Abstract
Dynamin, the product of the shibire gene of Drosophila, is a GTPase critically required for endocytosis. Some studies have suggested a functional link between dynamin and the actin cytoskeleton. This link is of special interest, because there is evidence implicating actin dynamics in endocytosis. Here we show that endogenous dynamin 2, as well as green fluorescence protein fusion proteins of both dynamin 1 and 2, is present in actin comets generated by Listeria or by type I PIP kinase (PIPK) overexpression. In PIPK-induced tails, dynamin is further enriched at the interface between the tails and the moving organelles. Dynamin mutants harboring mutations in the GTPase domain inhibited nucleation of actin tails induced by PIPK and moderately reduced their speed. Although dynamin localization to the tails required its proline-rich domain, expression of a dynamin mutant lacking this domain also diminished tail formation. In addition, this mutant disrupted a membrane-associated actin scaffold (podosome rosette) previously shown to include dynamin. These findings suggest that dynamin is part of a protein network that controls nucleation of actin from membranes. At endocytic sites, dynamin may couple the fission reaction to the polymerization of an actin pool that functions in the separation of the endocytic vesicles from the plasma membrane.
Dynamin is a GTPase critically required for endocytosis (reviewed in refs. 1–3). In Drosophila, temperature-sensitive mutations of the shibire gene, which encodes dynamin, produce a temperature-sensitive paralysis resulting from a block of the internalization of synaptic vesicle membranes (4). Similarly, overexpression of dominant negative mutants of dynamin in mammalian cells inhibits several forms of endocytosis (5–9). Ultrastructural studies have shown that dynamin assembles into rings at the neck of deeply invaginated endocytic pits (4, 10, 11). These rings are thought to play an important role in fission, although their precise mechanism of action remains unclear (3). They may mediate a constriction of the bud neck and then its scission via a GTP-hydrolysis-dependent conformational change (10–12). Alternatively, they may act indirectly by recruiting and/or activating other proteins that function as the final effectors in fission (13).
Dynamin comprises several distinct modules: an NH2-terminal GTPase domain, a pleckstrin homology (PH) domain, a predicted coiled-coil domain [GTPase effector domain (GED)] that may function as a GTPase activating protein for the GTPase domain, and a COOH-terminal proline-rich domain (PRD). In the cytoplasm, dynamin shuttles between a cytosolic state and a membrane-bound state. Binding to the membrane is mediated by its PH domain, which binds PI(4,5)P2 (14, 15), and by a region upstream of this domain that can directly penetrate into the lipid bilayer (16). In the assembled dynamin ring, the GED domain is thought to form an intermolecular contact with the GTPase domain of adjacent dynamin molecules, thus providing an explanation for the stimulation of GTPase activity produced by dynamin self-assembly (17). The PRD represents a site of interaction with a variety of SH3-containing proteins (18) and was proposed to be responsible for the recruitment of dynamin to sites of endocytosis (19–21). PRDs of dynamin oligomers may be engaged in multiple interactions with different SH3-containing proteins, thus mediating the formation of a protein network. In such a way, PRDs not only may participate in dynamin recruitment to endocytic sites but also may contribute to the recruitment of other cytoplasmic proteins or protein scaffolds to assembled dynamin oligomers.
Early studies of Drosophila shibire mutants revealed defects in growth cone motility at the restrictive temperature (22). A similar effect was produced by the disruption of dynamin function in vitro by using antisense oligonucleotides (23). These results had raised the possibility that dynamin could play a role in actin dynamics in addition to its role in endocytosis. Recently, this hypothesis received further support from the observation of direct and indirect interactions of dynamin with actin regulatory proteins and from the localization of dynamin at actin-rich sites at cell periphery. Affinity chromatography on profilin, a protein that facilitates actin polymerization (24), identified dynamin as a main binding protein (25). In addition, some of the major SH3 domain interactors of dynamin's PRD are members of evolutionary conserved protein families implicated in actin function. These include syndapin/pacsin, amphiphysin/Rvs167, intersectin/DAP160, cortactin, Abp1, and Grb2 (reviewed in refs. 26 and 27). Via proteins like Grb2, syndapin, and intersectin, dynamin can associate with the neural Wiskott–Aldrich syndrome protein (N-WASP) (28–30), a major regulator of actin polymerization (31, 32). Furthermore, via the dbl domain of intersectin, dynamin is linked to activation of cdc42 (30), a rho family small GTPase that cooperates with N-WASP in actin nucleation (reviewed in ref. 33). Dynamin colocalizes with actin at peripheral ruffles in fibroblasts (34) and at adhesion sites (podosome rosettes) between cells and the substrate in baby hamster kidney cell line (BHK21) cells transformed with Rous sarcoma virus (BHK/RSV) (35). In macrophages, dynamin is present at phagocytic cups, and a mutant form of dynamin (K44A) blocks phagocytosis (9).
A connection of dynamin to actin function is of significant interest because genetic studies in yeast have shown an important link between endocytosis and the actin cytoskeleton (36, 37). In addition, morphological and physiological studies have implicated actin in various forms of endocytosis, including receptor-mediated endocytosis (reviewed in ref. 26). In nerve terminals, where a dynamin-dependent endocytic reaction plays a critical role in the recycling of synaptic vesicles, endocytic sites are enriched in actin (38, 39). Finally, actin comets originating form endocytic vesicles have been reported (40, 41).
