Abstract
Focal gene amplifications are among the most common cancer-associated mutations1 but have proven challenging to engineer in primary cells and model organisms. Here we describe a general strategy to engineer large (more than 1 Mbp) focal amplifications mediated by extrachromosomal DNAs (ecDNAs)2 in a spatiotemporally controlled manner in cells and in mice. By coupling ecDNA formation with expression of selectable markers, we track the dynamics of ecDNA-containing cells under physiological conditions and in the presence of specific selective pressures. We also apply this approach to generate mice harbouring Cre-inducible Myc- and Mdm2-containing ecDNAs analogous to those occurring in human cancers. We show that the engineered ecDNAs spontaneously accumulate in primary cells derived from these animals, promoting their proliferation, immortalization and transformation. Finally, we demonstrate the ability of Mdm2-containing ecDNAs to promote tumour formation in an autochthonous mouse model of hepatocellular carcinoma. These findings offer insights into the role of ecDNA-mediated gene amplifications in tumorigenesis. We anticipate that this approach will be valuable for investigating further unresolved aspects of ecDNA biology and for developing new preclinical immunocompetent mouse models of human cancers harbouring specific focal gene amplifications.
Subject terms: Oncogenes, Chromosomes
Large extrachromosomal DNAs are engineered using a CRISPR- and Cre–loxP-based approach and shown to drive cancer in mouse models, with potential applications in determining the role of oncogene amplifications in human cancers.
Main
Engineering cancer-associated genetic lesions in cells and in model organisms is essential to defining their contribution to tumour initiation and progression and to the development of accurate preclinical models of human cancers. Over the past few decades, advances in germline and somatic gene editing methods have substantially improved our ability to model a wide range of loss- and gain-of-function mutations in a temporally and spatially controlled fashion. However, focal amplifications—a common mechanism of oncogene activation in human cancers1—have so far resisted efforts to model them in primary cells or in vivo. Although approaches relying on the ectopic expression of individual oncogenes by means of transgenes are useful for modelling transcriptional activation, they fail to reproduce the complexity, intratumoural heterogeneity and evolution of naturally occurring gene amplifications.
Two main classes of amplification have been described: chromosomal and non-chromosomal. The latter are characterized by the presence of multiple copies of circular DNAs that are thought to originate from the fragmentation and subsequent circularization of segments of chromosomes3,4. These large (0.5–3.0 Mbp) extrachromosomal DNAs (ecDNAs) are also known as ‘double minutes’ owing to their paired appearance in metaphase spreads5 (reviewed in refs. 2,6,7). ecDNAs have been detected in several cancer types and are associated with unfavourable prognosis8,9. At least two dozen oncogenes have been reported in circular amplicons, with mouse double minute 2 (MDM2) and cellular myelocytomatosis (MYC) among the most common8. A distinguishing feature of ecDNAs is the absence of centromeres, which results in their random segregation at mitosis10–12. This enables rapid accumulation of large numbers of ecDNAs, promotes intratumoural heterogeneity and facilitates tumour evolution8,13,14. Genes residing on ecDNAs are also transcribed more efficiently than those residing on a linear chromosome, probably owing to a more accessible chromatin configuration, a lack of higher-order compaction15,16 and the co-option of DNA regulatory elements that are active in the relevant cell of origin17. Finally, ecDNAs can act as mobile enhancers, regulating gene expression in trans11,18.
Despite these advances, several fundamental questions remain unresolved. For example, although recent studies have demonstrated the presence of ecDNAs in precancerous oesophageal lesions19, the precise roles of ecDNAs in tumour initiation and progression, as well as their interactions with the host immune system, remain poorly understood. It is also unknown whether normal cells possess mechanisms to protect against the formation or propagation of ecDNAs, and whether the presence of ecDNAs confers therapeutically actionable vulnerabilities. Progress in this field has been hampered by current limitations in our ability to precisely engineer focal amplifications. Here we report a strategy to induce and track the formation of specific ecDNAs in primary cells and whole organisms and apply it to engineer the formation of oncogene-containing ecDNAs in cells and mice.
An inducible system to engineer ecDNAs
To engineer ecDNAs analogous in size and behaviour to those found in human cancers, we leveraged the ability of the Cre recombinase to excise and circularize any region flanked by two loxP sites with the same orientation20,21. We designed two ‘circularization cassettes’ to be inserted in cis at the desired circularization breakpoints (Fig. 1a). Cre-mediated recombination between the two loxP sites contained in the cassettes circularizes the intervening genomic region and reconstitutes a functional H2B–GFP transgene that is encoded by the resulting ecDNA (Fig. 1a). The 3′ cassette contains a second fluorescence marker (mScarlet) whose expression is also induced upon recombination but remains linked to the linear chromosome. This dual colour system allows tracking of cells harbouring the engineered ecDNAs and discriminates between cells that have undergone circularization but have subsequently lost the ecDNA(s) owing to random segregation (GFP−mScarlet+) (Extended Data Fig. 1a) and cells harbouring a tandem duplication caused by Cre-mediated recombination between sister chromatids (GFP+mScarlet−).
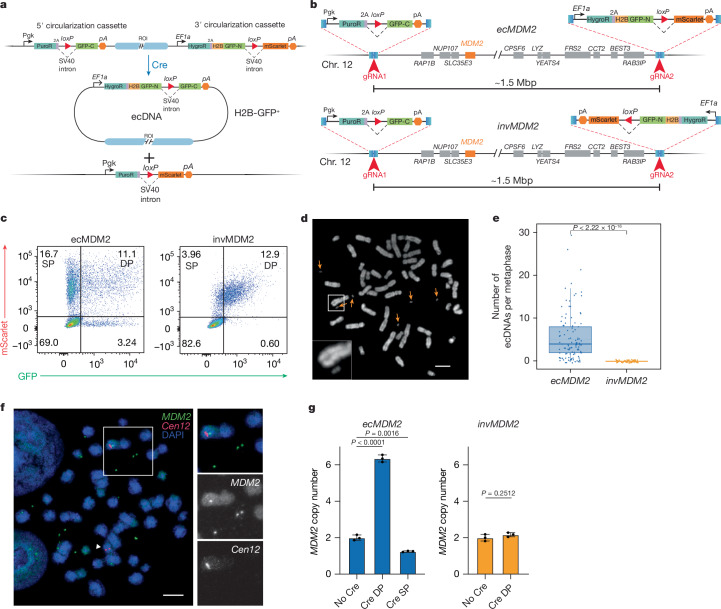
Fig. 1. A general strategy for ecDNA engineering.
a, Schematic of the circularization strategy. b, Schematics of the ecMDM2 and invMDM2 alleles generated in HCT116 cells. The circularization cassettes are not drawn to scale. c, Flow cytometry scatter plot of representative ecMDM2 and invMDM2 clones 6 days after AdCre infection. SP, single positive (mScarlet+GFP−); DP, double positive (mScarlet+GFP+). d, Metaphase spread obtained from sorted double-positive ecMDM2 cells. Orange arrows indicate double minutes (ecDNAs). Inset shows a magnified ecDNA next to a chromosome. Repeated three times in independent clones. e, Numbers of ecDNAs observed in metaphases from double-positive ecMDM2 and invMDM2 cells (P value: two-tailed Fisher t-test; at least 50 metaphases per genotype were analysed). Boxes indicate upper quartile, median and lower quartile. Whiskers extend to ±1.5 × IQR (interquartile range). f, Representative DNA FISH using MDM2 (green) and Chr12 centromere (red) probes performed on metaphase spreads from double-positive ecMDM2 cells. Numerous MDM2-positive double minutes are observed (insets), with concomitant loss of MDM2 signal from one of the two chromosomes 12 (white arrowhead). Representative of three independent experiments. g, Relative MDM2 copy number determined by qPCR in the different ecMDM2 and invMDM2 cell populations (P values are indicated; analysis of variance for multiple comparison and two-tailed Student’s t-test; n = 3). Error bars indicate mean ± s.d. ROI, region of interest. Scale bars, 5 μm.
