Abstract
The basic leucine zipper transcription factor CCAAT/enhancer binding protein-β (C/EBPβ) is expressed in many cell types, including keratinocytes. C/EBPβ activity can be increased by phosphorylation through pathways stimulated by oncogenic Ras, although the biological implications of Ras-C/EBPβ signaling are not currently understood. We report here that C/EBPβ-nullizygous mice are completely refractory to skin tumor development induced by a variety of carcinogens and carcinogenesis protocols, including 7,12-dimethylbenz[a]anthracene-initiation/12-O-tetradecanoylphorbol 13-acetate promotion, that produce tumors containing oncogenic Ras mutations. No significant differences in TPA-induced epidermal keratinocyte proliferation were observed in C/EBPβ-null versus wild-type mice. However, apoptosis was significantly elevated (17-fold) in the epidermal keratinocytes of 7,12-dimethylbenz[a]anthracene-treated C/EBPβ-null mice compared with wild-type mice. In v-Ha-ras transgenic mice, C/EBPβ deficiency also led to greatly reduced skin tumor multiplicity and size, providing additional evidence for a tumorigenesis pathway linking Ras and C/EBPβ. Oncogenic Ras potently stimulated C/EBPβ to activate a C/EBP-responsive promoter-reporter in keratinocytes and mutating an ERK1/2 phosphorylation site (T188) in C/EBPβ abolished this Ras effect. Finally, we observed that C/EBPβ participates in oncogenic Ras-induced transformation of NIH 3T3 cells. These findings indicate that C/EBPβ has a critical role in Ras-mediated tumorigenesis and cell survival and implicate C/EBPβ as a target for tumor inhibition.
The Ras family of GTP binding proteins function as intracellular mediators of extracellular signals to regulate cell proliferation, apoptosis, survival, senescence, and differentiation (1–5). Ras protooncogenes are frequently mutated in tumors, and ≈25% of human cancers contain transforming mutations in ras. Therefore, understanding oncogenic Ras-signaling pathways is critical for elucidating the mechanisms that underlie cellular transformation and for designing effective therapeutic strategies to prevent the development or block the growth of many classes of tumors. Ras has numerous effectors, and its pathways are multifaceted (3, 6, 7). Ras activation by growth factors or oncogenic mutations elicits activation of several transcription factors, which in turn regulate the expression of genes that control the cellular responses to Ras signaling, including oncogenesis. The transcription factors Ets, c-jun, c-myc, and NF-κB are known to have roles in oncogenic ras-induced cellular transformation (8–11).
The basic leucine zipper (bZIP) transcription factor CCAAT/enhancer binding protein-β (C/EBPβ, also known as NF-IL6, IL-6DBP, LAP, CRP2, and NF-M) is expressed in a variety of cell types (12, 13) including keratinocytes (14, 15), where it plays a role in squamous differentiation (16). C/EBPβ is also involved in regulating differentiation of specific mesenchymal, epithelial, and hematopoietic cell types (17–21). C/EBPβ activity can be activated or derepressed by phosphorylation through pathways stimulated by oncogenic Ras in fibroblasts, erythoblasts, and P19 embryonal carcinoma cells (22, 23), suggesting a role for C/EBPβ as a nuclear effector of Ras signaling. However, the physiological functions of C/EBPβ as a downstrean target of Ras are unclear.
The fact that C/EBPβ is present in numerous epithelial and hematopoietic cells, and that some of these cell types give rise to human and rodent tumors containing mutant Ras (24, 25), prompted us to investigate whether C/EBPβ has a role in oncogenic Ras-mediated tumorigenesis and transformation. To address this question, we have examined C/EBPβ-nullizygous mice in the mouse skin model of multistage carcinogenesis. The mouse skin model is one of the best-defined in vivo paradigms of experimental epithelial carcinogenesis (26, 27), and there is ample evidence that the mutational activation of Ras plays a central role in skin tumor development induced by a variety of carcinogens (25, 27–29). For example, initiation with a single dose of the carcinogen 7,12-dimethylbenz[a]anthracene (DMBA), followed by repetitive treatment with the tumor promoter 12-O-tetradecanoylphorbol 13-acetate (TPA), results in the appearance of squamous papillomas, 95–100% of which contain an A → T182 mutation in Ha-ras (25, 28, 29). Furthermore, transgenic mice that express an oncogenic Ras transgene in their epidermis develop skin tumors, demonstrating a causal role for activated Ras in squamous papilloma development (30, 31).
Here we report that C/EBPβ-null mice are completely refractory to carcinogen-induced skin tumors involving mutant Ras and that v-Ha-ras transgenic mice that carry the C/EBPβ-null mutation also show a significant reduction in tumorigenesis. Apoptosis was significantly elevated in C/EBPβ-null mice in response to DMBA but not to ultraviolet-B (UVB) treatment. These findings reveal a previously uncharacterized role for C/EBPβ in tumorigenesis and cell survival.
Methods
Tumor Experiments in Wild-Type and C/EBPβ Mutant Mice.
The C/EBPβ-deficient mice used in our studies have been described (32). The mutant and wild-type mice were generated by mating heterozygous 129/Sv females to heterozygous males from the sixth- to eighth-generation backcross into the C57BL/6 strain. No significant sex difference in tumor response was observed.
Reporter Assays.
DNA fragments encoding mouse C/EBPβ [p35 and liver inhibitory protein (LIP) isoforms] were inserted into pcDNA3.1. The T188A mutation was generated by using the QuickChange system (Stratagene). BALB/MK2 keratinocytes (a gift from B. E. Weissman, Univ. North Carolina, Chapel Hill) at ≈25–30% confluence were transfected by using Lipofectin reagent (GIBCO/BRL) with pcDNA3–C/EBPβ (0.5 μg) and/or pcDNA3-Ha-ras(12V) (0.5 μg) (gift from C. J. Der, Univ. North Carolina) and 1.0 μg of the specified C/EBP-dependent promoter/reporter as described in the text. The total amount of DNA among all groups was kept constant by using empty vector. After 4 h, cultures were washed and grown in low-calcium Eagle's minimal essential medium containing 4 ng/ml epidermal growth factor, 0.05 mM calcium chloride, and 8% chelex treated FBS. Forty-eight hours later, cells were harvested and luciferase activity was determined (16). Primary keratinocytes were isolated from C/EBPβ−/− or C/EBPβ+/+ newborn littermates (2–3 days old) and cultured as described (14, 16). At ≈100% confluence, cells were transfected by using the PerFect Lipid, Pfx-3 (Invitrogen). Cells were processed for reporter assays as described above.