The goal of this study was to explore further a functional relationship between dynamin and actin. To this aim, we have focused on actin comets, a powerful model system for the study of regulatory mechanisms in actin nucleation in vivo (41–43). For our analysis, we have used prominent actin tails induced by infection with Listeria or by overexpression of type I PIP kinase (PIPK), a PI(4,5)P2 generating enzyme (43, 44).
Materials and Methods
DNA Constructs.
Dynamin 2-aa green fluorescence protein (GFP) (a kind gift from M. McNiven) and K44A dynamin 2-GFP (in pEGFP-N1 vector, CLONTECH; EGFP, enhanced GFP) were previously described (35). Dynamin 1-aa GFP in the same vector was a kind gift from V. Slepnev. Other dynamin 2-aa mutant constructs were generated from dynamin 2-aa GFP by using the quickchange kit from Stratagene and Pwo Polymerase (Roche). A mammalian expression vector of type I PIPK was generated by placing hemagglutinin (HA)-tagged type I PIPK γ isoform (PIPKIγ) from pBluescript-HA-PIPKIγ (a kind gift from Y. Oka) into pCDNA3 (Invitrogen). The dynamin 2-aa PRD-GFP construct was generated by inserting amino acids 735–869 of dynamin 2-aa fused to GFP into pCDNA3. The dynamin 2-aa ΔPRD-GFP construct was generated by PCR amplification of the dynamin 2 region corresponding to amino acids 1–745 (34) and insertion of this fragment into the pEGFP-N1 vector.
Transfection and Listeria Infection.
HeLa cells were transfected with wild-type and mutant dynamin–GFP constructs by Lipofectamine 2000 (Life Technology, Gaithersburg, MD). After 12–16 h, they were infected with Listeria monocytogenes as described (42) and observed after 10 h. A wild-type (kindly provided by G. Galan) and a mutant strain of Listeria that is deficient in d-alanine synthesis was used (a generous gift from D. Portnoy) (45). In the latter case, the culture medium was supplemented with 100 μg/ml of d-alanine to allow normal actin-based motility of the bacteria, as described (45).
PIPK transfection of several cell types revealed that BHK/RSV (a kind gift of P. C. Marchisio) (35) generated the best actin tails. These cells were cotransfected with PIPK and dynamin–GFP constructs. Wild-type and mutant dynamins were transfected in parallel by using the same concentration of DNA, and cells with comparable degree of fluorescence were analyzed for measurements of tail speed. Cells were typically observed 10–15 h after transfection to minimize cell damage induced by expression of PIPK or mutant dynamin proteins.
Cell Labeling and Immunostaining.
Cells were fixed and processed for immunocytochemistry as described (35). Actin was stained with tetramethylrhodamine B isothiocyanate (TRITC)-phalloidin (Sigma) after paraformaldehyde fixation. For immunostaining of endogenous dynamin 2 in Listeria tails, rat epithelial PtK2 cells were used instead of HeLa cells, because our anti-rat dynamin 2 antibodies did not detect endogenous human dynamin 2. Anti-dynamin 2 antibody (a kind gift from G.-C. Ochoa) was generated in rabbits by injecting amino acids 761–787 (SHSPTPQRRPVSSVHPPGRPPAVRPHT) of rat dynamin 2, i.e., the same immunogen used to generate the so-called dyn2 antibodies (35, 46). Affinity purification was performed on the same peptide coupled to SulfoLink Coupling Gel (Bio-Rad) according to the manufacturer's instruction. Primary antibodies were revealed with Oregon green-conjugated goat anti-rabbit IgGs (Molecular Probes).
To label endocytic vesicles, cells were incubated with 1 mg/ml of TRITC-dextran (Sigma) for 30 min and washed twice before fixation. For Golgi staining, Bodipy Texas red ceramide (Molecular Probes) was used, following the manufacturer's instruction. For transferrin uptake, cells were incubated with 20 μg/ml of Cy3-transferrin for 10 min at 37°C before fixation. Specimens were observed by confocal microscopy.
The number of actin tails was counted from phalloidin staining in fixed GFP-positive cells. The tail number in cells expressing GFP fusions of mutant dynamins is given as percentage of tails observed in cells expressing wild-type dynamin 2–GFP. On the basis of counterstaining for the HA tag of PIPK, more than 90% of GFP fluorescent cells coexpressed PIPK.
Microscopy of Living Cells.
All experiments were performed at room temperature unless indicated otherwise, and movies (see Movies 1 and 2, which are published as supporting information on the PNAS web site, www.pnas.org) were generated from 5- to 10-min observations. For Listeria motility, live cells were observed with a Bio-Rad MRC 1024 confocal microscope by using a 63× water immersion lens (N.A. 1.25, Zeiss). Movies were imported into NIH image 1.62 (developed at the National Institutes of Health and available on the internet at http://rsb.info.nih.gov/nih-image/). Movies of PIPK-induced vesicles were generated by capturing images with a charge-coupled device camera (Micromax 1300 YHS, Roper Scientific). Organelle speed was measured by calculating the distance traveled in consecutive frames using metamorph software (Universal Imaging).
