Abstract
Structured RNAs achieve their active states by traversing complex, multidimensional energetic landscapes. Here we probe the folding landscape of the Tetrahymena ribozyme by using a powerful approach: the folding of single ribozyme molecules is followed beginning from distinct regions of the folding landscape. The experiments, combined with small-angle x-ray scattering results, show that the landscape contains discrete folding pathways. These pathways are separated by large free-energy barriers that prevent interconversion between them, indicating that the pathways lie in deep channels in the folding landscape. Chemical protection and mutagenesis experiments are then used to elucidate the structural features that determine which folding pathway is followed. Strikingly, a specific long-range tertiary contact can either help folding or hinder folding, depending on when it is formed during the process. Together these results provide an unprecedented view of the topology of an RNA folding landscape and the RNA structural features that underlie this multidimensional landscape.
RNAs that adopt specific functional structures face the exacting challenge of attaining these specific structures from a myriad of unfolded conformations. An early and critical step in understanding the folding process is the development of a quantitative kinetic and thermodynamic description. For the Tetrahymena group I ribozyme, work over the past decade has established a complex pathway for folding. Intermediates have been identified (1–3), structurally characterized and shown to represent kinetic traps (4–6), and a folding model that unifies many results has been presented (7).
A newer view of folding, first proposed for proteins and explored with lattice models, describes a folding funnel (8–10). In its most rudimentary form, the folding funnel represents a statement that any complex molecular process with many degrees of freedom can be described in terms of a multidimensional energy landscape. For functional macromolecules, kinetic and thermodynamic considerations provide general constraints on the shape of the landscape, necessitating some degree of overall funnel-like shape. That is, folding intermediates leading to the final structure must progress downhill energetically, in general, to allow folding on observed time scales, and the final folded state must sit in a relatively narrow region at the bottom of the energy landscape to ensure that the specific functional state is favored over the ensemble of all other possible conformations (11, 12).
Beyond these requirements, there are many models and disagreements about the topology and nature of barriers in folding landscapes (9, 10, 13, 14), but little experimental data that describe actual folding landscapes beyond individual pathways. For example, it is generally held that folding landscapes for RNAs are more rugged than those for proteins (6, 15, 16), and we believe this to be likely from consideration of the molecular properties of RNA (17, 18). Nevertheless, exploration of folding landscapes will be required to directly address the fundamental question of landscape shape and to provide a framework for revealing potential intermediates and the structural features responsible for “choosing” among the multitude of potential folding pathways (19–21).
The visualization of single molecules provides a powerful means of identifying and characterizing multiple folding pathways (22). However, the ability of this or any other approach to broadly explore features of a folding landscape may be limited by conformational biases present in the starting unfolded conformation (23–25). Such biases may be particularly severe for RNA, which typically has considerable secondary structure present in the starting state. Because there is insufficient time during folding for all unfolded and partially folded conformations on the landscape to be explored (12), a bias in the starting state must give a bias in the portion of the landscape explored during folding. Similarly, a change in the bias of the starting state, leading to population of a different portion of the top of the folding funnel, must lead to exploration of a different part of the landscape during folding. The extent to which folding through different portions of the landscape, or sides of the funnel, leads to gross changes in the folding process remains an open question.
We have therefore turned to a protocol that allows us to populate different ensembles of starting states for the Tetrahymena ribozyme, and then rapidly change conditions so that folding proceeds in all cases on the same energy landscape. We follow the folding pathways of the ribozyme by using single-molecule fluorescence methods and demonstrate the presence of discrete folding pathways from the different ensembles of starting states. Structural studies and mutagenesis are then carried out to uncover the structural differences between the starting ensembles and the specific structural features that determine which pathways are traversed.
Materials and Methods
Materials.
DNA encoding 5′- and 3′-extended ribozymes was prepared by PCR and cloned into the plasmid pT7L-21 (26). Ribozymes were prepared as described (27). Cy3 and Cy5 (Amersham Pharmacia) labeling of RNA oligonucleotides (Dharmacon, Lafayette, CO) was performed as described (22).
Single-Molecule Fluorescence Resonance Energy Transfer (FRET).
Experiments were performed essentially as described (22). For FRET experiments the ribozyme was extended by 5 nucleotides at the 5′ end with the sequence GGUUU. This allowed tight binding of an oligonucleotide substrate (S) capable of base-pairing to the 5′-extension and to the internal guide sequence of the ribozyme (S-CCCUCUAAACC), thereby minimizing dissociation of S from the immobilized ribozyme during experiments. These modifications have no detectable effect on the enzymatic activity of the ribozyme (data not shown).
Small-Angle X-Ray Scattering (SAXS) Measurements.
Data were collected on beamline 4–2 at the Stanford Synchrotron Radiation Laboratory essentially as described (ref. 28; see SAXS Measurements, which is published as supporting information on the PNAS web site, www.pnas.org).
Dimethyl Sulfate (DMS) Footprinting.
Footprinting was performed essentially as described (29). Only residues that reproducibly gave protections of ≥2-fold are shown as giving protections in Fig. 3. A196, A304, and A306 gave increased DMS accessibility in the presence of Na+, but these enhancements were not reproducibly larger than 2-fold.