Detection of Apoptotic Cells.
Wild-type and C/EBPβ-null mice were treated with a single dose of 400 nmol DMBA, and 24 h later the treated dorsal skin was excised and fixed for 24 h in a 10% neutral buffered formalin, processed, and embedded in paraffin. Hematoxylin and eosin-stained sections (5 μm) were examined. Apoptotic keratinocytes in the stratum basale were scored by using the following criteria: dark pyknotic nuclei, cytoplasmic eosinophilia, and absence of cellular contacts.
NIH 3T3 Cell Focus Assay.
NIH 3T3 cells (gift from C. J. Der) were plated at 5 × 105 cells per 60-mm dish in DMEM containing 10% calf serum. One day later the cells were transfected by using a calcium phosphate precipitation method (33). Transformed foci were identified 14 days after transfection as defined by Clark et al. (33).
Western Analysis.
BALB/MK2 keratinocytes were transfected with pcDNA3-C/EBPβ (0.5 μg) and/or pcDNA3-Ha-ras(12V) (0.5 μg) as described above. Forty-eight hours later, lysates were prepared, equal amounts of each protein sample were loaded on 10% polyacrylamide Tris⋅glycine gels (Novex) and separated by electrophoresis, and Western analysis was conducted with a rabbit polyclonal IgG raised against C/EBPβ or Ras (1:2,000; Santa Cruz Biotechnology).
Tg.AC × C/EBPβ−/− Crosses.
Female Tg.AC mice (R. Cannon, National Institute on Environmental Health Sciences, RTP, NC) were crossed with C/EBPβ−/− mice, and F1 mice were backcrossed to C/EBPβ−/+ mice. Mice were genotyped by Southern analysis for v-Ha-ras and C/EBPβ. All v-Ha-ras-positive mice were also genotyped to verify that they were of the responder genotype (34).
Results
Effect of C/EBPβ Deficiency on Skin Tumor Development.
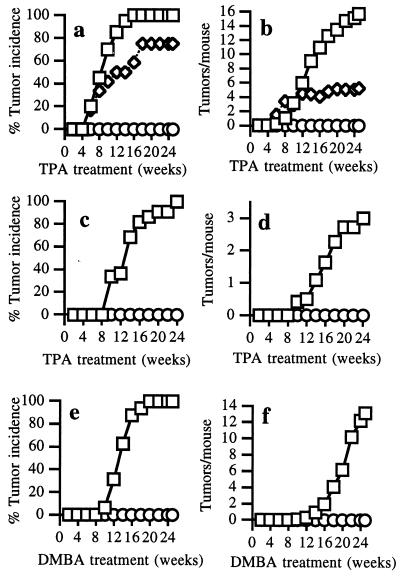
Initiation with a single dose of DMBA followed by TPA treatment produces squamous papillomas, 95–100% of which contain an A → T (182) mutation in Ha-ras (25, 28, 29). Therefore, C/EBPβ nullizygous and wild-type littermates were initiated with 200 nmol DMBA, and 1 week later these mice were promoted thrice weekly with 5 nmol TPA for 25 weeks. Wild-type mice developed an average of 15 squamous papillomas per mouse and exhibited a 100% incidence of papillomas (Fig. 1 a and b). In contrast, C/EBPβ nullizygous mice were completely refractory to papilloma development and no papillomas appeared after 25 weeks of promotion. In some groups of mutant mice, TPA promotion was continued for 35 weeks, but no tumors developed within this time (data not shown). C/EBPβ heterozygous mice express a level of C/EBPβ protein in keratinocytes that is intermediate between that of wild-type and C/EBPβ-deficient animals (14). C/EBPβ heterozygous mice were partially resistant to DMBA/TPA-induced carcinogenesis (Fig. 1 a and b), indicating that the tumor-modifying effect of C/EBPβ is gene-dosage dependent.
Figure 1.
C/EBPβ-null mice are completely refractory to carcinogen-induced skin tumorigenesis. C/EBPβ-null (○), wild-type (□), or heterozygous (◊) mice littermates (7–8 weeks old) were treated with a single application of 200 nmol DMBA followed 1 week later with thrice weekly treatment with 5 nmol of TPA (n = 20 C/EBPβ+/+, 21 C/EBPβ−/−, 12 C/EBPβ+/−) (a and b); a single application of 2.5 μmol MNNG followed 1 week later with thrice weekly application of 5 nmol (n = 22 C/EBPβ+/+, 19 C/EBPβ−/−) (c and d); or 100 nmol of DMBA once a week for 25 weeks (n = 16 C/EBPβ+/+, 18 C/EBPβ−/−) (e and f). All agents were applied in 200 μl of acetone, and all experiments were repeated twice with similar results.
Because C/EBPβ can be phosphorylated via a protein kinase C pathway (35), and because this event is required for TPA-induced mitogenesis in hepatocytes (36), we examined whether TPA-induced keratinocyte proliferation was altered in epidermis of C/EBPβ-null mice. No significant differences were observed between wild-type and C/EBPβ nullizygous mice after single or multiple treatment with TPA (Table 1). C/EBPβ has been implicated in the regulation of cyclooxygenase 2 (COX2; ref. 37) and tumor necrosis factor (TNF-α) (38) expression and both TNF-α-null (39) and COX2-null (40) mice are partially resistant to DMBA/TPA-induced carcinogenesis. However, TNF-α mRNA and COX2 protein expression were not different in untreated or TPA-treated C/EBPβ-deficient mice compared with similarly treated wild-type mice (data not shown). These results indicate that TNF-α and COX2 expression, as well as TPA-induced proliferative responses in the epidermis of C/EBPβ-null mice, are normal and thus are not responsible for the resistance of C/EBPβ-null mice to DMBA/TPA-induced tumorigenesis.