Results
Dynamin Is Present in Listeria Tails.
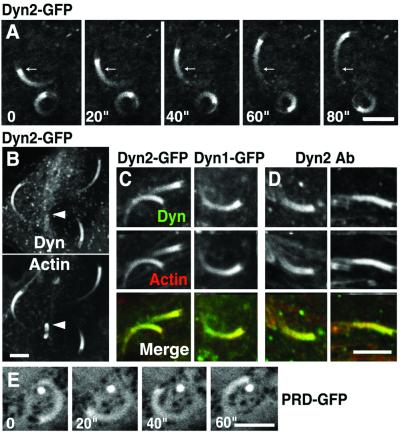
When HeLa cells were transfected with dynamin 1- or dynamin 2-GFP and then infected with L. monocytogenes, strong GFP fluorescence was observed at the tails of moving bacteria [Fig. 1A, supporting information on the PNAS web site (www.pnas.org), and data not shown]. As revealed by counterstaining with phalloidin, the GFP fluorescence pattern had a distribution very similar to that of filamentous actin (Fig. 1 B and C). Immunostaining of Listeria-infected PtK2 cells with anti-rat dynamin 2 antibodies showed a similar localization of endogenous dynamin 2 in actin tails (Fig. 1D). Interestingly, dynamin staining was barely detectable on nonmoving bacteria, despite the presence of a shell of phalloidin staining on their surface (arrowheads in Fig. 1B). It was previously shown that some components of the actin polymerization machinery, such as profilin, are present in Listeria-associated actin only when the bacterium moves within the cell (47). Thus, the absence of dynamin from nonmoving bacteria indicates this protein is selectively associated with a dynamic actin pool. A GFP-fusion construct of the PRD of dynamin 2 (PRD-GFP) was also found at Listeria tails, although it was less efficiently concentrated in these structures than the full length protein (Fig. 1E).
Figure 1.
Localization of full length dynamin and dynamin-PRD at actin tails of Listeria. (A) HeLa cells expressing dynamin 2-GFP were infected with Listeria and then observed by confocal microscopy at 20-sec time intervals. Arrows mark the same position in each frame to demonstrate the movement of a bacterium. (B) HeLa cells processed as in A were fixed and counterstained for F-actin with phalloidin. Note the lack of dynamin staining on nonmoving Listeria, which are surrounded only by an actin shell (arrowheads). (C) Both dynamin 1-GFP and dynamin 2-GFP (green) colocalize with phalloidin staining (red) on Listeria tails in HeLa cells. (D) PtK2 cells were infected with Listeria and then processed by dual labeling for endogenous dynamin 2 immunoreactivity (green) and for actin with phalloidin staining (red). (E) HeLa cells transfected with PRD-GFP were observed by confocal microscopy at 20-sec time intervals. (Bars = 5 μm for all fields.)
Dynamin Is Present at Tails Induced by Overexpression of PIPK.
Actin tails are likely to play an important role in the motility of endogenous cell organelles (40, 41). However, tails easily detectable by light microscopy are seldom observed in normal cells. They can be induced by several treatments besides pathogen infection, such as overexpression of type I PIPK (43), which stimulates PI(4,5)P2 production, or expression of the active form of arf6 (48), which may act, at least partially, via the recruitment of type I PIPK (49). PI(4,5)P2, in turn, may act as a cofactor in the recruitment and/or activation of actin regulatory proteins (50). To determine whether dynamin is also recruited to the tails of these endogenous organelles, targeting of dynamin 1- and dynamin 2-GFP was investigated in cells transiently cotransfected with the HA-tagged γ isoform of type I PIPK (PIPKIγ) (44, 51).