Figure 3.
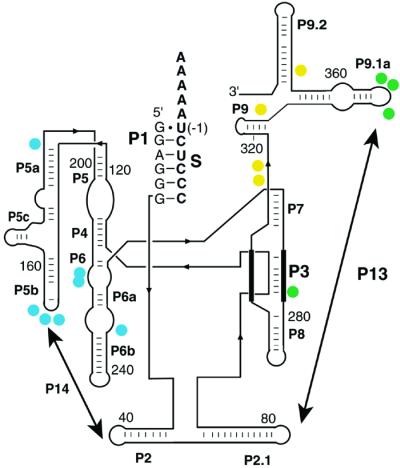
Structural changes in the starting ensemble monitored by DMS accessibility. Residues that increase in protection from DMS in the presence of NaCl are indicated with filled circles. Residues that gave an Na+ concentration for half-maximal protection (K1/2) < 100 mM are colored yellow (C315, C316, C332, C371), residues that gave K1/2 values between 100 mM and 300 mM Na+ are green (C278, A347, A351, A352), and residues with K1/2 > 300 mM Na+ are blue (A151–153, A196, C216, C217, A246). Results are for the standard L-21 ScaI ribozyme in the presence of bound S. The extended version used for single-molecule experiments gave an indistinguishable Na+ dependence of DMS protection (data not shown), suggesting that the structural changes induced by Na+ are similar for the two versions of the ribozyme. The strands that pair to form Alt P3, which can replace P3 (3), are shown with thick lines.
Results
Exploring the Folding Landscape for the Tetrahymena Ribozyme.
We wanted to initiate folding with RNA molecules having different starting structures. In the language of energy landscapes, we wanted to populate different regions within the unfolded portions of the landscape and then determine what effect, if any, these different starting states had on the folding process. To do this we incubated the Tetrahymena group I ribozyme with different concentrations of Na+ ion, with the idea that varying the cation concentration would change the structure, by ameliorating electrostatic repulsion, without allowing folding to occur, as formation of the native state requires divalent cations (30, 31). We then wanted to add divalent metal ions to observe individual RNA molecules that initiate folding from different structures. If, however, the variable Na+ concentrations were maintained after addition of Mg2+, the intent of the experiment would be defeated; molecules starting from different structures would also be folding under different conditions, undermining direct comparison. It was therefore critical to rapidly adjust the solution to a constant Na+ concentration simultaneous with divalent metal ion addition. With this protocol, all of the molecules face the identical energy landscape, and differences in folding behavior can provide information about the topology of the energy landscape.
We followed the folding of individual ribozyme molecules with FRET after preincubation with 20–620 mM Na+; folding was always performed with 5 mM Mg2+ and 20 mM Na+. Individual ribozyme molecules on a glass surface were visualized by fluorescence emission of Cy3 and Cy5 dyes attached to the S of the ribozyme and to the oligonucleotide tether used in immobilization of the ribozyme (22). For each molecule, we recorded the time at which the FRET value increased to 0.9, indicating native state formation [the ribozyme molecules acquire enzymatic activity concomitantly with the FRET value of 0.9 (22)]. We then plotted the number of native ribozyme molecules as a function of time.
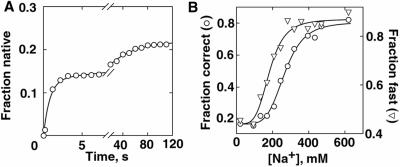
Fig. 1A shows results from a representative experiment, with folding following preincubation with 170 mM Na+. As observed with 20 mM Na+ present both before and after Mg2+ addition (22), folding to the native state is well described by the sum of two exponential curves with rate constants of ≈1 s−1 and ≈0.02 s−1. A third folding phase, conversion of a long-lived misfolded state to the native state, does not occur significantly on the time scale of these experiments (7, 27). These two folding phases are collectively referred to as “correct” folding, giving a fraction of the population that folded correctly (i.e., avoided the long-lived misfolded form).
Figure 1.
Exploring the folding landscape. (A) Folding was initiated by flowing buffer containing 5 mM Mg2+ over the surface-attached ribozyme, replacing the 170 mM Na+ solution present initially. The fraction of molecules that attained FRET values of ≈0.9, indicating native-state formation, is plotted against folding time. The curve represents a fit by two exponentials with rate constants of 1.21 ± 0.07 s−1 and 0.025 ± 0.001 s−1. The combined amplitude of these two folding phases, 0.219 ± 0.003, represents the fraction of ribozyme that folded to the native state without forming the long-lived misfolded state. (B) The fraction of ribozyme that folded correctly (○), and the fraction of the correct-folding molecules that folded fast (≈1 s−1, ▿), are plotted against initial Na+ concentration. Only the correct-folding molecules were analyzed for the rate of folding because the molecules that misfold do not give a detectable change in FRET value after formation of an early intermediate (22). The dependence of fast folding on Na+ concentration gave a half-maximal value for Na+ (K1/2) of 183 ± 9 mM and a Hill coefficient (n) of 4.4 ± 0.9. For correct folding, these values were K1/2 = 267 ± 8 mM, n = 5.0 ± 0.7.