Table 1.
Effect of TPA treatment on epidermal cell proliferation in wild-type and C/EBPβ-null mice
| Treatment | Nucleated cell layers | BrdUrd-positive cells, % |
|---|---|---|
| Single | ||
| Acetone | ||
| Wild type | 1.3 ± 0.1 | 4.6 ± 1.3 |
| C/EBPβ−/− | 1.5 ± 0.1 | 7.4 ± 3.7 |
| TPA | ||
| Wild type | 1.8 ± 0.3 | 39.7 ± 8.5 |
| C/EBPβ−/− | 2.0 ± 0.9 | 43.9 ± 3.7 |
| Multiple | ||
| Acetone | ||
| Wild type | 1.3 ± 0.1 | 6.0 ± 1.2 |
| C/EBPβ−/− | 1.7 ± 0.2 | 10.9 ± 5.6 |
| TPA | ||
| Wild type | 3.8 ± 1.6 | 32.2 ± 7.7 |
| C/EBPβ−/− | 3.7 ± 0.6 | 30.6 ± 8.7 |
Mice were treated with a single application or thrice weekly for 1 month with 5 nmol TPA/200 μl acetone or with acetone alone. BrdUrd labeling was conducted by a single-dose i.p. injection of BrdUrd 18 h after the last TPA treatment; 1 h later the animals were euthanized, and immunochemical staining of BrdUrd-positive cells was performed (14, 16). Data are expressed as the mean ± SD from at least three mice. Each value for wild-type mice and similarly treated C/EBPβ-null mice within each category was not significantly different (P > 0.05) as determined by the Student's t test.
To test the possibility that C/EBPβ nullizygous mice are refractory to DMBA initiation because of their inability to convert DMBA to the carcinogenic species, we subjected the mice to the direct-acting carcinogen N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) followed by TPA promotion. The MNNG/TPA carcinogenesis protocol produces papillomas, 85–90% of which contain oncogenic mutations in the 12th codon of either Ha-ras (25, 41) or Ki-ras (41). Wild-type mice displayed a 100% incidence of papillomas with ≈3 papillomas per mouse, whereas C/EBPβ null littermates did not develop any tumors (Fig. 1 c and d). Because both MNNG- and DMBA-initiated mice were treated with TPA, it was possible that the inability of C/EBPβ-null mice to develop tumors resulted from a defective response to TPA or to an initiation/promotion protocol. Therefore, we used a complete carcinogenesis protocol in which wild-type and C/EBPβ-null mice were treated weekly with DMBA. All of the wild-type mice developed papillomas with an average of 12 papillomas/mouse whereas C/EBPβ mutant mice were again completely resistant to carcinogenesis (Fig. 1 e and f). We also observed that C/EBPβ-null mice were refractory to DMBA-initiation followed by promotion with the non-phorbol ester tumor promoter mirex (data not shown). Thus, C/EBPβ nullizygous mice are fully resistant to tumorigenesis induced by a variety of carcinogens, tumor promoters, and carcinogenesis protocols that, in normal mice, cause tumors that contain mutant oncogenic Ha-ras or Ki-ras (25, 27–29, 41). These results suggest that C/EBPβ is an essential downstream mediator of oncogenic Ras tumorigenesis.
The lack of tumor development in carcinogen-treated C/EBPβ-null mice could be caused by apoptosis of C/EBPβ-deficient keratinocytes that have acquired oncogenic Ha-ras lesions. To examine this possibility, we treated mice with DMBA and scored the number of apoptotic keratinocytes in C/EBPβ-null and wild-type epidermis by using the cytological parameters described in Methods. Compared with wild-type mice, C/EBPβ-null mice exhibited a 17-fold increase in the number of basal apoptotic keratinocytes (Table 2), indicating that C/EBPβ functions as a survival factor in DMBA/Ras-induced oncogenesis. Similar fold increases in apoptotic cells were observed by using terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) staining (data not shown). To determine whether C/EBPβ-null mice also display increased apoptosis in response to UVB irradiation, a potent inducer of apoptosis and DNA damage, wild-type and C/EBPβ-null mice were irradiated with UVB doses of 50, 200, and 500 mJ/cm2. Although all UVB doses increased the number of apoptotic epidermal keratinocytes, there was no difference between wild-type and mutant mice (data not shown). These results show that the enhanced apoptosis in DMBA-treated C/EBPβ-null mice is stimulus specific and that DNA damage alone is not sufficient to elicit increases in apoptosis in epidermal keratinocytes of C/EBPβ-null mice.
Table 2.
Apoptosis is significantly elevated in epidermal keratinocytes of DMBA-treated C/EBPβ-null mice
| Apoptotic keratinocytes, %
|
||
|---|---|---|
| Wild type | C/EBPβ−/− | |
| Acetone treated | 0.02 ± 0.02 | 0.04 ± 0.01 |
| DMBA treated | 0.10 ± 0.02* | 1.73 ± 0.14*† |
Mice (three per group) were treated with a single application of 400 nmol DMBA/200 μl acetone or acetone alone. More than 4,000 basal keratinocytes were counted for each individual mouse. Data are expressed as mean ± SD.
, Significantly different from acetone-treated group (P < 0.01) as determined by Student's t test.
, Significantly different from wild-type DMBA-treated group (P < 0.01) as determined by Student's t test.
Impaired Skin Tumorigenesis in C/EBPβ−/− v-Ha-ras Transgenic Mice.