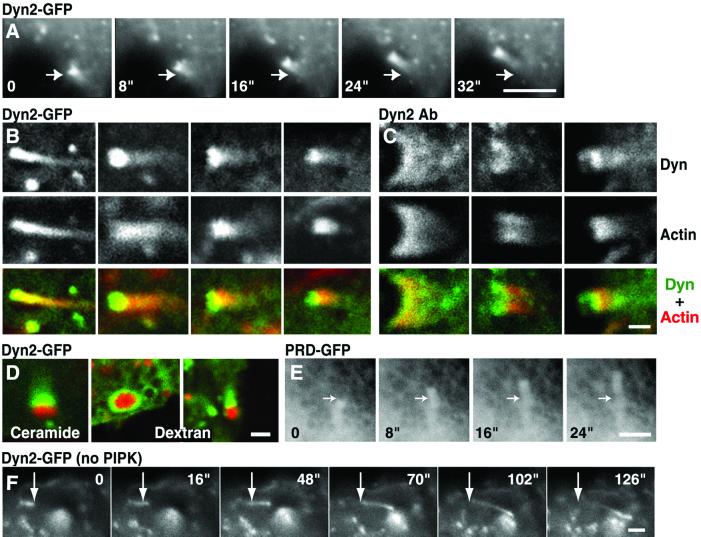
As previously shown in cells overexpressing α and β isoforms of type I PIPK, many cells overexpressing PIPKIγ contained numerous actin tails (43) (Fig. 2 A–D and supporting information on the PNAS web site). At least some of them originated from vesicles of the endocytic pathway, because after a 30-min incubation with TRITC–dextran (a fluid phase endocytic marker), 33% (15/45) of the comets originated from dextran-containing vesicles. Some of the other tails may originate from Golgi-derived vesicles, because after 1-h labeling with Bodipy Texas red-ceramide, 13% (10/76) of them had a Texas red-positive organelle at their tips. Bodipy Texas red–ceramide is a membrane-permeable fluorescent precursor of sphingolipids that becomes incorporated into Golgi-derived vesicles destined to reach the plasma membrane via the secretory pathway (52). Dynamin 1-GFP (data not shown), dynamin 2-GFP (Fig. 2 A, B, and D), and endogenous dynamin 2 (Fig. 2C) were concentrated at these comets, irrespective of the cargo present at their tips (Fig. 2D). Both endogenous and full length dynamin-GFP were further enriched in close proximity of the moving organelle relative to phalloidin staining (Fig. 2 B and C). In contrast, PRD-GFP was present at low concentration throughout the tail (Fig. 2E).
Figure 2.
Presence of dynamin in actin tails of BHK/RSV cells. A–E are from cells cotransfected with dynamin constructs and with PIPK; F is from a cell not cotransfected with the kinase. (A) A series of images from a time-lapse movie of a cell cotransfected with dynamin-GFP and HA-PIPK (8-sec time intervals). Arrows point to the same position in each frame to demonstrate the movement of a vesicle. (B and C) Dynamin 2-GFP (B) and endogenous dynamin 2 immunoreactivity (C) (green) are compared with phalloidin stain (red) in tails induced by PIPK. Note the highest concentration of dynamin relative to actin at the heads of the comets. (D) Cells cotransfected with dynamin 2-GFP (green) and PIPK were imaged by confocal microscopy after a pulse–chase labeling with Bodipy Texas red-ceramide (red) or an incubation with TRITC-dextran (red) to label Golgi-derived vesicles and endocytic vesicles, respectively. (E) A series of images from a time-lapse movie of a cell cotransfected with PRD-GFP and HA-PIPK (8-sec time intervals). The PRD of dynamin 2 is present at low concentration throughout actin tails. The arrow indicates the same position in all frames. (F) Images from a time-lapse movie of a cell from a separate culture transfected with dynamin-GFP but not with PIPK (8-sec time intervals). A small tail-like structure moves across the field. The arrow indicates the same position in all frames. (Bars = 5 μm in A and D; 1 μm in B, C, E, and F.)
Although well recognizable tails were visible only in PIPK overexpressing cells, high-power observation often revealed very small tail-like structures positive for dynamin 2-GFP even in cells that are not cotransfected with this lipid kinase (Fig. 2F). The speed of these structures, which were typically localized in the cortical region of the cell, was about half compared with tails induced by PIPK (2.6 μm/min versus 4.9 μm/min). Whether they represent bona fide tails of endogenous organelles deserves further investigation.
Mutant Dynamins Inhibit Tail Formation and Reduce Tail Speed.
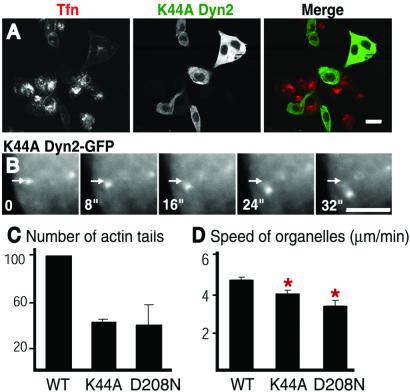
Next, we investigated whether the GTPase cycle of transfected dynamin affects the formation of actin tails or their dynamics. Several dynamin mutants containing amino acid changes in the GTPase domain have a powerful dominant negative effect on endocytosis (5–7). Two such mutants of dynamin 2-GFP were tested: the K44A mutation, which results in lower affinity for guanylnucleotides, and the D208N mutation, which is predicted to have reduced affinity for GTP (5, 6). Both mutants inhibited transferrin uptake as expected (Fig. 3A and data not shown). Furthermore, both mutants greatly reduced, by 59 and 61% respectively, the number of tails induced by PIPK overexpression (Fig. 3C). They also reduced the number of Listeria tails (data not shown), although this effect could have resulted, at least partially, from an inhibitory effect of the mutant dynamins on bacteria infection. Both K44Adynamin 2-GFP and D208Ndynamin 2-GFP, nonetheless, were associated with Listeria and PIPK-induced tails in the corresponding cells (Fig. 3B and data not shown). The speed of these tails was moderately reduced but in a statistically significant way, relative to the tails of cells transfected with wild-type dynamin2-GFP (Fig. 3D). Collectively, these findings suggest that dynamin, via its GTPase domain, may contribute to the nucleation of actin tails at specific intracellular sites.
Figure 3.