Extended Data Fig. 1. Generation of ecMDM2 and invMDM2 cells.
a. Schematic and predicted outcomes of the circularization strategy. Upon Cre-mediated recombination, ecDNAs encoding GFP are generated, while mScarlet is expressed from the linear chromosome harboring the corresponding deletion. Random segregation of the ecDNAs will lead some cells to acquire extra copies of the ecDNAs and therefore become more strongly positive for GFP, while other cells will lose the ecDNAs and become GFP-negative. b. Schematic and predicted outcome of the inversion strategy. In this allele, the 3’ circularization cassette is inserted at the same location as in the ecDNA allele, but with opposite orientation. Upon Cre-mediated recombination, the entire region is inverted resulting in the expression of both GFP and mScarlet reporters. Because both reporters remain on the chromosome, the resulting cells are predicted to remain double positive for mScarlet and GFP indefinitely. c. ecMDM2 and invMDM2 cells were infected with AdCre, and then sorted for GFP + ;mScarlet+ population. The mScarlet vs. GFP scatter plots of post-sorted double positive cells are overlaid, and the density plots for each fluorescence are shown on their respective axes. d. Box-and-whiskers plots comparing mScarlet or GFP fluorescence of sorted double positive ecMDM2 and invMDM2 cells, respectively, with indicated ratio of variances as determined by the F-test. n = 43760 cell for ecMMD2 and 48911 cells for invMDM2. Boxplot represents upper quartile, median, lower quartile. Whiskers extend to ±1.5 × IQR. Illustrations in a and b were created using BioRender (https://biorender.com).
As a proof of concept for this strategy, we chose the HCT116 colorectal cancer cell line, because it is near-diploid and chromosomally stable and harbours no ecDNAs22. Using CRISPR–Cas9-mediated genome editing, we inserted two cassettes flanking a 1.5 Mbp region on chromosome 12. This region is recurrently amplified in human cancers23–25 and includes the human MDM2 oncogene (Fig. 1b), a key negative regulator of the p53 pathway26,27. As a control, we generated a companion HCT116 line in which the orientation of the 3′ cassette was flipped, so that the two loxP sites were in the opposite orientation (Fig. 1b). With this configuration, the genomic region flanked by the cassettes was inverted rather than excised upon Cre expression and therefore, both fluorescent reporters remained linked to the linear chromosome (Extended Data Fig. 1b). We refer to these two engineered cell lines as ecMDM2 and invMDM2, respectively.
Cells expressing both GFP and mScarlet (double positive) were readily detected in both invMDM2 and ecMDM2 clones infected with Cre-expressing recombinant adenoviruses (AdCre) (Fig. 1c; gating strategy and gel source data for all figures are provided in Supplementary Fig. 1). As predicted, in ecMDM2 clones, we also observed a large fraction of cells expressing mScarlet but not GFP (single positive), as well as a much smaller population of cells expressing GFP but not mScarlet (Fig. 1c). The presence of single-positive cells was consistent with loss of ecDNAs, whereas the small population of GFP+mScarlet− ecMDM2 cells was probably a product of Cre-mediated recombination between loxP sites on sister chromatids, which is predicted to lead to tandem duplication.
To confirm that concomitant mScarlet and GFP expression in ecMDM2 cells reflected the presence of engineered ecDNAs, we generated metaphase spreads from sorted double-positive ecMDM2 and invMDM2 cells. Most metaphases from ecMDM2 cells—but none from invMDM2 cells—contained MDM2-positive ecDNAs. As expected, loss of MDM2 signal from one of the two copies of chromosome 12 was observed in metaphases from ecMDM2 cells (89 of 91, 98% of metaphases examined; Fig. 1d–f). Genomic quantitative PCR (qPCR) confirmed amplification of the MDM2 locus in double-positive ecMDM2 cells as well as loss of one copy of MDM2 in the single-positive population, whereas MDM2 copy number was unchanged in double-positive invMDM2 cells (Fig. 1g). Notably, the variance in GFP expression within the double-positive population was substantially greater in ecMDM2 cells compared with invMDM2 cells, reflecting the diversity in ecDNA copy number caused by random segregation at mitosis (Extended Data Fig. 1c,d).
Tracking ecDNA dynamics in cells
To assess whether GFP expression could be used to track ecDNA abundance, we sorted AdCre-infected ecMDM2 cells into four bins on the basis of GFP expression (Fig. 2a). As shown in Fig. 2b, the average number of ecDNAs per metaphase was positively correlated with GFP intensity, and shallow whole-genome sequencing (sWGS) identified a corresponding focal amplification precisely spanning the 1.5 Mbp region demarcated by the two circularization cassettes (Fig. 2c). As expected, no amplification of this region was present in double-positive invMDM2 cells (Fig. 2c).
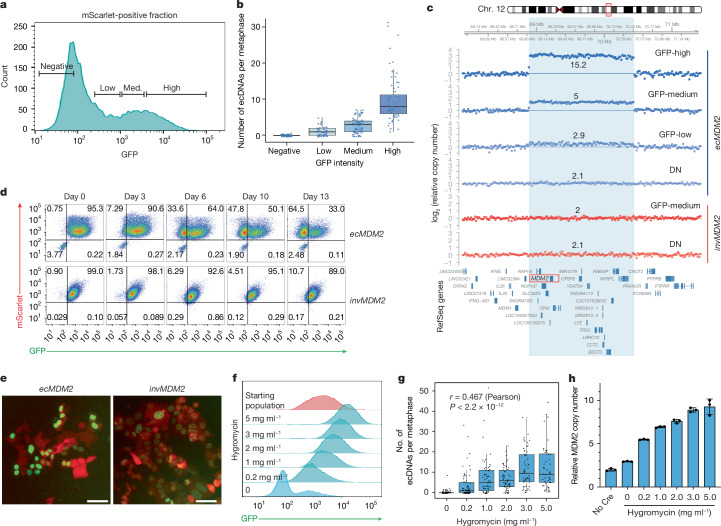
Fig. 2. Dynamics of engineered ecDNAs in HCT116 cells.