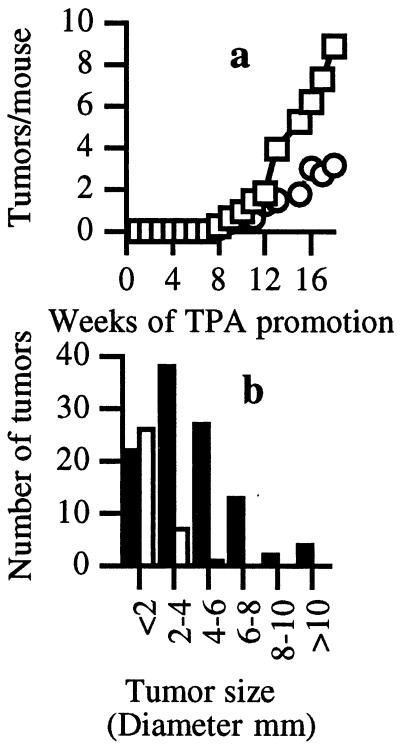
To provide additional evidence for a relationship between Ras and C/EBPβ in skin tumorigenesis, we crossed C/EBPβ-null mice with Tg.AC transgenic mice. Tg.AC mice contain a v-Ha-ras transgene under the control of a partial ζ-globin promoter and are susceptible to skin tumor development in the absence of carcinogen exposure (31). Tumorigenesis in Tg.AC mice does require a promoting stimulus, such as wounding, or treatment with a tumor promoter. As shown in Fig. 2a, TPA-treated C/EBPβ-deficient mice carrying the v-Ha-ras transgene developed ≈65% fewer skin tumors than C/EBPβ+/+ transgene-positive mice, and the tumor size (Fig. 2b) was significantly reduced by 60% in the C/EBPβ-null mice (4.1 ± 2.4 mm C/EBPβ+/+ vs 1.7 ± 1.0 mm C/EBPβ−/−, P < 0.01, Student's t test). Although there was not a complete ablation of tumor development in the C/EBPβ-null mice carrying the v-Ha-ras transgene, it is clear that C/EBPβ significantly affects the development and growth of Ras-induced papillomas. These results support a direct role for C/EBPβ as a nuclear effector of Ras-mediated tumorigenesis.
Figure 2.
C/EBPβ-deficient v-Ha-ras transgenic mice display decreased tumor multiplicity and tumor size. v-Ha-ras+/− C/EBPβ+/+ mice (n = 16) and v-Ha-ras+/− C/EBPβ−/− mice (n = 14) were treated twice weekly with 5 nmol of TPA in 200 μl of acetone. (a) Tumor multiplicity in v-Ha-ras+/− C/EBPβ−/− mice (○) is decreased compared v-Ha-ras+/− C/EBPβ+/+ mice (□; P < 0.05, F test). (b) Tumor size distribution in v-Ha-ras+/− C/EBPβ+/+ mice (closed bars) and v-Ha-ras+/− C/EBPβ−/− mice (open bars; P < 0.05, Fisher's Exact test).
C/EBPβ Activation by Ha-ras Signaling in Keratinocytes.
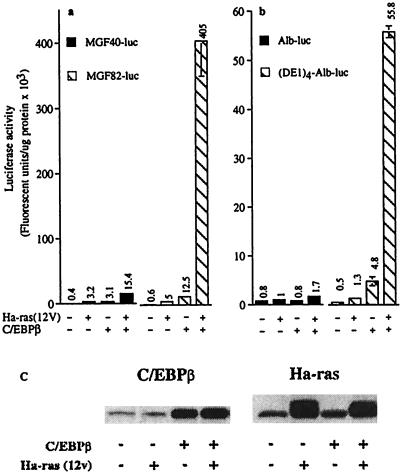
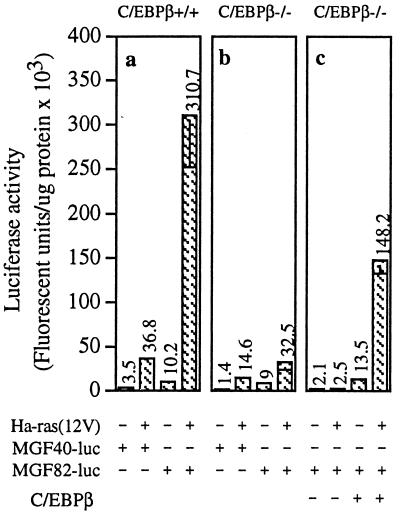
To ascertain whether oncogenic Ha-ras signaling can stimulate C/EBPβ activity in epidermal keratinocytes, we transfected BALB/MK2 keratinocytes with C/EBPβ and/or Ha-ras(12V) and a luciferase reporter gene fused to different lengths of the C/EBP-dependent myelomonocytic growth factor (MGF) promoter (16, 42). pMGF-40 contains a 40-bp portion of the MGF promoter that lacks C/EBP sites, whereas pMGF-82 contains an additional 42-bp region of the promoter harboring two C/EBP-binding sites. Cotransfection of Ha-ras(12V) and C/EBPβ resulted in 30- and 80-fold increases, respectively, in pMGF-82 reporter activity over that observed with C/EBPβ or Ha-ras(12V) alone (Fig. 3a). In contrast, cotransfection of Ha-ras(12V) and C/EBPβ caused only a 5-fold increase in transcription from the MGF-40 promoter, demonstrating that C/EBP binding sites are required for the synergistic response. Similar results were obtained by using a minimal albumin promoter with four tandem C/EBP sites [(DE1)4-Alb-luc] (Fig. 3b and ref. 43). Western blot analysis of cell lysates from BALB/MK2 cells cotransfected with C/EBPβ and Ha-ras(12V) demonstrated that the observed synergistic effect on C/EBP-responsive promoter–reporter activity is not caused by increased Ha-ras(12V) or C/EBPβ expression (Fig. 3c).
Figure 3.