Effect of GTPase domain dynamin mutants on endocytosis and actin comets. (A) K44A dynamin 2-GFP inhibits transferrin uptake. K44Adynamin 2-GFP transfected cells were incubated with Cy3-transferrin for 10 min before fixation. Note the strong inhibition of transferrin uptake (red) in cells that express K44Adynamin (green). (B) K44Adynamin 2-GFP is present at actin tails of cells cotransfected with PIPK and does not block tail motility. A series of images from a time-lapse movie (8-sec time intervals). Arrows mark the same position in each frame to demonstrate vesicle movement. (Bars = 20 μm in A and 5 μm in B.) (C) K44Adynamin 2-GFP and D208Ndynamin 2-GFP inhibit actin tail formation in PIPK-overexpressing cells. The data represent the number of tails per transfected (or GFP-fluorescent) cell expressed as percentage of the value observed with wild-type dynamin. The average and standard deviation of at least three separate experiments (total cells counted for each condition >180) are shown. Actin tails were counted in fixed cells after phalloidin stain. Cotransfection efficiency, as accessed by anti-HA staining for PIPK expression, was more than 90%. (D) K44Adynamin 2-GFP and D208Ndynamin 2-GFP slightly reduce motility of tails induced by PIPK cotransfection. The average tail speed in these cells was 4.9 μm/min for wild-type dynamin 2 (n = 120), 4.2 μm/min for K44Adynamin 2 (n = 124), and 3.5 μm/min (n = 49) for D208Ndynamin 2. Standard errors are indicated. The difference is statistically significant: P < 0.001 for K44Adynamin 2 and P < 0.000002 for D208Ndynamin 2.
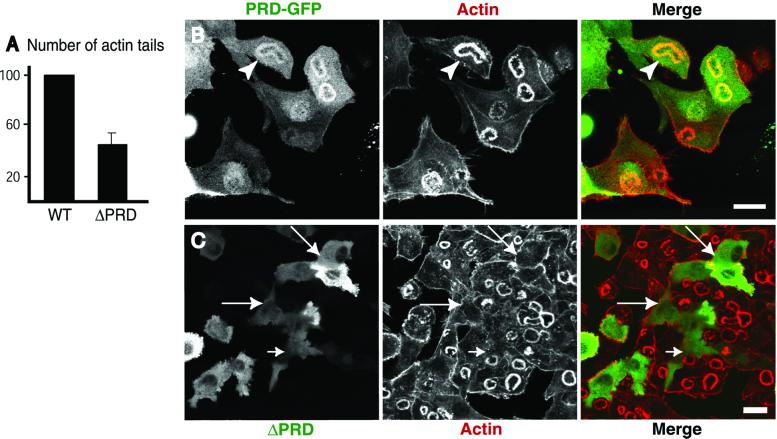
The presence of both wild-type and GTPase domain mutants of dynamin in actin tails may reflect interactions of the PRD because, when expressed alone, this domain is enriched in tails (see above). Dynamin interacting proteins, such as profilin, cortactin, and Abp1 (25, 34, 53), may be responsible for this localization of the PRD, because these dynamin interactors were recently described in actin tails (refs. 24, 48, and 54, and M. Kessels, personal communication). To elucidate further the role of this domain in the targeting of dynamin, we also expressed a dynamin 2-GFP mutant in which the entire PRD (the last 124 amino acids) had been deleted (ΔPRD dynamin 2-GFP). In contrast to dynamin GTPase domain mutants, ΔPRD dynamin 2-GFP was not detectable in the tails (data not shown), confirming a critical role of the PRD in targeting full length dynamin 2 to actin tails. Surprisingly, this fusion protein as well, which potently blocks endocytosis (data not shown), inhibited tail formation. The number of tails was reduced by 55% compared with the cells expressing wild-type dynamin 2 (Fig. 4A).
Figure 4.
ΔPRD dynamin 2-GFP disrupts podosome rosettes of BHK/RSV cells. (A) Expression of ΔPRD dynamin 2-GFP (ΔPRD) in cells cotransfected with PIPK inhibits actin tail formation. Actin tails were counted in phalloidin-stained fixed cells and their number was calculated as in Fig. 3C. Values represent average and standard deviations derived from three separate experiments in which more than 250 cells were counted. (B) The PRD of dynamin is targeted to the podosome rosettes (arrowheads). BHK/RSV cells were transfected with a GFP fusion protein of the PRD of dynamin 2 (green) and stained with phalloidin (red). (C) Expression of ΔPRD dynamin 2-GFP (green) in BHK/RSV cells (arrows) results in loss of the podosome rosette previously shown to be positive for endogenous dynamin (35). (Bar = 20 μm.)