a, Double-positive cells from an AdCre-infected ecMDM2 line were cultured in the absence of hygromycin and sorted into four bins on the basis of GFP intensity (negative, low, medium (med.) and high) as indicated. b, Box-and-whiskers plot showing the number of ecDNAs per metaphase in each bin. Box plot represents upper quartile, median and lower quartile. Whiskers extending to ±1.5 × IQR are shown (n = 60, 57, 78 and 68 metaphases). c, ecMDM2 and invMDM2 cells were infected with AdCre, sorted according to mScarlet and GFP intensity, and analysed by sWGS. The panel shows the log2 ratio of relative copy number, as inferred from quantitative DNA sequencing (QDNAseq) analysis, across the region of chromosome 12 surrounding the expected ecDNA (defined by the light blue highlight). The number above each track indicates the amplicon copy number. d, Sorted double-positive cells from AdCre-infected ecMDM2 and invMDM2 clones were cultured in the absence of hygromycin and analysed by flow cytometry at the indicated time points. e, mScarlet and GFP expression in double-positive sorted ecMDM2 (left) and invMDM2 (right) cells maintained in media without hygromycin for 1 week. Independently repeated twice. f, Histogram plot of GFP fluorescence. AdCre-infected and sorted double-positive ecMDM2 cells were expanded for 13 days in medium containing the indicated concentration of hygromycin and analysed by flow cytometry. g, Box-and-whiskers plot showing the number of ecDNAs per metaphase observed in the cells described in e. Boxes indicate upper quartile, median and lower quartile. Whiskers extend to ±1.5 × IQR. Pearson correlation coefficients and associated P values are shown (n = 58, 65, 56, 56, 54 and 51 metaphases). h, MDM2 copy number as determined by genomic qPCR on cells from f. Error bars indicate mean ± s.d. n = 3 technical replicates. Scale bars, 50 μm.
Having established that GFP intensity could be used as a semiquantitative surrogate for ecDNA abundance, we then examined the dynamics of the engineered ecDNAs in HCT116 cells. We infected ecMDM2 and invMDM2 cells with AdCre and measured the fraction of double-positive and single-positive cells at different time points. For both ecMDM2 and invMDM2 cells, the fraction of double-positive cells peaked approximately 5–6 days postinfection (Extended Data Fig. 2a). However, whereas the double-positive population of invMDM2 cells remained constant throughout the rest of the experiment, that of ecMDM2 cells progressively decreased, accompanied by a corresponding increase in the fraction of single-positive (GFP−mScarlet+) cells. To test whether the single-positive cells derived from initially double-positive cells, we first sorted double-positive ecMDM2 and invMDM2 cells. Upon serial passage, ecMDM2 cells gradually lost GFP expression, becoming single positive, whereas invMDM2 cells remained uniformly double positive (Fig. 2d,e). This result indicates that the engineered ecDNAs are gradually lost in HCT116 cells, probably because extra copies of the genes on the amplicon do not provide a fitness advantage to these cancer cells in vitro14. If this interpretation is correct, providing an artificial selective pressure should be sufficient to promote accumulation of the engineered ecDNAs. To test this hypothesis, we leveraged the hygromycin resistance gene encoded by the engineered ecDNAs (Fig. 1a and Extended Data Fig. 1a). When double-positive ecMDM2 cells were cultured in the presence of increasing concentrations of hygromycin (0–5 mg ml−1), we observed a corresponding increase in GFP intensity in the population (Fig. 2f and Extended Data Fig. 2b), accompanied by progressive accumulation of MDM2-containing ecDNAs (Fig. 2g and Extended Data Fig. 2c) and an increase in MDM2 copy number (Fig. 2h).
Extended Data Fig. 2. Characterization of ecDNAs in ecMDM2 cells.
a. ecMDM2 and invMDM2 cells were infected with AdCre, expanded in the absence of hygromycin, and analyzed by flow cytometry at the indicated timepoints. Clones were propagated for 6 days in the presence or absence of hygromycin (200 µg/ml) and analyzed by flow cytometry. Pseudocolor scatter plots of GFP and mScarlet fluorescence at each time point are shown. Notice the progressive disappearance of double positive cells and the concomitant increase in GFP-;mScarlet+ cells in the ecMDM2 samples. b. Violin plots showing GFP intensity of sorted double positive ecMDM2 and invMDM2 cells expanded in the presence of the indicated concentration of hygromycin for 13 days and analyzed by flow cytometry (see also Fig. 2F–H). Median GFP intensity for each sample is also indicated. Boxes indicated interquartile range. **** indicates p-value < 0.0001 as determined by the Wilcoxon–Mann–Whitney two-side test. c. Representative DNA FISH on metaphase spreads from ecMDM2 cells maintained at the indicated hygromycin concentration. Scale bar: 7.5 µm. n = 15 (No Hygro), 14 (0.2 mg/ml), 16 (1 mg/ml), 14 (5 mg/ml) metaphases analyzed. d. Presence of MDM2-positive HSRs in representative metaphase spreads (n = 6 HSR containing metaphases out of 91 analyzed). Scale bar: 7.5 µm.
While analysing the results of these experiments, we also noticed labelling of homogeneously staining regions instead of ecDNAs by the MDM2 probe in approximately 6% (6 of 91) of metaphases generated from sorted double-positive ecMDM2 cells (Extended Data Fig. 2d). In all cases, one of the two chromosomes 12 showed loss of endogenous MDM2 staining, confirming that these cells had previously circularized and excised the ecMDM2 allele. This indicates that the engineered ecDNAs can occasionally reintegrate into a different chromosome, a poorly understood phenomenon that has been reported for naturally occurring ecDNAs3,4,28.
Generation of mice with inducible ecDNAs
We next generated genetically engineered mice harbouring loxP sites spanning a 1.7 Mbp genomic region on chromosome 15 that contained, among other genes, the Myc locus (Fig. 3a and Extended Data Fig. 3a). This engineered allele will be referred to hereafter as Mycec. Mycec/+ and Mycec/ec mice were obtained at Mendelian frequency, were viable and fertile, had no obvious phenotype (Extended Data Fig. 3b–d) and showed excision and circularization of the region flanked by the loxP sites upon Cre expression (Extended Data Fig. 3e,f).