Varying the Na+ concentration in the preincubation from 20 to 620 mM increased the fraction that folded correctly from 0.2 to ≈0.8 (Fig. 1B) even though folding was always under the same conditions (5 mM Mg2+, 20 mM Na+). Also, the fraction of the correct-folders that folded fast (1 s−1) increased from ≈0.45 to ≈0.9. With high Na+ concentration in the preincubation (>400 mM), nearly the entire population folded rapidly and to the native state. Replacing Na+ with other monovalent cations (Li+, K+, NH4+) gave similar changes in folding properties (not shown). The changes in folding properties, despite folding under identical conditions, suggest that the initial Na+ concentration affects the starting conformation. Then, from these different starting conformations (or, more precisely, ensembles of starting conformations), different portions of the landscape are traversed during folding.
If Na+ were shifting the ribozyme between only two conformational ensembles, one of which gave slow and largely incorrect folding whereas the other gave fast and correct folding, the dependencies of both fast and correct folding on initial Na+ concentration would be the same. However, the changes in fast and correct folding give distinct Na+ concentration dependencies (Fig. 1B), suggesting the presence of at least three conformational ensembles. These three conformational ensembles are as follows. (i) One, populated with low Na+ in the preincubation, gives mostly slow and incorrect folding. (ii) With intermediate Na+ in the preincubation, the populated ensemble gives mostly incorrect folding, but the molecules that fold correctly also fold rapidly. (iii) With high Na+ in the preincubation, the populated ensemble gives mostly fast and correct folding.
Although in principle Na+ dilution to 20 mM is instantaneous with Mg2+ addition and the onset of folding, in practice there is a finite mixing time. Thus, the differences in folding properties could arise if, instead of or in addition to changing the starting conformation, the Na+ still present only milliseconds after Mg2+ addition affected very early folding steps. To test this possibility, we performed an experiment in which Na+ was washed out with variable time delays before the addition of Mg2+. Although the bulk and RNA-associated Na+ ions are expected to exchange on the submillisecond time scale (32), the time dependencies for the changes in folding behavior upon removal of Na+ were on the scale of seconds (Fig. 6, which is published as supporting information on the PNAS web site). These slow transitions provide strong evidence that the changes in folding behavior arise from changes in starting conformation rather than Na+ effects on very early events in folding. Further, the losses of correct folding and fast folding gave distinct rate constants (0.6 s−1 and 0.09 s−1, respectively), confirming the existence of at least two discrete structural transitions induced by Na+ (i.e., at least three conformational ensembles).
The experiments above build on previous results by Woodson and colleagues (33, 34), who showed that preincubation with Na+ allows the ribozyme to fold to the native state in the presence of Mg2+ within the time required to enter a native gel (≈30 s). Here, we have used the increased time resolution and decreased mixing time in our single-molecule approach to directly demonstrate and follow a fast-folding phase. Further, we have shown that the observed change in folding behavior results from at least two structural transitions; one transition gives fast folding and the other gives correct folding. Below we use these results to further our understanding of the folding landscape, first by providing evidence that the starting states induced by Na+ give folding along discrete pathways through the folding landscape and then by identifying structural features that determine which pathways are followed.
Determination of Multiple Pathways on the Folding Landscape.
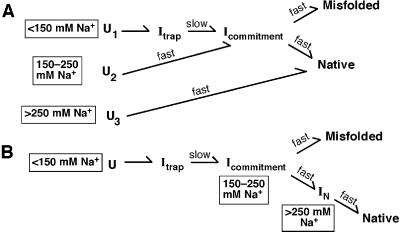
Scheme S1 depicts two models that can account for the data described above. [The models also incorporate previous results (7, 22)**.] In Model 1, different Na+ concentrations cause three different starting states to be differentially populated. These different starting states then fold along discrete pathways through the landscape (Scheme S1A). With low concentrations of Na+ in the preincubation, a significant fraction (0.55) starts folding from U1. These molecules proceed through the late folding intermediate Itrap and then slowly escape this intermediate to partition between the native and long-lived misfolded forms (1, 2, 7). With intermediate Na+ in the preincubation, folding starts from U2; Itrap is avoided but the partitioning between the native and long-lived misfolded forms is the same as at low Na+. This identical partitioning suggests the presence of the common folding intermediate Icommitment (22). With high Na+ in the preincubation, U3 is populated and essentially all of the ribozyme folds fast and correctly; thus, both Itrap and Icommitment are avoided.
Scheme 1.
In Model 2, the changes in folding properties arise because increasing Na+ concentration causes the ribozyme to fold part of the way along a single pathway; i.e., when Mg2+ is added the ribozyme is already partially folded (Scheme S1B). Folding with <150 mM Na+ begins with U, proceeds through a rate-limiting step escaping from the intermediate Itrap, and then, after formation of Icommitment, partitions between formation of the native and long-lived misfolded states. Folding is faster after preincubation with 150–250 mM Na+ because an intermediate (e.g., Icommitment) situated after the rate-limiting step is populated at the start of folding. With still higher Na+ in the preincubation both Itrap and Icommitment are avoided so that folding is fast and gives exclusively native ribozyme.