Oncogenic Ha-ras stimulates C/EBPβ transactivation function. (a and b) BALB/MK2 keratinocytes were transfected with pcDNA3-C/EBPβ (0.5 μg) and/or pcDNA3-Ha-ras(12V) (0.5 μg) and 1.0 μg of the specified C/EBP-dependent promoter/reporter as described in the text. Luciferase activity is expressed as fluorescent units per μg of protein, and each value represents the mean ± SD of triplicate dishes per treatment. Similar results were obtained from two repeat experiments. Inclusion of pSV-β-galactosidase and subsequent normalization of luciferase to β-galactosidase activity produced similar results to those normalized to protein levels. (c) BALB/MK2 keratinocytes were transfected with Ha-ras(12V) and/or C/EBPβ, and 48 h later, lysates were prepared and Western analysis was conducted.
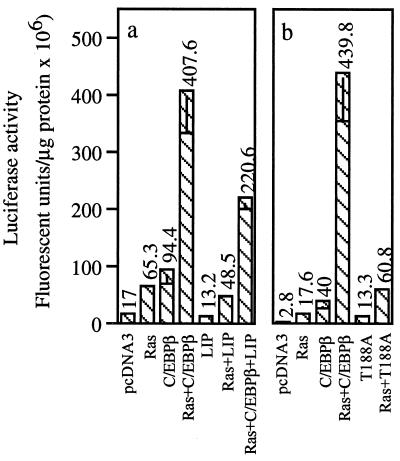
Cotransfection of Ha-ras(12V) with a truncated form of C/EBPβ (LIP) that lacks the N-terminal activation domain but retains the bZIP DNA-binding and leucine zipper domain (44) did not increase the activity of the pMGF-82 reporter (Fig. 4a). In fact, LIP inhibited the activation of wild-type C/EBPβ by Ha-ras(12V) by ≈50%, which is consistent with its known role as a dominant negative inhibitor of C/EBPβ (44) (Fig. 4a). Studies have identified an ERK1/2 phosphorylation site (T188) in C/EBPβ, and substituting T188 with alanine diminished Ras activation of C/EBPβ (22). Therefore, we tested the Ras-responsiveness of a C/EBPβ mutant containing the T188A substitution. Oncogenic Ha-ras-induced stimulation of C/EBPβ activity was abolished in this mutant (Fig. 4b). Thus, an oncogenic Ha-ras pathway can activate C/EBPβ in keratinocytes, and this activation depends on T188 of C/EBPβ.
Figure 4.
Activation of C/EBPβ by oncogenic Ha-ras involves a T188 and requires the presence of the C/EBPβ transactivation domain. BALB/MK2 keratinocytes were transfected with 1.0 μg of the promoter/reporter MGF82-luc and 0.5 μg of one or more of the specified vectors. The experimental procedures were carried out as described in the legend to Fig. 3. Each value represents the mean ± SD of triplicate dishes per treatment. Similar results were obtained from two repeat experiments. Inclusion of pSV-β-galactosidase and subsequent normalization of luciferase to β-galactosidase activity produced similar results to those normalized to protein levels.
To determine whether endogenous C/EBPβ can mediate Ras signaling, we transfected primary keratinocytes isolated from C/EBPβ-nullizygous and wild-type mice with oncogenic Ha-ras and the C/EBP promoter-reporter constructs. Transfection of Ha-ras(12V) into wild-type keratinocytes resulted in a 30-fold increase in pMGF-82 reporter activity whereas in C/EBPβ-nullizygous keratinocytes Ha-ras(12V) caused less than a 4-fold increase (Fig. 5 a and b). The Ras-induced increase in promoter activity required C/EBP binding sites (Fig. 5a). Similar results were obtained with the (DE1)4-Alb-luc reporter (data not shown). Ectopic expression of C/EBPβ in C/EBPβ-null keratinocytes restored responsiveness to oncogenic Ras (Fig. 5a). Thus, endogenous C/EBPβ is a downstream mediator of oncogenic Ha-ras signaling in primary keratinocytes.
Figure 5.
Endogenous C/EBPβ is a downstream mediator of oncogenic Ha-ras signaling in keratinocytes. Primary keratinocytes were isolated from C/EBPβ−/− or C/EBPβ+/+ newborn littermates (2–3 days old) and cultured as described (14, 16). Primary keratinocytes were transfected with the specified vector (0.5 μg each) and 1.0 μg of the C/EBP-dependent promoter/reporter vector and were processed as described in the legend to Fig. 3. Each value represents the mean ± SD of triplicate dishes per treatment. Similar results were obtained from two repeat experiments.
C/EBPβ Augments Ras-Induced Transformation of NIH 3T3 Cells.
The NIH 3T3 focus assay has been widely used to identify pathways and genes that cooperate with Ras to induce transformation (33). To examine the role of C/EBPβ in NIH 3T3 transformation, we first confirmed that oncogenic Ras could stimulate C/EBPβ to activate a C/EBP-responsive promoter-reporter in NIH 3T3 cells (ref. 22 and data not shown). Next we examined whether C/EBPβ has the capacity to transform cells and/or cooperate with oncogenic Ha-ras to increase its transforming potential in the NIH 3T3 focus formation assay. Transfection of C/EBPβ alone did not induce NIH 3T3 transformation (Table 3), showing that this transcription factor does not possess intrinsic transforming activity. Cotransfection of 5 or 10 ng of C/EBPβ enhanced the transformation potential of oncogenic Ha-ras(12V), producing a ≈1.3- and 1.7-fold increase, respectively, in the number of transformed foci compared with Ha-ras(12V) alone (Table 3). Paradoxically, cotransfection of 50 ng of C/EBPβ with Ha-ras(12V) inhibited transformation by 20%, suggesting that the enhancing effect of C/EBPβ is not only saturable, but that high levels of C/EBPβ can inhibit ras transformation, perhaps because of a nonspecific effect of transfecting too high a level of C/EBPβ. Importantly, we observed that cotransfection of 10 ng LIP or 10 ng C/EBPβ T188A inhibited Ha-ras(12V)-induced transformation, indicating an important role for endogenous C/EBPβ in ras-induced transformation of NIH 3T3 cells. C/EBPβ also enhanced the transforming potential of oncogenic Raf, lending further support for a Ras-Raf-ERK-C/EBPβ pathway. In contrast to C/EBPβ, neither C/EBPα or C/EBPδ enhanced the transforming activity of oncogenic Ha-ras(12V) (Table 3). Thus, not all C/EBP family members are capable of augmenting Ras transformation.