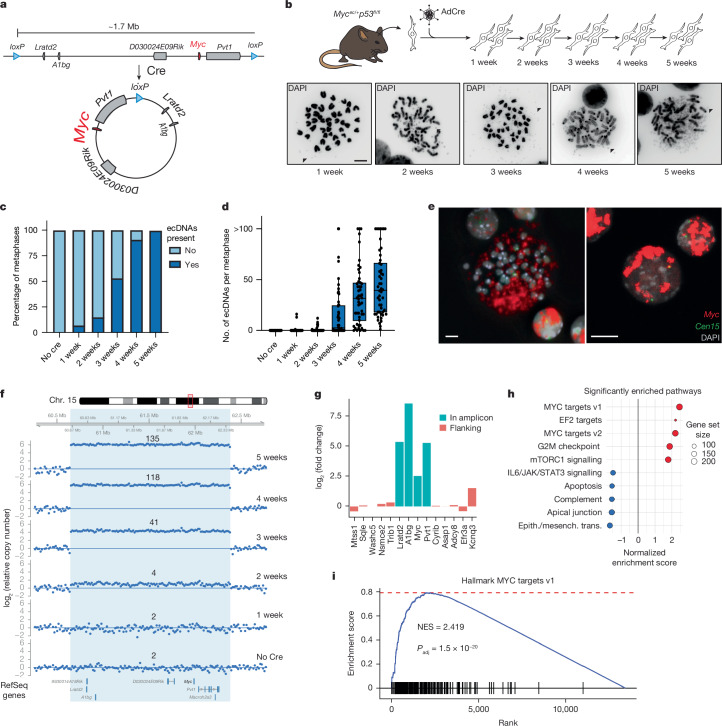
Fig. 3. Generation of Myc-containing ecDNAs in primary cells from genetically engineered mice.
a, Schematic of the Mycec allele. b, aNSCs derived from Mycec/+p53fl/fl mice were infected with AdCre and propagated in vitro for 5 weeks. Metaphase spreads (lower panels) at different time points were collected, and representative micrographs are shown. Arrowheads indicate double minutes (ecDNAs). c, Bar plot showing the fraction of metaphases with or without ecDNAs. d, Box-and-whiskers plot showing the number of ecDNA per metaphase in Mycec/+p53fl/fl aNSC at different times upon AdCre infection. n = 50 metaphases analysed per condition. Boxes indicate upper quartile, median and lower quartile. Whiskers extend to ±1.5 × IQR. e, Metaphase (left) and interphase FISH (right) performed on aNSCs from Mycec/+p53fl/fl 5 weeks after AdCre infection. The Myc signal is shown in red, and a probe labelling a pericentromeric region of chromosome 15 is shown in green. Representative of two independent experiments. f, sWGS analysis of Mycec/+p53fl/fl aNSCs at different time points after AdCre infection. The panel shows relative copy number, as inferred from QDNAseq analysis, across the region of chromosome 15 surrounding the predicted Myc-containing ecDNA (light blue). Notice the progressive increase in copy number of a focal amplification matching the predicted ecDNA boundaries. g, log2 fold change of mRNA expression of genes included in (blue) or flanking (red) the engineered Myc amplicon in Mycec/+p53fl/fl versus p53fl/fl aNSCs 5 weeks after AdCre infection, as determined by RNA-seq analysis. h, Gene set enrichment analysis showing the top enriched hallmark pathways in Mycec/+p53fl/fl versus p53fl/fl aNSCs at 5 weeks after AdCre infection. i, Gene set enrichment plot of hallmark gene set MYC Targets v.1, the most enriched gene set in Mycec/+p53fl/fl aNSCs. NES, normalized enrichment score; Padj, adjusted P value. NES and Padj were calculated as per the fgsea package in R. Scale bars, 5 μm.
Extended Data Fig. 3. Generation and characterization of Mycec/+ mice.
a. Schematic of the Mycec allele. Upon AdCre transduction, Cre recombinase promotes the excision and circularization of the genomic region flanked by the two loxP sites. Genes are indicated in gray boxes. Myc gene is highlighted in red. Arrows indicate primers to detect the insertion of loxP sites for genotyping and Sanger sequencing. b. Sanger sequencing results of inserted loxP sites in the F1 progeny. LoxP sequences in the correct orientation are highlighted in light blue. c. Breeding schemes to test viability and fertility of Mycec/+ mice. Expected and observed genotypes in the F2 generation are reported. Representative genotyping PCR results are shown. All animals were genotyped. gDNA-PCR analysis to check for the insertion of both loxP sites (A-B and C-D) is shown on the left. d. Breeding schemes to test viability and fertility of Mycec mice. Expected and observed genotypes in the F2 generation are reported. e. gDNA-PCR analysis with primers designed to detect the circularized allele (Primers C-B) and scar on the linear chromosome (Primers A-D) on the linear chromosome upon Cre-expression performed on DNA extracted from different tissues of a Mycec/+;Actin-Cre mouse (1 month old). Amplification of an unrelated genomic region is included as a PCR control. Quantification by digital droplet PCR (ddPCR) of the circularized allele (Probes c-b) and the scar in linear chromosome (Probes a-d) is shown at the bottom. Heart, kidney, large bowel, pancreas, and spleen of a single Myc+/+;Actin-Cre mouse are used as controls. f. gDNA-PCR analysis (top) and quantification by ddPCR (bottom) of the circularized allele and the scar in linear chromosome in brain and large intestine of mice expressing the Cre recombinase under the control of the tissue-specific promoter of Nestin and Villlin1 (Vil1), respectively. The genotype of each mouse is indicated.
To determine whether the engineered ecDNAs could form and accumulate in primary cells, we isolated adult neural stem cells (aNSCs) from Mycec/+p53 fl/fl mice and p53fl/fl littermate control mice and infected them with AdCre. Following infection, these cells were propagated in vitro and evaluated weekly for the presence of ecDNAs (Fig. 3b). Initially, only a small fraction of Mycec/+p53fl/fl aNSCs contained ecDNAs, but the number of metaphases with ecDNAs and the number of ecDNAs per metaphase increased markedly over the ensuing weeks, such that by week 5, every metaphase contained multiple Myc-positive ecDNAs (Fig. 3c–e). Detection of a circular amplicon exactly matching the region flanked by the loxP sites and reaching an average of around 135 copies per cell by week 5 further corroborated these findings (Fig. 3f and Extended Data Fig. 4a). Notably, in interphase nuclei, the Myc DNA fluorescence in situ hybridization (FISH) signal was often localized into discrete clusters, rather than being randomly distributed (Fig. 3e and Extended Data Fig. 4b), reminiscent of the ‘enhancer hubs’ previously described for naturally occurring MYC-containing ecDNAs11,15,18.
Extended Data Fig. 4. Characterization of ecDNAs in Mycec/+ NSCs.