The models in Scheme S1 A and B were distinguished by performing SAXS on the conformational ensembles induced by Na+. Model 2 predicts that Na+ allows the ribozyme to form folding intermediates that are also formed in Mg2+ after the rate-limiting step for overall folding (after Itrap in Scheme S1B). Previous SAXS experiments showed that Itrap is nearly as compact as the native state (28) and the simplest expectation is that intermediates after Itrap would also be compact (see Determination of Discrete Pathways by SAXS, which is published as supporting information on the PNAS web site). As the concentration of Na+ was increased in the range that gives changes in the folding properties, SAXS profiles showed appearance of a peak in Kratky plots (Fig. 2) (35). This peak indicates that the ribozyme becomes somewhat more compact and globular as the concentration of Na+ is increased. However, even the highest concentration of Na+ used (820 mM), sufficient to give essentially all fast and correct folding, gave a Kratky peak much less pronounced than that of the intermediate Itrap or the native ribozyme.
Figure 2.
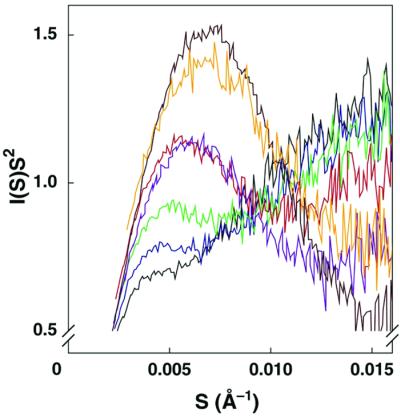
Structural changes in the starting ensemble monitored by SAXS. Kratky plots are shown for the standard L-21 ScaI ribozyme at 25°C in buffer containing 20 mM Na+ (black), 70 mM Na+ (blue), 120 mM Na+ (green), 420 mM Na+ (red), 820 mM Na+ (purple), the intermediate Itrap (orange, formed at 15°C), and the native ribozyme in buffer containing 15 mM Mg2 + (brown). Kratky plots have the form of I(S)S2 against S, where I(S) is scattering intensity and S is related to the scattering angle [S = 2sinθ/λ, where λ is the x-ray wavelength, 1.54 Å, and 2θ is the scattering angle (35)].
These results indicate that the ribozyme is substantially less compact and globular than Itrap and the native ribozyme across the range of Na+ concentrations, providing strong evidence against Model 2. Further, solution radical protection experiments gave no indication of native tertiary structure formation in Na+ concentrations that give fast and correct folding (ref. 31; M. Brenowitz, personal communication). Thus, the results strongly favor Model 1 (Scheme S1A), in which folding starts from different conformational ensembles that depend on the initial Na+ concentration and proceeds through discrete pathways.
Structural Features That Determine Which Pathway Is Traversed.
To identify the structural changes that might be responsible for the different observed folding behavior after preincubation with different concentrations of Na+, protection of adenosine and cytosine residues from DMS was monitored. As the concentration of Na+ was increased, residues in P9, P3, and P4-P6 became protected from DMS modification with distinct concentration dependencies (Fig. 3). Interestingly, two sets of protections were observed at Na+ concentrations in the range that gives changes in folding properties (100–300 mM; Fig. 1B). The observed protection of residue C278 in P3 suggests the formation of P3 (3), and the protection of residues in loop L9.1 suggests the formation of P13, a long-range base-pairing contact between residues in L9.1 and L2.1. This interpretation was supported by the finding that a variant ribozyme with a three-nucleotide substitution in L2.1 sequence to disrupt P13 [ΔP13 (36)] does not give protection of L9.1 (data not shown). These results raise the possibility that initial formation of P3 and/or P13 in the presence of Na+ is responsible for the observed fast and correct folding.
P3 formation was particularly appealing as a structural determinant for folding pathways in light of previous studies showing that an alternative local duplex termed Alt P3 replaces P3 in the long-lived misfolded state (3). Further, a variant ribozyme, U273A, in which P3 is stabilized relative to Alt P3, has been observed to fold both fast and correctly (37). These previous results suggested a model in which formation of the correct secondary structure early in folding is critical: if P3 is formed correctly, folding is fast and correct, whereas if Alt P3 is formed early, the ribozyme gets trapped in misfolded species (37, 38).
Our results confirming that correct folding is observed with high initial Na+ concentration (33) and showing P3 is formed initially under these conditions provides further support for the model that P3 formation is responsible for the correct folding (37, 38). On the other hand, the observation that the conformational transitions that give fast folding and correct folding are distinct (Fig. 1B) calls into question the previous conclusion that P3 formation is also responsible for fast folding.