Table 3.
C/EBPβ enhances oncogenic Ha-ras-induced transformation of NIH-3T3 cells
| Transformed foci per dish | |
|---|---|
| 10 ng pcDNA3 | 0.0 ± 0.0 |
| 10 ng C/EBPβ | 0.0 ± 0.0 |
| 10 ng Ha-ras(12V) | 35.3 ± 3.5 |
| + 5 ng C/EBPβ | 46.0 ± 7.0* |
| + 10 ng C/EBPβ | 57.7 ± 1.5* |
| + 50 ng C/EBPβ | 28.0 ± 5.6 |
| 10 ng Ha-ras(12V) | 34.7 ± 3.5 |
| + 10 ng C/EBPβ | 58.9 ± 3.8* |
| + 10 ng LIP | 20.7 ± 2.4* |
| + 10 ng C/EBPβ (T188A) | 14.0 ± 2.0* |
| 10 ng Ha-ras | 25.0 ± 1.0 |
| + 10 ng C/EBPα | 25.0 ± 4.4 |
| + 10 ng C/EBPδ | 22.3 ± 2.3 |
| 100 ng Raf(22W) | 29.3 ± 4.5 |
| + 10 ng C/EBPβ | 47.0 ± 1.7* |
Data are expressed as transformed foci per plate, and each value represents the mean ± SD of triplicate dishes per treatment. All experiments were repeated at least two times, and similar results were obtained in each experiment. C/EBPβ, C/EBPβ (T188A), or LIP did not produce any transformed foci at all doses examined (1–1,000 ng/plate).
, Significantly different from the value of cells transfected with Ha-ras(12V) or Raf-22W (a gift from C. J. Der) alone as determined by the Student's t test, P < 0.01.
Discussion
We have identified C/EBPβ as a critical gene in two established in vivo models of Ras-mediated epithelial tumorigenesis as well as in Ras-induced transformation of NIH 3T3 fibroblasts. C/EBPβ was found to be essential for skin tumorigenesis induced by a variety of carcinogens that are known to cause the mutational activation of Ha-ras and K-ras. Moreover, C/EBPβ deficiency in oncogenic v-Ha-ras transgenic mice inhibited v-Ha-ras-induced tumorigenesis by 60%. Substitution of C/EBPβ T188 with alanine, which disrupts an ERK1/2 phosphorylation site, blocked the ability of Ras to stimulate C/EBPβ transactivation function and also inhibited Ras-induced NIH 3T3 transformation. Although our studies cannot rule out the existence of a Ras-independent pathway responsible for the observed responses in the C/EBPβ-deficient mice, our collective results do implicate C/EBPβ as a critical component of a Ras-dependent tumorigenesis/transformation pathway. Future studies using of C/EBPβ−/− MEFs may provide further insight into the exact role of C/EBPβ in Ras-induced transformation and apoptosis.
Depending on the cellular context, strength of signal, and pathways engaged, oncogenic Ras can regulate cell proliferation, differentiation, senescence, apoptosis, or survival (3, 45, 46). Ras-induced apoptosis is suppressed by its activation of pro-survival pathways involving NF-κB (2) or Rac GTPase (46); if these pathways are blocked, apoptosis results. Moreover, in PC-12 cells the MEK/ERK pathway promotes cell survival (47). We have observed that C/EBPβ-null mice display a 17-fold increase in the number of apoptotic keratinocytes in DMBA-treated epidermis compared with similarly treated wild-type epidermis. Studies in mouse epidermis have indicated that 0.1–5% of Ha-ras genes sustain a codon-61 mutation within 1–3 days of topical carcinogen application (48). The observed increase in apoptotic cells in DMBA-treated C/EBPβ-deficient epidermis is consistent with a survival/antiapoptotic role for C/EBPβ in oncogenic Ras-expressing cells. Previously we demonstrated that C/EBPβ positively regulates the program of squamous differentiation in the epidermis and in isolated keratinocytes (16). Therefore, we suggest that in normal keratinocytes C/EBPβ regulates differentiation as well as survival, which is required to complete the differentiation program. However, in the presence of oncogenic Ha-ras the C/EBPβ prosurvival response may predominate over the differentiation pathway and clonal expansion occurs, ultimately resulting in tumor formation. When the C/EBPβ prosurvival signaling pathway is deleted as in the C/EBPβ-deficient mice, cells containing oncogenic Ras undergo apoptosis and tumorigenesis is blocked.
In addition to Ras, there is evidence for additional DMBA target genes that cooperate with oncogenic Ras to induce skin tumorigenesis, and it is conceivable that C/EBPβ is also a component of non-ras oncogenic circuitry (49). Such a notion is consistent with our observation that C/EBPβ deficiency in the v-Ha-ras transgenic mice did not result in the complete inhibition of tumorigenesis. Alternatively, it is possible that high levels of expression of v-Ha-ras transgene in Tg.AC epidermal keratinocytes and/or the expression of the transgene within a different subpopulation of keratinocytes could account for the differences between the degree of inhibition of tumorigenesis in v-Ha-ras and carcinogen-treated C/EBPβ-deficient mice. C/EBPβ-null keratinocytes could also have a defective DNA repair mechanism that contributes to the elevated levels of DMBA-induced apoptosis. However, such a mechanism would not involve all forms DNA damage, as we did not observe differences in UVB-induced apoptosis in wild-type versus C/EBPβ-null keratinocytes. Moreover, because both UVB-induced pyrimidine dimers and DMBA bulky adducts are repaired by the same nucleotide excision repair mechanism, it is unlikely that defects in nucleotide excision repair account for the elevated level of DMBA-induced apoptosis in C/EBPβ-null mice. MNNG-induced DNA damage is repaired by a base excision repair mechanism, suggesting that activation of ras and not altered DNA repair is the common link between the two carcinogens. Collectively, our results tend to support a role for C/EBPβ in the survival of oncogenic Ha-ras cells.