a. sWGS data from the 5 week time points (see Fig. 3B) were analyzed using AmpliconArchitect to identify structural variants. The structural variant plot reveals a structural variant closing the left-and-right endpoints of the amplified region forming an ecDNA-like cycle spanning the region flanked by the loxP sites. b. Representative interphase-nuclei FISH (right) on aNSC from Mycec/+; p53fl/fl 5 weeks upon AdCre infection showing clustering inside the nucleus. Myc probe is in red. Control probe labeling a pericentromeric region of chromosome 15 is in green. Scale bar: 5 μm. c. ATAC-seq fragment size distribution of ecDNA and chromosomal DNA regions. n = 2 replicate per group. d. Location of active enhancers contained within and immediately outside the Myc ecDNA. Enhancers from the indicated mouse tissues at birth (P0) were obtained from the Encyclopedia of DNA elements (ENCODE) (ref. 59). The two bottom tracks show ATAC-Seq reads count across the same region generated from NSCs with the indicated genotypes. The region corresponding to the ecDNA is highlighted in light red. e. ATAC-seq read counts in Mycec region normalized by sequencing depth and copy number in p53fl/fl;Mycec/+ and p53fl/fl cells (5,001,487 and 58,957 reads, respectively). Boxes indicate upper quartile, median, and lower quartile. Whiskers extend to ±1.5 × IQR. Two-sided Wilcoxon test. P value = 8.3e-10. f. Relative copy number and mRNA expression were determined by qPCR in Mycec/+;p53fl/fl neuronal stem cells at 1 week, 3 weeks, and 5 weeks post Ad-Cre infection. Only genes within the amplicon detectable at baseline are shown. Note that A1bg and Pvt1 expression increases more than predicted based on copy number (dashed line), while Myc expression increase is lower than predicted. g. Immunoblotting of MYC expression levels in Mycec/+; p53fl/fl and p53fl/fl aNSC 5 weeks upon AdCre Infection. h. Volcano plot of Mycec/+; p53fl/fl vs p53fl/fl aNSC 5 weeks upon AdCre infection. Genes with log2 Fold Change > 1, and adjusted P-value calculated using the DESeq2 R package <0.01 are indicated in red. Genes located in the Mycec amplicon are labeled. i. Growth curves of Mycec/+; p53fl/fl (black) and control p53fl/fl (grey) aNSC 5 weeks after AdCre infection. n = 10 biological replicates, 3 fields for each replicate have been acquired.
To define the transcriptional status of the engineered ecDNAs, we performed assay for transposase-accessible chromatin with sequencing (ATAC-seq) and RNA sequencing (RNA-seq) analyses of p53fl/fl and Mycec/+p53fl/fl aNSCs at 5 weeks after AdCre infection (Supplementary Table 1). ATAC-seq indicated that the chromatin on the engineered ecDNAs was largely open or mononucleosomal (Extended Data Fig. 4c,d). After normalization for copy number, the region on the ecDNAs contained slightly more ATAC-seq reads than the same region on the linear chromosome (Extended Data Fig. 4e). Accordingly, all genes included in the amplicon were upregulated, albeit to different extents (Fig. 3g and Extended Data Fig. 4f–h), and gene set enrichment analysis showed strong activation of MYC targets (Fig. 3h,i). In analogous experiments, we detected spontaneous accumulation of the engineered ecDNAs upon Cre expression in three further primary cell types derived from these mice: cerebellar progenitors, mouse embryonic fibroblasts (MEFs) and hepatocytes (Extended Data Fig. 5).
Extended Data Fig. 5. Generation of Myc-containing ecDNAs in different primary cell types.
Mycec/+;p53fl/fl and Myc+/+;p53fl/fl primary mouse embryo fibroblasts (a-d), cerebellar stem cells (e-h), and primary hepatocytes (i-l) were infected with Cre recombinase, passaged for 5-8 weeks and analyzed by PCR to detect circularization and excision of the ecDNA allele (a, e, i). (b,f,j) Metaphase spreads were examined by DNA-FISH using a Myc probe. Scale bar = 5 µm (c,g,k) Total genomic DNA was used to determine mean Myc copy number. Each bar corresponds to a biological replicate. (Error bars: mean ± SD). (d,h,l) Stacked bar plot showing the fraction of metaphases with (dark color) or without (light color) detectable Myc positive double minutes (ecDNAs). n = 50 metaphases per bar. Illustrations in a, b and i were created using BioRender (https://biorender.com).
These results demonstrate the generation of a targeted focal oncogene amplification mediated by replication-competent and transcriptionally active engineered ecDNAs in several primary cell types.
ecDNAs immortalize primary cells
To investigate the oncogenic potential of engineered ecDNAs, we generated a second mouse strain containing loxP sites flanking a 1.3 Mb segment on chromosome 10 (Mdm2ec, Fig. 4a and Extended Data Fig. 6a–c). This region contains, among other genes, the Mdm2 oncogene and is syntenic to a region recurrently amplified in ecDNAs in human cancers. Mdm2ec/+ and Mdm2ec/ec mice are viable and fertile and do not show any obvious abnormality (Extended Data Fig. 6d,e and data not shown).
Fig. 4. Engineered Mdm2-containing ecDNAs promote immortalization and transformation of primary mouse cells.
a, Schematic of the Mdm2ec allele. b, Mdm2ec/+ MEFs approximately 5 weeks after being infected with AdCre or left untreated. Repeated on three independent MEF lines. c, Metaphase spread and DNA FISH showing the presence of multiple Mdm2-positive ecDNAs in AdCre-infected Mdm2ec/+ MEFs (inset). Repeated twice. d, Number of ecDNAs per metaphase in AdCre-infected Mdm2ec/+ MEFs transduced or not with HRASG12V and in tumours (n = 4) derived from injecting the transduced cells into the flanks of nude mice. Boxes indicate upper quartile, median and lower quartile. Whiskers extend to ±1.5 × IQR. P values, pairwise Wilcoxon rank-sum test corrected for multiple testing. n = 57, 54 and 106 metaphases. e, sWGS analysis of AdCre-infected Mdm2ec/+ MEFs at the indicated time points after AdCre infection showing progressive accumulation of the Mdm2 ecDNA over time. ‘Tumour’ indicates sWGS of a tumour obtained by injecting the HRAS-infected cells into the flank of a nude mouse. The absence of signal in the region immediately downstream of the amplicon is due to QDNASeq ignoring low-mappability regions. Amplicon copy number is indicated above each track. f, Representative image of a tumour developing in a nude mouse injected subcutaneously with AdCre- and HRASG12V-infected Mdm2ec/+ MEFs. g, Metaphase spread showing numerous ecDNAs (arrows) in cells isolated from the tumour shown in f. h, MDM2 immunoblot of lysates from tumours generated as indicated above. Mdm2-p53 double-knockout tumours served as negative controls for the MDM2 antibody. Asterisks indicate non-specific bands. Repeated on three independent tumours. i, Bar plot showing the log2 fold change of genes included within the Mdm2 amplicon (light orange) and immediately flanking it (grey) in Cre-treated Mdm2ec/+HRASG12V versus p53fl/flHRASG12V MEFs as determined by RNA-seq. Mdm2 is highlighted in blue. Scale bars, 350 μm (b), 10 μm (c,g).
Extended Data Fig. 6. Generation and characterization of Mdm2ec/+ mice.
a. Schematic of the Mdm2ec allele, with the two loxP sites flanking a 1.3Mbp region on chromosome 12. Arrowheads indicate the PCR primers used to identify successful loxP insertion. b. Zygotes were injected with Cas9-gRNA complexes and donor DNA containing loxP sites. Genotyping of F0 mice shows that Mouse #2 has loxP sites inserted at both the upstream and downstream locations. Note that the upstream wildtype band for Mouse #2 is lost, in this case indicating homozygous insertion of the loxP. After initial validation by sequencing, all mice were genotyped by PCR. c. Chromatograms showing correct insertion of the two loxP sites. d. A heterozygote F1 progeny of Mouse #2 crossed to a wildtype mouse shows that the loxP bands for the upstream and downstream integration sites co-segregate in the resulting F2 progeny, indicating that the loxP sites were inserted in cis e. Mdm2ec/+ F1 mice are bred to each other and resulting expected and observed F2 progeny ratio are described. f. AdCre-treated Mdm2ec/+ MEFs develop recombination of the loxP sites, resulting in the circularization of the intervening region as indicated by the amplification product of primers B and C. Repeated in three independent experiments. g. Chromatogram obtained by Sanger sequencing of the circularization product from E) demonstrated expected recombination sequence surrounding the loxP site.