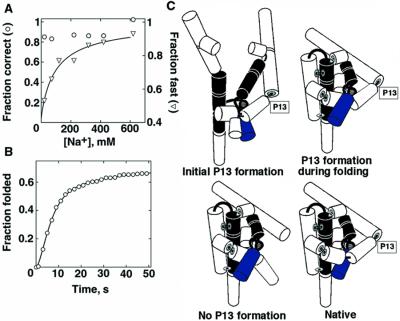
To determine whether early P3 formation is responsible for both fast and correct folding we examined folding of the variant U273A by single-molecule FRET as a function of initial Na+ concentration. As expected (3, 37), folding of the U273A variant ribozyme was largely correct at all initial Na+ concentrations (Fig. 4A). Further, DMS footprinting revealed that P3 is formed initially in the U273A variant across the entire range of Na+ concentration (20–820 mM; data not shown). However, with low initial Na+ concentrations, two rate constants for folding were observed as with the wild type; about half of the variant ribozyme folded slowly (0.012 s−1), approximately the same fraction of slow folding as for the wild type. The concentration of Na+ required to give fast folding of the U273A variant was similar to that for the wild type. Thus, although initial P3 formation correlates strongly with correct folding, it is not sufficient to give fast folding. It is also suggested that P3 formation is not necessary for fast folding, as fast folding with the wild-type ribozyme occurs at Na+ concentrations lower than required to give correct folding.
Figure 4.
Structural changes responsible for distinct folding properties. (A) Folding of the U273A variant ribozyme, in which P3 is stabilized. The fraction of ribozyme molecules that folded correctly (○) and fast (▿) are plotted against initial Na+ concentration. A fit by the Hill equation gave values for fast folding of K1/2 = 100 ± 50 mM Na+ and n = 0.9 ± 0.2. (B) Folding of the ΔP13 variant (U75A, G76C, C77G), in which P13 is disrupted. The ribozyme also contained the U273A substitution, allowing it to avoid the long-lived misfolded form. An increase in rate at early times (a kinetic lag) was observed, necessitating two rate constants of 0.30 ± 0.04 s−1 and 0.29 ± 0.04 s−1 to describe the fast phase of folding [620 mM Na+ initially; similar results were obtained with an initial Na+ concentration of 20 mM. A small fraction (10%) folded slowly, giving a third rate constant of 0.038 ± 0.005 s−1]. These two larger rate constants are the same within error as that for docking of the P1 duplex (0.23 s−1; unpublished results). As docking is required for efficient FRET (22), the other rate constant of 0.3 s−1 is assigned to overall folding. (C) Model for the effects of P13 formation on folding (see text). The model reflects folding in the context of preformed P3, shown as two thick lines connected by thin lines. Other secondary structure elements are shown as cylinders, and long-range contacts are shown as thin connecting lines.
For the U273A variant, as for the wild type, residues in P13 became protected from DMS in the Na+ concentration range that gave fast folding (not shown). We therefore tested the effect of P13 on fast folding, monitoring the folding of the variant ribozyme in which P13 is disrupted (ΔP13). This substitution was made in the background of the U273A variant to simplify analysis by allowing predominantly correct folding at all Na+ concentrations. We reasoned that if P13 formation were responsible for fast folding, the ΔP13 variant would give slow folding at all initial concentrations of Na+ because of the inability to form P13 (36).
Contrary to this prediction, predominantly fast folding was observed after preincubation with both low and high Na+ concentrations (Fig. 4B). This surprising result indicates that the order of tertiary structure formation can be important. That is, if P13 is initially formed, fast folding is observed. Likewise, if P13 cannot form, fast folding is also observed. However, if P13 is not formed initially but has the ability to form during folding, folding is slow for a fraction of the ribozyme.
One general possibility for the origin of these effects is that formation of P13 at the “wrong” time can lead to topological barriers in folding (Fig. 4C). According to this model, if P13 is formed initially, as it is with high initial Na+ concentration for the U273A variant and the wild-type ribozyme, folding is fast because constraints imposed by the preformed P13 give a ribozyme core that is prearranged to fold rapidly. In contrast, if P13 is not formed initially but has the potential to form, as is the case with low Na+ concentration for both the U273A and wild-type ribozymes, folding of a large fraction is slow, consistent with a barrier to rearrangement that is created after P13 formation, as suggested (6, 39). Fig. 4C shows one specific possibility: after formation of P13, the P8 duplex becomes trapped outside the ring composed of P2, P2.1, and P9. Finally, if P13 is prevented from forming by mutagenesis (ΔP13), barriers created by P13 formation would not be present, allowing the observed fast folding (see Additional Results from P13 Disruption, which is published as supporting information on the PNAS web site).
Discussion
When a macromolecule folds to an active structure, it traverses a multidimensional energetic landscape; thus, understanding the energetic topology of the landscape is essential for understanding the folding process for a given molecule. The landscape is a representation of intermediates and their energetic connections with each other and with the native and unfolded states. Beyond this energetic description, structural descriptions of intermediate forms are needed. Further, we want to know what specific parts of the structure are important for defining the different intermediates and the energy barriers for interconversion between them.
To obtain new information about the topology of the landscape for the Tetrahymena ribozyme, we used a procedure that allowed us to broadly explore the landscape. Preincubation with different concentrations of Na+ allows folding to be initiated from different starting structures, i.e., different regions of the folding landscape. To probe the same landscape, folding was then followed under identical conditions in all experiments. With this procedure, we found that the folding properties strongly depend on the region of the landscape from which the molecules start to fold. With increasing concentrations of Na+ present in the preincubation, the fraction of ribozyme molecules that folded fast increased, as did the fraction that avoided a long-lived misfolded conformation.