The complete inhibition of tumorigenesis in carcinogen-treated C/EBPβ-deficient mice is striking and is, to our knowledge, the most profound inhibitory effect of a single gene deletion on skin tumorigenesis reported to date. The potent effect of C/EBPβ in an experimental mouse model raises the question of whether C/EBPβ plays a similar role in human cancers. Interestingly, C/EBPβ levels are strongly increased in human colorectal tumors (50) and are also associated with human ovarian epithelial tumor progression (51). Thus, the induction of C/EBPβ may be an important event in the development epithelial tumors, perhaps by providing an essential antiapoptotic signal.
Recent studies have implicated another C/EBP family member, C/EBPα, in myeloid leukemogenesis. However, in contrast to C/EBPβ, C/EBPα functions as a tumor suppressor by promoting granulocytic differentiation and growth arrest. Thus, C/EBPα inactivation by mutation (52) or by its association with the oncoprotein AML-1-ETO (53, 54) contributes to myeloid leukemogenesis by maintaining cells in an extended proliferative state. It is notable that, despite their structural relatedness, C/EBPβ and C/EBPα apparently play very different roles in transformed cells and have opposite effects on tumorigenesis.
Although further studies are required to discern the downstream pathways and genes through which C/EBPβ regulates tumor development, our study reveals a function for C/EBPβ as a critical component of the tumorigenesis pathway initiated by activated Ras. C/EBPβ may therefore represent an attractive target for antineoplastic pharmacological agents.
Acknowledgments
We thank Ron Cannon and Michael Kim for their help with the Tg.AC tumor experiments. We also thank Carrie Cantwell and Sara Parkin for constructing the murine C/EBPβ expression vectors. This work was supported by National Cancer Institute Grant CA46637 (to R.C.S.).
Abbreviations
- TPA
12-O-tetradecanoylphorbol-13-acetate
- bZIP
basic leucine zipper
- C/EBPβ
CCAAT/enhancer binding protein-β
- DMBA
7,12-dimethylbenz[a]anthracene
- LIP
liver inhibitory protein
- MNNG
N-methyl-N′-nitro-N-nitrosoguanidin
- MGF
myelomonocytic growth factor
- UVB
ultraviolet-B
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Serrano M, Lin A W, McCurrach M E, Beach D, Lowe S W. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 2.Mayo M W, Wang C Y, Cogswell P C, Rodgers-Graham K S, Lowe S W, Der C J, Baldwin A S. Science. 1997;278:1812–1815. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 3.Cambell S L, Khosravi-Far R, Rossman K L, Clark G J, Der C J. Oncogene. 1998;17:1395–1413. doi: 10.1038/sj.onc.1202174. [DOI] [PubMed] [Google Scholar]
- 4.Gille H, Downward J. J Biol Chem. 1999;274:22033–22040. doi: 10.1074/jbc.274.31.22033. [DOI] [PubMed] [Google Scholar]
- 5.Bonni A, Brunet A, West A E, Datta S R, Takasu M A, Greenberg M E. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 6.Katz M E, McCormick F. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 7.Marshall C J. Curr Opin Cell Biol. 1996;8:197–204. doi: 10.1016/s0955-0674(96)80066-4. [DOI] [PubMed] [Google Scholar]
- 8.Sklar M D, Tompson E, Welsh M J, Liebert M, Harney J, Grossman H B, Smith M, Prochownik E V. Mol Cell Biol. 1991;11:3699–3710. doi: 10.1128/mcb.11.7.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langer S L, Bortner D M, Roussel M F, Sherr C J, Ostrowski M C. Mol Cell Biol. 1992;12:5355–5362. doi: 10.1128/mcb.12.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson R, Spiegelman B, Hanahan D, Wisdom R. Mol Cell Biol. 1994;16:4501–4511. doi: 10.1128/mcb.16.8.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finco T S, Westwick J K, Norris J L, Beg A A, Der C J, Baldwin A S. J Biol Chem. 1997;272:24113–24116. doi: 10.1074/jbc.272.39.24113. [DOI] [PubMed] [Google Scholar]
- 12.Williams S C, Cantwell C A, Johnson P F. Genes Dev. 1991;5:1553–1567. doi: 10.1101/gad.5.9.1553. [DOI] [PubMed] [Google Scholar]
- 13.Lekstrom-Himes J, Xanthopoulos K G. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 14.Oh H-S, Smart R C. J Invest Dermatol. 1998;110:939–945. doi: 10.1046/j.1523-1747.1998.00199.x. [DOI] [PubMed] [Google Scholar]
- 15.Maytin E V, Habener J F. J Invest Dermatol. 1998;110:238–246. doi: 10.1046/j.1523-1747.1998.00123.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhu S, Oh H S, Shim M, Sterneck E, Johnson P F, Smart R C. Mol Cell Biol. 1999;19:7181–7190. doi: 10.1128/mcb.19.10.7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao Z, Umek R M, McKnight S L. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 18.Natsuka S, Akira S, Nishio Y, Hashimoto S, Sugita T, Isshiki H, Kishimoto T. Blood. 1992;79:460–466. [PubMed] [Google Scholar]
- 19.Scott L M, Civin C I, Roth P, Friedman A D. Blood. 1992;80:1725–1735. [PubMed] [Google Scholar]
