19 diagnosis. Elife. 2020;9:e58227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zheng L, Wang O, Hao S, et al. Development of an early-warning system for high-risk patients for suicide attempt using deep learning and electronic health records. Transl Psychiatry. 2020;10:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dorr DA, Phillips WF, Phansalkar S, Sims SA, Hurdle JF. Assessing the difficulty and time cost of de-identification in clinical narratives. Methods Inf Med. 2006;45:246-252. [PubMed] [Google Scholar]
- 7. Beckwith BA, Mahaadevan R, Balis UJ, Kuo F. Development and evaluation of an open source software tool for deidentification of pathology reports. BMC Med Inform Decis Mak. 2006;6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Douglass M, Clifford GD, Reisner A, Moody GB, Mark RG. Computer-assisted de-identification of free text in the MIMIC II database. In: Computers in Cardiology. IEEE; 2004:341-344.
- 9. Friedlin FJ, McDonald CJ. A software tool for removing patient identifying information from clinical documents. J Am Med Inform Assoc. 2008;15:601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neamatullah I, Douglass MM, Lehman L-wH, et al. Automated de-identification of free-text medical records. BMC Med Inform Decis Mak. 2008;8:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sweeney L. Replacing personally-identifying information in medical records, the Scrub system. Proc AMIA Annu Fall Symp. 1996:333-337. [PMC free article] [PubMed] [Google Scholar]
- 12. Jian Z, Guo X, Liu S, et al. A cascaded approach for Chinese clinical text de-identification with less annotation effort. J Biomed Inform. 2017;73:76-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee HJ, Wu Y, Zhang Y, Xu J, Xu H, Roberts K. A hybrid approach to automatic de-identification of psychiatric notes. J Biomed Inform. 2017;75S:S19-S27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Norgeot B, Muenzen K, Peterson TA, et al. Protected Health Information filter (Philter): accurately and securely de-identifying free-text clinical notes. NPJ Digit Med. 2020;3:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meystre SM, Friedlin FJ, South BR, Shen S, Samore MH. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC Med Res Methodol. 2010;10:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stubbs A, Uzuner Ö. Annotating longitudinal clinical narratives for de-identification: the 2014 i2b2/UTHealth corpus. J Biomed Inform. 2015;58:S20-S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uzuner O, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc. 2007;14:550-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nadeau D, Sekine S. A survey of named entity recognition and classification. LI. 2007;30:3-26. [Google Scholar]
- 19. Heider S, Meystre SM. An extensible evaluation framework applied to clinical text deidentification natural language processing tools: multisystem and multicorpus study. J Med Internet Res. 2024;26:e55676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yeh A. 2000. More accurate tests for the statistical significance of result differences. In: COLING 2000 Volume 2: The 18th International Conference on Computational Linguistics, Saarbrücken, Germany.
- 21. Eyre HAB, Chapman KS, Peterson J, et al. Launching into clinical space with medspaCy: a new clinical text processing toolkit in Python. AMIA Annu Symp Proc. 2022;2021:438. [PMC free article] [PubMed]
- 22. Sundrelingam V. medspaCy tutorial with pyDeid. 2024. Accessed December 13, 2024. https://github.com/GEMINI-Medicine/pyDeid/blob/master/docs/tutorials/using_spacy_with_pydeid.ipynb
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Research ethics board approval was obtained from all participating hospitals. This study qualifies for exception from informed consent as outlined in Article 3.7A & B of the Tri-Council Policy Statement: Ethical Conduct for Research Involving Humans, 2nd Edition (TCPS2). Data from this manuscript can be accessed upon request to the corresponding author, to the extent that is possible in compliance with local research ethics board requirements and data sharing agreements. The code is publicly available, and the open source is available at https://github.com/GEMINI-Medicine/pyDeid.