Two models were considered to explain this behavior: the different starting structures lead to folding along discrete pathways or the different starting structures instead represent successive intermediates along a single folding pathway. It was shown that the folding properties of the ribozyme depend on starting conditions (33, 34), but the models above were not distinguished. Here, SAXS experiments and our knowledge of the compactness of folding intermediates provided strong evidence for discrete folding pathways. Two discrete folding pathways have also been demonstrated for a circularly permuted variant of RNase P (40).
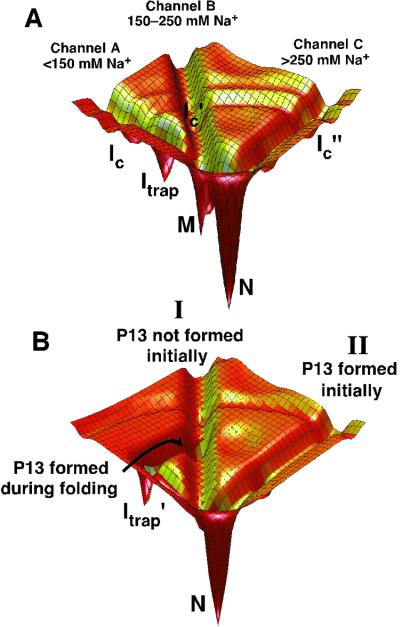
These results extend our understanding of the folding landscape for the Tetrahymena ribozyme (Fig. 5A). The landscape contains pathways that lie in deep channels, with barriers, likely with both enthalpic and entropic components, sufficient to prevent interconversion of molecules on the different pathways (see also refs. 41 and 42 for further discussions of discrete pathways). The solution conditions before Mg2+-induced folding determine the starting RNA structures and, thus, which pathways are entered. After incubation in low Na+ (<150 mM), pathway A (which lies in Channel A in Fig. 5A) is predominantly traversed, leading to slow folding by means of Itrap and partitioning between N and M, the native and long-lived misfolded states, respectively. Pathways B and C give fast folding by avoiding the intermediate Itrap, and pathway C gives correct folding by avoiding the region of the landscape that drains into the long-lived misfolded form M (this corresponds to avoiding Icommitment in Scheme S1A).
Figure 5.
RNA folding pathways contained in channels. (A) A schematic of channels for the wild-type ribozyme. The dependence of folding properties on initial conditions indicates the presence of free energy barriers between the channels, whereas the figure is contoured by internal free energy (9, 10). At least some of these barriers are likely to be present when considering internal free energy because interconversion between the starting states requires base pairs to exchange. Therefore, “walls” are shown separating the channels. The simplest model is shown, in which there are three pathways, A–C, that lie in channels A, B, and C, respectively. Pathways A and B predominate at low initial Na+ concentration, pathway B at intermediate Na+ concentration, and pathway C at high Na+ concentration. The fast-folding fraction under low Na+ conditions could instead arise from an additional pathway that branches from pathway A during folding before the formation of Itrap (not shown). Ic is a collapsed intermediate that forms along pathway A (R.R., I.S.M., M. Tate, L. Kwok, B. Nakatani, V. Pande, S. Gruner, S. Mochrie, S.D., D.H., and L. Pollack, unpublished data), and analogous intermediates are postulated along pathways B and C. (B) Effects of P13 formation during folding starting with a preformed P3 (see text). The landscape for folding of the U273A ribozyme is depicted for simplicity, in which the long-lived misfolded form is not formed significantly and is therefore not shown.
To probe the structural underpinnings of the discrete folding pathways, we used chemical protection to identify structural differences between the starting conformations and then mutagenesis to identify what structural changes are important for defining the different pathways. DMS footprinting showed that several regions became protected with increasing concentration of Na+ and identified the long-range base-pairing contact P3 as a strong candidate for involvement in the changes in folding properties (3, 37).
The involvement of early P3 formation in fast folding and correct folding was tested by following folding of a ribozyme variant in which P3 is stabilized. Folding of this variant showed that formation of P3 in the starting population is responsible for correct folding as suggested (37). On the other hand, we found that P3 formation in the starting population is not necessary or sufficient for fast folding. Thus, multiple structural changes are responsible for determining which pathways are traversed.
Interestingly, when P3 is formed initially in the variant, DMS protection experiments suggest that essentially all of the secondary structure is correctly formed (unpublished results); nevertheless, folding can be slow. This finding suggests that there are substantial energy barriers for RNA folding from preformed secondary structure, such as topological constraints on the dissolution of fortuitously formed nonnative tertiary interactions and on movements of subdomains relative to each other.
To further probe the structural features that determine which pathway is traversed, we explored the effects of disrupting the long-range contact P13, which was also shown by DMS footprinting to form in the presence of Na+. P13 formation is involved with fast folding but in a surprising manner: forming the right structure, P13 can either help or hinder folding, depending on when it is formed during the folding process.