We next examined whether Cre-mediated induction of ecDNAs containing the Mdm2 locus could lead to Mdm2 amplification and suppression of the p53 pathway in primary cells. Freshly isolated Mdm2ec/+ and wild-type MEFs were either left untreated or infected with AdCre (Extended Data Fig. 6f,g). As expected, untreated Mdm2ec/+ and wild-type MEFs stopped proliferating after approximately 3–4 weeks in culture and acquired the characteristic morphology of senescent cells (Fig. 4b). By contrast, AdCre-infected Mdm2ec/+ MEFs became immortalized, accumulated ecDNAs containing the Mdm2 locus (Fig. 4c,d) and showed strong upregulation of Mdm2 RNA and protein expression (Extended Data Fig. 7a,b). sWGS of these cells confirmed the presence of a circular focal gene amplification matching the region flanked by the two loxP sites (Fig. 4e and Extended Data Fig. 7c). Inactivation of the p53 pathway is also required for the transformation of MEFs by ectopically expressed oncogenic HRASG12V29. We transduced early passage wild-type and Cre-treated Mdm2ec/+ MEFs with retroviruses encoding the HRASG12V oncogene. Control cells became senescent after an initial burst of proliferation, whereas AdCre-infected Mdm2ec/+ cells continued proliferating and formed tumours when injected into immunocompromised mice (all seven mice, two independent MEF lines, Fig. 4f). No tumours developed in the five mice injected with HRASG12V-transduced wild-type MEFs or in the four mice injected with HRASG12V-transduced Mdm2ec/+ MEFs that had not been previously infected with AdCre.
Extended Data Fig. 7. Mdm2 expression in Mdm2ec/+ MEFs.
a. Western blot for MDM2 and tubulin of wildtype, Mdm2ec/+, and p53fl/fl MEFs with or without treatment by Cre and HRASG12V show significant accumulation of Mdm2 only in Cre-treated Mdm2ec/+ MEFs. Biological replicates from different Mdm2ec/+ MEF lines were analyzed. b. qPCR in Cre-treated Mdm2ec/+, and p53fl/fl MEFs show preferential upregulation of Mdm2 transcripts in Mdm2ec/+ MEFs. p-value: 0.0018, two-tailed t-test, Error bars: mean ± SD. N = 3 technical replicates. c. sWGS data from AdCre- and HRAS infected Mdm2ec/+ MEFs (see Fig. 4) were analyzed using AmpliconArchitect to identify structural variants. The coverage and structural variant plot reveals a structural variant closing the left-and-right endpoints of the amplified region forming an ecDNA-like cycle spanning the region flanked by the loxP sites.
The resulting tumours were characterized by marked circular amplification of the region flanked by the loxP sites (Fig. 4e) and contained on average approximately 60 Mdm2 ecDNAs per cell (Fig. 4d,e,g). The circular amplicons were characterized by open chromatin and drove increased expression of Mdm2 and the other genes they contained (Fig. 4h,i, Extended Data Fig. 8a–d and Supplementary Table 2), indicating that the engineered ecDNAs were transcriptionally active and could propagate in vivo. Histologically, the tumours had a high-grade phenotype, with spindle and pleomorphic cells arranged in short fascicles, showing infiltrative growth within skeletal muscle (Extended Data Fig. 8e–h). The tumour showed a high mitotic count and in areas showed single or multi-intracytoplasmic fat vacuoles with focal nuclear indentation, a characteristic feature of lipoblasts. The alternating high-grade undifferentiated component with areas with adipocytic differentiation closely resembled the features of dedifferentiated liposarcoma, a tumour whose genetic hallmark is the amplification of MDM2 (ref. 30).
Extended Data Fig. 8. Characterization of Mdm2ec/+;HRASG12V sarcomas.
a. Location of active enhancers contained within and immediately outside the Mdm2 ecDNA. Enhancers from the indicated mouse tissues at birth (P0) were obtained from the Encyclopedia of DNA elements (ENCODE) (ref. 59). The two bottom tracks show ATAC-Seq reads count across the same region generated from NSCs with the indicated genotypes. The region corresponding to the ecDNA is highlighted in light red. b. ATAC-seq fragment size distribution of ecDNA and chromosomal DNA regions in Mdm2ec/+ and p53fl/fl MEFs transduce with HRASG12V. n = 4 replicates for Mdm2ec/+ and 3 replicates for p53fl/fl. c. ATAC-seq read counts in Mdm2ec region normalized by sequencing depth and copy number in Mdm2ec/+;p53fl/fl and p53fl/fl cells. (3,693,906 and 108,897 reads, respectively). Boxes indicate upper quartile, median, and lower quartile. Whiskers extend to ±1.5 × IQR. Two-sided Wilcoxon test. P value = 5.2e-10. d. RNA-FISH using an Mdm2 probe on tumour tissues from Mdm2ec/+;HRASG12V and p53fl/fl;HRASG12V MEFs at both low (20x, scale bar = 50 µm) and high (100x, scale bar = 10 µm) magnifications. e. Low magnification view showing a high-grade spindle cell sarcoma arranged in short fascicles and infiltrating into skeletal muscle. f. The lesional cells show increased nuclear pleomorphism, with scattered multinucleated forms (arrows) and increased mitotic activity (arrowheads). g. Higher magnification shows solid sheets of epithelioid to ovoid cells with distinct single or multi- intra-cytoplasmic fat vacuoles consistent with signet ring lipoblasts. Focal nuclear indentation, a characteristic feature of lipoblasts, is also noted. h. Increased mitotic activity and pleomorphic spindle cells with amphophilic cytoplasm and ovoid nuclei with clumped chromatin and prominent nucleoli in keeping with a high-grade sarcoma. Analysis repeated in tumours from 3 mice.
To verify the causative role of Mdm2 amplification in the immortalization and transformation of these cells, we treated them with increasing concentrations of MDM2 antagonist milademetan31. This treatment reactivated the p53 pathway and potently inhibited proliferation of the transformed Mdm2ec/+ cells at nanomolar concentrations, whereas it was largely ineffective against the Cre-treated HRAS-transformed p53fl/fl MEFs used as controls (Extended Data Fig. 9a–c).