An effect of ΔPRD dynamin 2-GFP was also observed on other actin-containing structures. We have recently reported a colocalization of dynamin with actin at podosome rosettes of RSV transformed BHK21 cells (arrowheads in Fig. 4B) (35). These rosettes represent sites of contact between cells and the substratum, where actin forms columnar arrays that are perpendicular to the plasma membrane and often contain a tubular invagination of this membrane (35). The PRD of dynamin plays an important role in targeting of dynamin to these structures, because a GFP fusion protein of the PRD colocalized with actin at podosome rosettes (Fig. 4B). Surprisingly, expression of ΔPRD dynamin 2-GFP almost completely disrupted the rosettes (in 146 of 152 cells counted, or 96%; see also Fig. 4C). Collectively, these results imply that ΔPRD dynamin 2 may sequester a critical factor for actin nucleation from membranes or for continued actin polymerization. Thus, an interplay the PRD and the other portions of dynamin is needed for the physiological function of this GTPase.
Discussion
The results reported in this study provide strong in vivo evidence for a functional link of dynamin to the actin cytoskeleton. They are convergent with growing evidence for a role of actin in endocytosis and suggest that dynamin itself has a function in the regulation of the actin cytoskeleton at endocytic sites. An attractive possibility is that dynamin may act at multiple steps in vesicle budding and may first help in generating and constricting the vesicle neck (4, 10–12). As the endocytic reaction proceeds to fission, oligomerized dynamin may also help to recruit an actin nucleating machinery via proteins that bind its PRD. After fission, this actin meshwork may help propelling the vesicle away from the endocytic site, either by comets or other actin-based transport mechanisms. Furthermore, a role of actin itself in fission cannot be ruled out.
Recent studies have revealed a central role of the Arp2/3 complex in the formation of crosslinked actin meshwork, such as those found in actin tails of Listeria and intracellular organelles (43, 55). The upstream regulators of the Arp2/3 complex, however, may vary at distinct sites of actin nucleation (56). We speculate that dynamin may be part of this regulatory machinery at sites of endocytosis. Its regulatory action may be achieved through an indirect interaction with the neural Wiskott–Aldrich syndrome protein and the Arp2/3 complex, for example via Grb2 or via SH3 domain-containing protein scaffolds, including syndapin/pacsin, Abp1, intersectin, and cortactin among other proteins (18, 26, 27, 30, 34, 53). Dynamin may also play a regulatory role in actin nucleation at nonendocytic sites, as suggested by the presence of this GTPase at attachment sites between cells and the substratum (35) and at Listeria tails.
On the basis of the modular structure of dynamin, we suggest the following model. The PRD of dynamin is a critical determinant for the interaction of this protein with the actin cytoskeleton, as revealed by its targeting to actin-rich sites when separated from the remaining portion of the protein. The PH domain, which binds PI(4,5)P2 (14, 15), and an amino acid stretch upstream of PH domain, which can insert into the lipid bilayer (16), help the protein to anchor at the membrane. Via this dual interaction with the membrane and the cytoskeleton, dynamin may facilitate nucleation of actin at specific membrane sites, such as the necks of coated pits. The GTPase domain, through yet unknown mechanisms, may play a regulatory role on this property of dynamin to connect the membrane to the actin cytoskeleton and on the intracellular sites where this adaptor function must be achieved. Intra- and intermolecular interactions within dynamin oligomers or interactions of the GTPase domain with other proteins may mediate this regulation. The importance of the GTPase domain is indicated by the inhibitory role on the formation of actin tails by expression of dynamins containing mutations in such domain.
Our results indicate that dynamin GTPase domain mutants inhibit primarily tail nucleation, with a moderate effect on tail motility. Thus, once tails have been formed, the function of dynamin GTPase in actin assembly becomes less critical. This finding is not surprising if one considers that dynamic actin tails have been reconstituted in vitro with purified components that do not include dynamin.
In conclusion, on the basis of the findings reported in this study, we propose that the link to the actin cytoskeleton may represent a critical aspect of dynamin function in endocytosis. It is also likely that dynamin plays a more general role in actin–membrane interactions.
Supplementary Material
Acknowledgments
We thank Dr. Vladimir Slepnev and Dr. Gian-Carlo Ochoa (our laboratory), Dr. George Galan (Yale University), Dr. Mark McNiven (Mayo Clinic), Dr. Yoshitomo Oka (Yamaguchi University, Yamaguchi, Japan), Dr. Pier Carlo Marchisio (San Raffaele Scientific Institute, Milan) and Dr. Daniel Portnoy (University of California, Berkeley) for the generous gift of reagents. We also thank Drs. Michele Solimena, Gianluca Cestra, and Gilbert Di Paolo for critical reading of the manuscript. E.L. is a postdoctoral fellow of American Cancer Society. This work was supported by grants from the National Institutes of Health (NS36251 and CA46128) and by a grant from the U. S. Army Medical Research and Development Command (to P.D.C.).