Forming P13 during the folding process is deleterious, leading to slow folding. This result is analogous to results showing that native interactions in P5abc can slow fold by stabilizing a kinetically trapped intermediate (4, 5) and can be rationalized by the location of P13 on the exterior of the ribozyme model structure (36). The deleterious effects of P13 could arise because a topological barrier to folding introduced by P13 formation prevents access of other structural elements to the molecule's core (Fig. 4C) or because P13 formation brings together regions of the molecule that have yet to establish their native interactions and are thus highly susceptible to formation of fortuitous nonnative interactions.
Remarkably, if P13 is formed early, at the outset of folding, it no longer hinders folding. This difference in the effect of P13 formation could arise because when P13 is formed in the starting state the core elements are preoriented to fold efficiently as depicted in Fig. 4C, or it could be because early P13 formation blocks a nonnative contact that otherwise forms during folding and is subsequently stabilized if P13 forms later. Regardless of the molecular origin, this result indicates that the presence of P13 is not alone responsible for and does not necessitate the formation of a kinetic trap. This finding is depicted in the simplified energy landscape in Fig. 5B. If P13 is not formed initially, pathway I is followed; after formation of P13, the RNA is shunted to form a trapped species (Itrap′). Thus, if P13 is mutated and cannot form, the trap is avoided and folding to N is fast. The avoidance of the trap when P13 is formed at the outset of folding indicates that a distinct folding pathway is followed (Fig. 5B, pathway II); the barriers separating pathway II from pathway I are sufficient to prevent significant formation of Itrap′.
Multiple kinetic phases are commonly observed for RNA folding (e.g., refs. 3, 27, 40, and 42–46), indicating the existence of multiple folding pathways. Thirumalai and Woodson have proposed that these pathways are separated from the earliest stages of folding (41). Results here provide direct evidence that some pathways are separated from the earliest stages of folding (Fig. 5A), and also demonstrate the existence of a slow-folding pathway that is separated by energy barriers only later in folding, after P13 formation (Fig. 5B). We showed that there is also a separation of pathways very late in folding that commits the ribozyme to fold to the native state or to a long-lived misfolded state (7). Thus, there are likely to be networks of folding pathways throughout the landscape that branch and reconnect at different points leading to formation of trapped species and the native ribozyme.
Recent work in protein folding has emphasized that the unfolded states can contain residual structure, and this residual structure is suggested to be important for the folding process (24, 47). For RNA it is particularly striking that the starting states are not simply “unfolded” but rather contain substantial structure. Further, results here show that the structural features in the starting states and structural differences between different starting states can have profound effects on folding. Future characterization of these folding pathways will clarify the relationship of these starting structures to intermediates on the folding pathways. Do the starting structures in Na+ give enhanced folding because they resemble late-folding intermediates, or are they conformations that are largely unfolded but are readily transformed into the late-folding intermediates? The methods used here should be valuable as we continue probing the pathways identified here, and continue exploration to uncover and understand new pathways and features of this landscape.
Supplementary Material
Acknowledgments
This research was supported by a Packard Interdisciplinary grant (to D.H. and S.C.), a National Science Foundation grant, and by an Air Force Office of Scientific Research grant (to S.C.). The U.S. Department of Energy and the National Institutes of Health support the Stanford Synchrotron Radiation Laboratory. R.R. and X.Z. were supported by National Institutes of Health postdoctoral fellowships. H.P.B. was supported in part by a Center on Polymer Interfaces and Macromolecular Assemblies grant.
Abbreviations
- DMS
dimethyl sulfate
- FRET
fluorescence resonance energy transfer
- S
oligonucleotide substrate
- SAXS
small angle x-ray scattering
Footnotes
References
- 1.Zarrinkar P P, Williamson J R. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 2.Sclavi B, Sullivan M, Chance M R, Brenowitz M, Woodson S A. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 3.Pan J, Woodson S A. J Mol Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- 4.Treiber D K, Rook M S, Zarrinkar P P, Williamson J R. Science. 1998;279:1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- 5.Rook M S, Treiber D K, Williamson J R. J Mol Biol. 1998;281:609–620. doi: 10.1006/jmbi.1998.1960. [DOI] [PubMed] [Google Scholar]