Extended Data Fig. 9. Mdm2ec/+;HRASG12V sarcomas response to Milademetan.
a. Dose-response curve of AdCre-treated p53fl/fl;HRASG12V and Mdm2ec/+;HRASG12V cells treated with the MDM2 antagonist milademetan. Each dot represents a technical replicate (n = 3). A representative plot of two independent experiments is shown. b-c. Cre-treated Mdm2ec/+;HRASG12V and p53fl/fl;HRASG12V MEFs were exposed to 1 µM milademetan, collected at indicated time points, and analyzed by RT-qPCR (b) and immunoblot (c). Error bars: mean ± SD. N = 3 technical replicates. Mdm2 and p21 mRNA and protein products are rapidly induced in Mdm2ec/+;HRASG12V cells in response to milademetan.
These results demonstrate that engineered Mdm2-containing ecDNAs rapidly and spontaneously accumulate in primary MEFs, promote their immortalization and cooperate with HRASG12V in inducing oncogenic transformation.
Engineered ecDNAs promote tumorigenesis
ecDNAs are frequently detected in advanced human cancers, but their contribution to tumour initiation and progression remains a key unanswered question. To assess whether the engineered ecDNAs could contribute to tumorigenesis in an autochthonous context, we induced widespread circularization of the Mdm2ec allele directly in vivo by crossing Actin–Cre32 and Mdm2ec/+ mice (Extended Data Fig. 10a). We then leveraged the fact that in mice, MYC-driven hepatocellular carcinoma requires a second genetic event, such as inactivation of the p53 pathway33,34, and reasoned that this could be achieved by Mdm2 amplification through engineered ecDNAs. We delivered a MYC transgene to the livers of adult Actin–Cre Mdm2ec/+ and control (Actin–Cre) mice by hydrodynamic tail-vein injection (Fig. 5a). Although none of the control animals (n = 6) developed tumours, three of the six Actin–Cre Mdm2ec/+ mice developed multiple liver tumours within 6–18 weeks postinjection (Fig. 5b).
Extended Data Fig. 10. Characterization of Mdm2ec/+;Myctg liver tumours.
a. gDNA-PCR analysis with primers designed to detect the circularized allele (Primers C-B) and allele excision (Primers A-D) on the linear chromosome upon Cre-expression performed on DNA extracted from different tissues of Mdm2ec/+;Actin-Cre mouse (1 month old). Amplification of an unrelated genomic region is included as a PCR control. Quantification by digital droplet PCR (ddPCR) of the circularized allele (Probes C-B) and excision from the linear chromosome (Probes A-D) is shown at the bottom. Heart, kidney, large bowel, pancreas, and spleen of Mdm2+/+;Actin-Cre mouse are used as controls. b. Lower magnification of a representative H&E of Mdm2ec/+ liver tumour, showing the primary tumour is a poorly differentiated hepatocellular carcinoma (scale bar = 500 µm). c. Representative CK19 staining with a CK19+ bile duct, indicating tumours are negative for the cholangiocyte marker CK19 (scale bar = 100 µm, n = tumours from 3 mice). d. qPCR analysis of Mdm2 mRNA expression levels in Mdm2ec/+tumours compared to normal liver. Error bars: mean ± SD. N = 3 technical replicates. e. sWGS data from a representative Mdm2ec/+ tumour were analyzed using Amplicon Architect to identify structural variants. The structural variant plot reveals a structural variant closing the left-and-right endpoints of the amplified region forming an ecDNA-like cycle spanning the region flanked by the loxP sites. All tumours analyzed from three mice showed the same circular amplicon. f. sWGS analysis of individual liver tumours from three Actin-Cre;Mdm2ec/+ mice.
Fig. 5. An autochthonous mouse model of cancer harbouring engineered ecDNAs.
a, Schematics of the experimental strategy. A MYC-IRES-luciferase-encoding transposon and the Sleeping Beauty transposase were delivered by hydrodynamic tail-vein injection to six Actin–Cre Mdm2ec/+ and six Actin–Cre mice. b, Macroscopic appearance of the liver of a MYC-injected Actin–Cre Mdm2ec/+ mouse showing multiple nodules. c,d, Representative haematoxylin and eosin staining (c) and anti-Hnf4a immunostaining (d) of a liver lesion arising in Actin–Cre Mdm2ec/+ mouse. Arrowheads in d point to mitotic figures. Representative of eight tumour nodules in three mice. e, Metaphase spreads of cells obtained from a dissociated liver tumour showing multiple double-minute chromosomes (left panel) containing the Mdm2 locus (DNA FISH, right panel). Representative of three independent experiments f, DNA FISH on a tumour section using an Mdm2-specific probe showing amplification of the Mdm2 locus. g, Sections of normal liver and of a tumour nodule were stained by RNA FISH using a probe specific to the Mdm2 mRNA. A marked increase in Mdm2 signal (red) is evident in the tumour section. Repeated on three independent tumours. h, sWGS analysis of tumours from three mice, showing the presence of a focal amplification spanning the Mdm2ec region (blue shaded area). The computed average copy number of the amplicon is indicated on top of each track. Scale bars, 100 μm (c,d,g), 10 μm (e), 20 μm (f).
The tumours had features of poorly differentiated hepatocellular carcinomas, characterized by broadly expanded trabeculae surrounded by endothelial cells, frequent mitoses and apoptotic figures (Fig. 5c and Extended Data Fig. 10b). Immunostaining showed strong expression of hepatocyte nuclear factor 4 alpha (HNF4α) and lack of expression of cytokeratin 19 (Fig. 5d and Extended Data Fig. 10c). Crucially, metaphase spreads obtained from freshly dissociated tumour cells showed the presence of dozens of Mdm2-positive ecDNAs (Fig. 5e), a result that was confirmed by DNA FISH on tumour sections (Fig. 5f) and was accompanied by strong upregulation of the Mdm2 messenger RNA (mRNA) (Fig. 5g and Extended Data Fig. 10d). Finally, sWGS and Amplicon Architect analysis of DNA obtained from various tumours revealed marked focal amplification of the Mdm2ec region in all cases, confirming that Mdm2 amplification had been initiated by Cre-mediated circularization (Fig. 5h and Extended Data Fig. 10e,f).
Together, these results offer direct experimental evidence that engineered ecDNAs can contribute to tumour formation in vivo and validate an autochthonous mouse model of cancer driven by an engineered focal gene amplification.
Discussion
We have described a general strategy to engineer focal gene amplifications mediated by ecDNAs in cells and in mice. To showcase its flexibility, we generated human cancer cell lines with Cre-inducible ecDNAs that can then be tracked using fluorescent reporters encoded by the ecDNAs and the linear chromosome from which they originate. We applied a similar strategy to engineer Mdm2- and Myc-containing ecDNAs in mice and showed that these ecDNAs confer a fitness advantage on primary cells harbouring them, accumulate over time in vitro, and promote cell proliferation and immortalization. Finally, we demonstrated that engineered Mdm2-containing ecDNAs promote the oncogenic transformation of mouse fibroblasts ex vivo when combined with oncogenic RAS and lead to the development of hepatocellular carcinomas when combined with ectopic MYC expression in vivo.