Abbreviations
- PRD
proline-rich domain
- PIPKIγ
type I phosphoinositide-4-phosphate kinase γ isoform
- GFP
green fluorescence protein
- PH
pleckstrin homology
- BHK/RSV
baby hamster kidney cell line (BHK21) transformed with Rous sarcoma virus
- TRITC
tetramethylrhodamine B isothiocyanate
- HA
hemagglutinin
References
- 1.De Camilli P, Takei K. Neuron. 1996;16:481–486. doi: 10.1016/s0896-6273(00)80068-9. [DOI] [PubMed] [Google Scholar]
- 2.McNiven M A, Cao H, Pitts K R, Yoon Y. Trends Biochem Sci. 2000;25:115–120. doi: 10.1016/s0968-0004(99)01538-8. [DOI] [PubMed] [Google Scholar]
- 3.Sever S, Damke H, Schmid S L. Traffic. 2000;1:385–392. doi: 10.1034/j.1600-0854.2000.010503.x. [DOI] [PubMed] [Google Scholar]
- 4.Kosaka T, Ikeda K. J Neurobiol. 1983;14:207–225. doi: 10.1002/neu.480140305. [DOI] [PubMed] [Google Scholar]
- 5.Herskovits J S, Burgess C C, Obar R A, Vallee R B. J Cell Biol. 1993;122:565–578. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Bliek A M, Redelmeier T E, Damke H, Tisdale E J, Meyerowitz E M, Schmid S L. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damke H, Baba T, Warnock D E, Schmid S L. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oh P, McIntosh D P, Schnitzer J E. J Cell Biol. 1998;141:101–114. doi: 10.1083/jcb.141.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gold E S, Underhill D M, Morrissette N S, Guo J, McNiven M A, Aderem A. J Exp Med. 1999;190:1849–1856. doi: 10.1084/jem.190.12.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinshaw J E, Schmid S L. Nature (London) 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- 11.Takei K, McPherson P S, Schmid S L, De Camilli P. Nature (London) 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- 12.Marks B, Stowell M H B, Vallis Y, Mills I G, Gibson A, Hopkins C R, McMahon H T. Nature (London) 2001;410:231–235. doi: 10.1038/35065645. [DOI] [PubMed] [Google Scholar]
- 13.Sever S, Muhlberg A B, Schmid S L. Nature (London) 1999;398:481–486. doi: 10.1038/19024. [DOI] [PubMed] [Google Scholar]
- 14.Lin H C, Barylko B, Achiriloaie M, Albanesi J P. J Biol Chem. 1997;272:25999–26004. doi: 10.1074/jbc.272.41.25999. [DOI] [PubMed] [Google Scholar]
- 15.Klein D E, Lee A, Frank D W, Marks M S, Lemmon M A. J Biol Chem. 1998;273:27725–27733. doi: 10.1074/jbc.273.42.27725. [DOI] [PubMed] [Google Scholar]
- 16.Burger K N, Demel R A, Schmid S L, de Kruijff B. Biochemistry. 2000;39:12485–12493. doi: 10.1021/bi000971r. [DOI] [PubMed] [Google Scholar]
- 17.Muhlberg A B, Warnock D E, Schmid S L. EMBO J. 1997;16:6676–6683. doi: 10.1093/emboj/16.22.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gout I, Dhand R, Hiles I D, Fry M J, Panayotou G, Das P, Truong O, Totty N F, Hsuan J, Booker G W, et al. Cell. 1993;75:25–36. [PubMed] [Google Scholar]
- 19.David C, McPherson P S, Mundigl O, De Camilli P. Proc Natl Acad Sci USA. 1996;93:331–335. doi: 10.1073/pnas.93.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wigge P, McMahon H T. Trends Neurosci. 1998;21:339–344. doi: 10.1016/s0166-2236(98)01264-8. [DOI] [PubMed] [Google Scholar]
- 21.Shupliakov O, Low P, Grabs D, Gad H, Chen H, David C, Takei K, De Camilli P, Brodin L. Science. 1997;276:259–263. doi: 10.1126/science.276.5310.259. [DOI] [PubMed] [Google Scholar]
- 22.Kim Y T, Wu C F. J Neurosci. 1987;7:3245–3255. doi: 10.1523/JNEUROSCI.07-10-03245.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torre E, McNiven M A, Urrutia R. J Biol Chem. 1994;269:32411–32417. [PubMed] [Google Scholar]
- 24.Theriot J A, Mitchison T J. Cell. 1993;75:835–838. doi: 10.1016/0092-8674(93)90527-w. [DOI] [PubMed] [Google Scholar]
- 25.Witke W, Podtelejnikov A V, Di Nardo A, Sutherland J D, Gurniak C B, Dotti C, Mann M. EMBO J. 1998;17:967–976. doi: 10.1093/emboj/17.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qualmann B, Kessels M M, Kelly R B. J Cell Biol. 2000;150:111–116. doi: 10.1083/jcb.150.5.f111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slepnev V, De Camilli P. Nat Neurosci Rev. 2000;1:161–172. doi: 10.1038/35044540. [DOI] [PubMed] [Google Scholar]
- 28.Qualmann B, Roos J, DiGregorio P J, Kelly R B. Mol Biol Cell. 1999;10:501–513. doi: 10.1091/mbc.10.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlier M F, Nioche P, Broutin-L'Hermite I, Boujemaa R, Le Clainche C, Egile C, Garbay C, Ducruix A, Sansonetti P, Pantaloni D. J Biol Chem. 2000;275:21946–21952. doi: 10.1074/jbc.M000687200. [DOI] [PubMed] [Google Scholar]