- 20.Seagroves T N, Krnacik S, Raught B, Gay J, Burgess-Beusse B, Darlington G J, Rosen J M. Genes Dev. 1998;12:1917–1928. doi: 10.1101/gad.12.12.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson G W, Johnson P F, Hennighausen L, Sterneck E. Genes Dev. 1998;12:1907–1916. doi: 10.1101/gad.12.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, Akira S. Proc Natl Acad Sci USA. 1993;90:2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kowenz-Leutz E, Twamley G, Ansieau S, Leutz A. Genes Dev. 1994;8:2781–2791. doi: 10.1101/gad.8.22.2781. [DOI] [PubMed] [Google Scholar]
- 24.Bos J L. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 25.Balmain A, Brown K. Adv Cancer Res. 1988;51:147–181. doi: 10.1016/s0065-230x(08)60222-5. [DOI] [PubMed] [Google Scholar]
- 26.Frame S, Crombie R, Liddell J, Stuart D, Linardopoulos S, Nagase H, Portella G, Brown K, Street A, Akhurst R, Balmain A. Philos Trans R Soc London B. 1998;353:839–845. doi: 10.1098/rstb.1998.0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuspa S H. Cancer Res. 1994;54:1178–1189. [PubMed] [Google Scholar]
- 28.Quintanilla M, Brown K, Ramsden M, Balmain A. Nature (London) 1986;322:78–80. doi: 10.1038/322078a0. [DOI] [PubMed] [Google Scholar]
- 29.Moser G J, Robinette C L, Smart R C. Carcinogenesis. 1993;14:1153–1160. doi: 10.1093/carcin/14.6.1155. [DOI] [PubMed] [Google Scholar]
- 30.Brown K, Strathsee D, Bryson S, Lambie W, Balmain A. Curr Biol. 1998;8:516–524. doi: 10.1016/s0960-9822(98)70203-9. [DOI] [PubMed] [Google Scholar]
- 31.Leder A, Kuo A, Cardiff R D, Sinn E, Leder P. Proc Natl Acad Sci USA. 1990;87:9178–9182. doi: 10.1073/pnas.87.23.9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sterneck E, Tessarollo L, Johnson P F. Genes Dev. 1997;11:2153–2162. doi: 10.1101/gad.11.17.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark G J, Cox A D, Graham S M, Der C J. Methods Enzymol. 1995;255:395–412. doi: 10.1016/s0076-6879(95)55042-9. [DOI] [PubMed] [Google Scholar]
- 34.Kantz D C, Lacks G D, Cannon R E. BioTechniques. 1999;27:278–280. doi: 10.2144/99272bm13. [DOI] [PubMed] [Google Scholar]
- 35.Trautwein C, van der Geer P, Karin M, Hunter T, Chojkier M. J Clin Invest. 1994;93:2554–2561. doi: 10.1172/JCI117266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buck M, Poli V, van der Geer P, Chojkier M, Hunter T. Mol Cell. 1999;4:1087–1092. doi: 10.1016/s1097-2765(00)80237-3. [DOI] [PubMed] [Google Scholar]
- 37.Reddy S T, Wadleigh D J, Herchman H R. J Biol Chem. 2000;275:3107–3113. doi: 10.1074/jbc.275.5.3107. [DOI] [PubMed] [Google Scholar]
- 38.Drouet C, Shakhov A N, Jongeneel C V. J Immunol. 1991;147:1694–1700. [PubMed] [Google Scholar]
- 39.Moore R J, Owens D M, Stamp G, Arnott C, Burke F, East N, Holdsworth H, Turner L, Rollins M B, Pasparakis M, et al. Nat Med. 1999;5:828–831. doi: 10.1038/10552. [DOI] [PubMed] [Google Scholar]
- 40.Langenbach R, Loftin C, Lee C, Tiano H. Biochem Pharmacol. 1999;58:1237–1246. doi: 10.1016/s0006-2952(99)00158-6. [DOI] [PubMed] [Google Scholar]
- 41.Rehman I, Lowry D T, Adams C, Abdel-Fattah R, Holly A, Yuspa S H, Hennings H. Mol Carcinog. 2000;27:298–307. [PubMed] [Google Scholar]
- 42.Sterneck E, Muller C, Katz S, Leuz A. EMBO J. 1992;11:115–126. doi: 10.1002/j.1460-2075.1992.tb05034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams S C, Baer M, Dillner A J, Johnson P F. EMBO J. 1995;14:3170–3183. doi: 10.1002/j.1460-2075.1995.tb07319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Descombes P, Schibler U. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 45.Shields J M, Pruit K, McFall A, Shaub A, Der C J. Trends Cell Biol. 2000;10:147–154. doi: 10.1016/s0962-8924(00)01740-2. [DOI] [PubMed] [Google Scholar]
- 46.Joneson T, Bar-Sagi D. Mol Cell Biol. 1999;19:5892–5901. doi: 10.1128/mcb.19.9.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 48.Chakravarti D, Mailander P, Franzen J, Higginbotham S, Cavalieri E L, Rogan E G. Oncogene. 1998;16:3201–3210. doi: 10.1038/sj.onc.1201853. [DOI] [PubMed] [Google Scholar]
- 49.Owens D M, Spalding J W, Tennant R W, Smart R C. Cancer Res. 1995;55:3171–3178. [PubMed] [Google Scholar]
- 50.Rask K, Thorn M, Poten F, Kraaz W, Sundfeldt K, Hedin L, Enerback S. Int J Cancer. 2000;86:337–343. doi: 10.1002/(sici)1097-0215(20000501)86:3<337::aid-ijc6>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 51.Sundfeldt K, Ivarsson K, Carlsson M, Enerback S, Janson P O, Brannstrom M, Hedin L. Br J Cancer. 1999;79:1240–1248. doi: 10.1038/sj.bjc.6690199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pabst T, Mueller B U, Zhang P, Radomska H S, Narravula S, Schnittger S, Behre G, Hiddemann W, Tenen D G. Nat Genet. 2001;27:263–270. doi: 10.1038/85820. [DOI] [PubMed] [Google Scholar]
- 53.Westendorf J J, Yamamoto C M, Lenny N, Downing J R, Selsted M E, Hiebert S W. Mol Cell Biol. 1998;18:322–333. doi: 10.1128/mcb.18.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pabst T, Muller B U, Harakawa N, Schoch C, Haferlach T, Behre G, Hiddemann W, Zhang D E, Tenen D G. Nat Med. 2001;7:444–451. doi: 10.1038/86515. [DOI] [PubMed] [Google Scholar]