- 6.Treiber D K, Williamson J R. Curr Opin Struct Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 7.Russell R, Herschlag D. J Mol Biol. 2001;308:839–851. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- 8.Leopold P E, Montal M, Onuchic J N. Proc Natl Acad Sci USA. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryngelson J D, Onuchic J N, Socci N D, Wolynes P G. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 10.Dill K A, Chan H S. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 11.Anfinsen C B, Haber E, Sela M, White F H J. Proc Natl Acad Sci USA. 1961;49:1309–1314. doi: 10.1073/pnas.47.9.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levinthal C. In: Proceedings of a Meeting Held at Allerton House, Monticello, IL. Debrunner P, Tsibris J C M, Münck E, editors. Urbana, IL: Univ. of Illinois Press; 1969. pp. 22–24. [Google Scholar]
- 13.Baldwin R L, Rose G D. Trends Biochem Sci. 1999;24:77–83. doi: 10.1016/s0968-0004(98)01345-0. [DOI] [PubMed] [Google Scholar]
- 14.Bilsel O, Matthews C R. Adv Protein Chem. 2000;53:153–207. doi: 10.1016/s0065-3233(00)53004-6. [DOI] [PubMed] [Google Scholar]
- 15.Woodson S A. Cell Mol Life Sci. 2000;57:796–808. doi: 10.1007/s000180050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treiber D K, Williamson J R. Curr Opin Struct Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 17.Sigler P B. Annu Rev Biophys Bioeng. 1975;4:477–527. doi: 10.1146/annurev.bb.04.060175.002401. [DOI] [PubMed] [Google Scholar]
- 18.Herschlag D. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 19.Jennings P A, Finn B E, Jones B E, Matthews C R. Biochemistry. 1993;32:3783–3789. doi: 10.1021/bi00065a034. [DOI] [PubMed] [Google Scholar]
- 20.Radford S E, Dobson C M, Evans P A. Nature (London) 1992;358:302–307. doi: 10.1038/358302a0. [DOI] [PubMed] [Google Scholar]
- 21.Kiefhaber T. Proc Natl Acad Sci USA. 1995;92:9029–9033. doi: 10.1073/pnas.92.20.9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhuang X, Bartley L E, Babcock H P, Russell R, Ha T, Herschlag D, Chu S. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 23.Srinivasan R, Rose G D, Leopold P E, Montal M, Onuchic J N. Proc Natl Acad Sci USA. 1999;96:14258–14263. doi: 10.1073/pnas.96.25.14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shortle D, Ackerman M S. Science. 2001;293:487–489. doi: 10.1126/science.1060438. [DOI] [PubMed] [Google Scholar]
- 25.Murthy V L, Srinivasan R, Draper D E, Rose G D. J Mol Biol. 1999;291:313–327. doi: 10.1006/jmbi.1999.2958. [DOI] [PubMed] [Google Scholar]
- 26.Zaug A J, Grosshans C A, Cech T R. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- 27.Russell R, Herschlag D. J Mol Biol. 1999;291:1155–1167. doi: 10.1006/jmbi.1999.3026. [DOI] [PubMed] [Google Scholar]
- 28.Russell R, Millett I S, Doniach S, Herschlag D. Nat Struct Biol. 2000;7:367–370. doi: 10.1038/75132. [DOI] [PubMed] [Google Scholar]
- 29.Inoue T, Cech T. Proc Natl Acad Sci USA. 1985;82:648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latham J A, Cech T R. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 31.Celander D W, Cech T R. Science. 1991;251:401–407. doi: 10.1126/science.1989074. [DOI] [PubMed] [Google Scholar]
- 32.Bleam M L, Anderson C F, Record M T. Biochemistry. 1983;22:5418–5425. [Google Scholar]
- 33.Heilman-Miller S L, Thirumalai D, Woodson S A. J Mol Biol. 2001;306:1157–1166. doi: 10.1006/jmbi.2001.4437. [DOI] [PubMed] [Google Scholar]
- 34.Heilman-Miller S L, Pan J, Thirumalai D, Woodson S A. J Mol Biol. 2001;309:57–68. doi: 10.1006/jmbi.2001.4660. [DOI] [PubMed] [Google Scholar]
- 35.Glatter O, Kratky O. Small Angle X-ray Scattering. London: Academic; 1982. [DOI] [PubMed] [Google Scholar]
- 36.Lehnert V, Jaeger L, Michel F, Westhof E. Chem Biol. 1996;3:993–1009. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
- 37.Pan J, Deras M L, Woodson S A. J Mol Biol. 2000;296:133–144. doi: 10.1006/jmbi.1999.3439. [DOI] [PubMed] [Google Scholar]
- 38.Thirumalai D, Woodson S A. RNA. 2000;6:790–794. doi: 10.1017/s1355838200000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan J, Woodson S A. J Mol Biol. 1999;294:955–965. doi: 10.1006/jmbi.1999.3298. [DOI] [PubMed] [Google Scholar]
- 40.Pan T, Fang X, Sosnick T. J Mol Biol. 1999;286:721–731. doi: 10.1006/jmbi.1998.2516. [DOI] [PubMed] [Google Scholar]
- 41.Thirumalai D, Woodson S A. Acc Chem Res. 1996;29:433–439. [Google Scholar]
- 42.Pan J, Thirumalai D, Woodson S A. J Mol Biol. 1997;273:7–13. doi: 10.1006/jmbi.1997.1311. [DOI] [PubMed] [Google Scholar]
- 43.Weidner H, Crothers D M. Nucleic Acids Res. 1977;4:3401–3414. doi: 10.1093/nar/4.10.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walstrum S A, Uhlenbeck O C. Biochemistry. 1990;29:10573–10576. doi: 10.1021/bi00498a022. [DOI] [PubMed] [Google Scholar]
- 45.Herschlag D, Cech T R. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- 46.Uhlenbeck O C. RNA. 1995;1:4–6. [PMC free article] [PubMed] [Google Scholar]
- 47.Brockwell D J, Smith D A, Radford S E. Curr Opin Struct Biol. 2000;10:16–25. doi: 10.1016/s0959-440x(99)00043-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.