- 30.Hussain N K, Jenna S, Glogauer M, Quinn C C, Wasiak S, Guipponi M, Antonarakis S E, Kay B K, Stossel T P, Lamarche-Vane N, McPherson P S. Nat Cell Biol. 2001;3:927–932. doi: 10.1038/ncb1001-927. [DOI] [PubMed] [Google Scholar]
- 31.Miki H, Miura K, Takenawa T. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- 32.Machesky L M, Insall R H. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 33.Bishop A L, Hall A. Biochem J. 2000;348:241–255. [PMC free article] [PubMed] [Google Scholar]
- 34.McNiven M A, Kim L, Krueger E W, Orth J D, Cao H, Wong T W. J Cell Biol. 2000;151:187–198. doi: 10.1083/jcb.151.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochoa G-C, Slepnev V I, Neff L, Ringstad N, Takei K, Daniell L, Cao H, McNiven M, Baron R, De Camilli P. J Cell Biol. 2000;150:377–390. doi: 10.1083/jcb.150.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geli M I, Riezman H. J Cell Sci. 1998;111:1031–1037. doi: 10.1242/jcs.111.8.1031. [DOI] [PubMed] [Google Scholar]
- 37.Wendland B, Emr S D, Riezman H. Curr Opin Cell Biol. 1998;10:513–522. doi: 10.1016/s0955-0674(98)80067-7. [DOI] [PubMed] [Google Scholar]
- 38.Brodin L. Biochimie. 1999;81:49. [Google Scholar]
- 39.Dunaevsky A, Connor E A. J Neurosci. 2000;20:6007–6012. doi: 10.1523/JNEUROSCI.20-16-06007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merrifield C J, Moss S E, Ballestrem C, Imhof B A, Giese G, Wunderlich I, Almers W. Nat Cell Biol. 1999;1:72–74. doi: 10.1038/9048. [DOI] [PubMed] [Google Scholar]
- 41.Taunton J, Rowning B A, Coughlin M L, Wu M, Moon R T, Mitchison T J, Larabell C A. J Cell Biol. 2000;148:519–530. doi: 10.1083/jcb.148.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Theriot J A, Mitchison T J, Tilney L G, Portnoy D A. Nature (London) 1992;357:257–260. doi: 10.1038/357257a0. [DOI] [PubMed] [Google Scholar]
- 43.Rozelle A L, Machesky L M, Yamamoto M, Driessens M H, Insall R H, Roth M G, Luby-Phelps K, Marriott G, Hall A, Yin H L. Curr Biol. 2000;10:311–320. doi: 10.1016/s0960-9822(00)00384-5. [DOI] [PubMed] [Google Scholar]
- 44.Ishihara H, Shibasaki Y, Kizuki N, Wada T, Yazaki Y, Asano T, Oka Y. J Biol Chem. 1998;273:8741–8748. doi: 10.1074/jbc.273.15.8741. [DOI] [PubMed] [Google Scholar]
- 45.Thompson R J, Bouwer H G, Portnoy D A, Frankel F R. Infect Immun. 1998;66:3552–3561. doi: 10.1128/iai.66.8.3552-3561.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henley J R, Krueger E W, Oswald B J, McNiven M A. J Cell Biol. 1998;141:85–99. doi: 10.1083/jcb.141.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geese M, Schulter K, Rothkegel M, Jockusch B M, Wehland J, Sechi A S. J Cell Sci. 2000;113:1415–1422. doi: 10.1242/jcs.113.8.1415. [DOI] [PubMed] [Google Scholar]
- 48.Schafer D A, D'Souza-Schorey C, Cooper J A. Traffic. 2001;1:892–903. doi: 10.1034/j.1600-0854.2000.011108.x. [DOI] [PubMed] [Google Scholar]
- 49.Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris A J, et al. Cell. 1999;99:521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- 50.Sechi A S, Wehland J. J Cell Sci. 2000;113:3685–3695. doi: 10.1242/jcs.113.21.3685. [DOI] [PubMed] [Google Scholar]
- 51.Wenk M R, Pellegrini L, Klenchin V A, Di Paolo G, Chang S, Daniell L, Arioka M, Martin T F, De Camilli P. Neuron. 2001;32:79–88. doi: 10.1016/s0896-6273(01)00456-1. [DOI] [PubMed] [Google Scholar]
- 52.Ktistakis N T, Roth M G, Bloom G S. J Cell Biol. 1991;113:1009–1023. doi: 10.1083/jcb.113.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kessels M M, Engqvist-Goldstein A E, Drubin D G, Qualmann B. J Cell Biol. 2001;153:351–366. doi: 10.1083/jcb.153.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaksonen M, Peng H B, Rauvala H. J Cell Sci. 2000;113:4421–4426. doi: 10.1242/jcs.113.24.4421. [DOI] [PubMed] [Google Scholar]
- 55.May R C, Hall M E, Higgs H N, Pollard T D, Chakraborty T, Wehland J, Machesky L M, Sechi A S. Curr Biol. 1999;9:759–762. doi: 10.1016/s0960-9822(99)80337-6. [DOI] [PubMed] [Google Scholar]
- 56.Frischknecht F, Way M. Trends Cell Biol. 2001;11:30–38. doi: 10.1016/s0962-8924(00)01871-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.