These results have notable biological implications because they demonstrate that oncogene-containing ecDNAs can form and rapidly accumulate in primary non-transformed cells derived from all three embryonic layers. This is relevant because until now, oncogene-containing ecDNAs had been observed only in established cancer cell lines and primary tumours, not in primary cells. Analogously, previous attempts to induce the formation of ecDNAs using CRISPR–Cas9 (refs. 14,35) or through drug selection4,36 had been performed in cancer cell lines but not in primary cells or whole organisms. It will be important to determine whether all cell types are equally permissive to ecDNA formation and propagation and to systematically explore the impact of gene regulatory elements contained in the engineered ecDNAs on their propagation, accumulation and tissue specificity, as recently suggested by studies in human cancers17. Equally relevant is the demonstration that engineered Mdm2 ecDNAs can cooperate with MYC in promoting autochthonous hepatocellular carcinomas in mice. This result provides direct experimental evidence that oncogene-containing ecDNAs can contribute to tumour progression in vivo and paves the way for future studies exploring the role of the immune system in the development and maintenance of ecDNA-driven tumours.
Besides the biological insights gained in this study, we expect that the general strategy we have developed and the reagents we have generated will prove useful in dissecting various aspects of ecDNA biology that are unresolved at present. For example, the presence of selectable markers and fluorescence reporters in the engineered ecDNAs make them ideally suited to use in large-scale screens to identify factors that are required for ecDNA propagation and maintenance and unique vulnerabilities conferred on cancer cells by the presence of ecDNAs. The ability to engineer ecDNAs in primary cells will also facilitate investigation of the mechanisms underlying chromatin changes that have been recently described in cancer-associated ecDNAs and the role of DNA regulatory elements present in human ecDNAs that have been proposed to affect gene expression in trans11,18. We note that although in the present study we used the Cre–lox system to induce circularization, the same results can be achieved using other site-specific recombinases (FlpO or Dre)37, further expanding the potential applications of this strategy to model tumour evolution in vivo.
Despite these benefits, the approach described here has some limitations. First, although our strategy recapitulates the subset of ecDNAs generated by two double-stranded DNA breaks followed by recircularization, ecDNAs can also result from chromothripsis4, for which Cre-induced recombination is not an accurate proxy. Second, the efficiency of Cre-induced circularization is known to decrease as the distance between the loxP sites increases. For the ecDNAs generated in this study, we observed an efficiency of recombination of 7–25% upon a single Cre pulse in cells and up to 80% in vivo using constitutive Cre expression. Although the size of the ecDNAs we have engineered (approximately 1.3–1.7 Mbp) is well within the range of many naturally occurring ecDNAs, it is likely that the circularization efficiency will be lower when attempting to model larger ecDNAs (3 Mbp or more) or when temporally restricting Cre expression. Third, for the in vitro experiments, we used H2B–GFP as a reporter for the presence of ecDNAs. Although H2B–GFP has the advantage of labelling the chromatin, allowing direct visualization of ecDNAs during mitosis, GFP variants with shorter half-lives may be better suited to the study of ecDNA dynamics in cell populations and may better facilitate pooled CRISPR screens.
Of note, we have not yet observed the development of autochthonous tumours harbouring amplified Myc-containing ecDNAs in Mycec/+ mice in tissues where MYC overexpression readily induces tumour formation, even upon concomitant p53 inactivation (Supplementary Table 3). This cannot be explained by poor Cre-mediated circularization of the Mycec allele in vivo (Extended Data Fig. 3e,f), or by an inability of the Myc-containing ecDNAs to propagate, as they readily accumulated in vitro in several cell types upon Cre expression (Fig. 3 and Extended Data Fig. 5). This resistance to tumour formation upon engineered Myc amplification might be in part related to a previously described negative autoregulatory feedback loop whereby ectopic MYC expression strongly represses transcription of the endogenous MYC gene in non-transformed cells38–40. This autoregulatory loop could serve as a tumour suppressive barrier, moderating MYC expression despite increased MYC copy number. Our finding that Myc mRNA levels were upregulated less than one would expect on the basis of the observed increase in copy number in aNSC-containing engineered Myc ecDNAs (Fig. 3 and Extended Data Fig. 4f) is consistent with this hypothesis.
Finally, it is notable that widespread in vivo Mdm2 circularization alone—as seen in Actin–Cre Mdm2ec/+ mice—did not lead to spontaneous tumour formation. This is in contrast to findings reported in mice harbouring Mdm2 transgenes41 or targeted deletion of p53 (refs. 42,43) and could reflect a unique feature of ecDNAs compared with other classes of cancer-associated mutations: the formation of an extrachromosomal copy of an oncogene—either through Cre-mediated engineering or spontaneously—is not per se oncogenic, as gene copy number is not affected by circularization. It is only after multiple rounds of cell division with random segregation and selection that the circular amplicon can exert oncogenic functions. This underappreciated feature of focal oncogene amplifications is likely to have clinical implications and needs to be taken into account in the design of other mouse models of human cancers harbouring ecDNAs. We expect that the tools and genetically engineered mouse models described here will enable the scientific community to address this and other key questions concerning the biology of ecDNAs and their roles in tumour initiation and tumour progression.
Methods
Plasmids and viral vectors
Plasmids containing the circularization cassettes were generated using a NEBuilder HiFi DNA Assembly Cloning Kit (New England BioLabs, E5520S) and KLD Enzyme Mix (New England BioLabs, M0554S) and validated by Sanger sequencing. Purified, high-titre recombinant adenoviruses encoding Cre (AdCre) were purchased from ViraQuest (VQ-Ad-CMV-Cre; 1 × 1012 particles per millilitre; catalogue no. 091317) and University of Iowa (Ad5CAGCre; VVC-U of Iowa-8193, University of Iowa). pBABE-Puro and pBABE-Puro-HRASG12V plasmids were obtained from Addgene. For retrovirus production, HEK-293T cells were seeded in high-glucose Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) without antibiotics in 100 mm petri dishes. The next day, cells at approximately 60–70% confluence were transfected with 20 µg of retroviral vector carrying the HRASG12V cDNA or a control vector, 5 µg of packaging plasmid and 1 µg of envelope plasmid. After 24 h, the medium was replaced with DMEM with antibiotics. Cells were incubated for 48 h with two consecutive collections of the medium containing the retroviral particles at 24 and 48 h. Medium collected at 24 and 48 h was filtered using a 0.45 µm filter unit and used to transduce MEF cells. Retroviral infection was performed, incubating cells with viral supernatant supplemented with polybrene (0.2 µl ml−1; Millipore Sigma, TR-1003G).
Cell culture
The culture medium for HCT116 cells (ATCC, CCL-247) was McCoy’s 5a Medium Modified (Thermo Fisher Scientific, catalogue no. 16600108) supplemented with 10% heat-inactivated FBS (Sigma-Aldrich, F2442), penicillin–streptomycin (100 U l−1; Gemini Bio-Products, catalogue no. 400-109). For selection of targeted cells engineered with the circularization cassettes, the following antibiotics were used: puromycin (2 μg ml−1; Sigma-Aldrich, P9620), hygromycin B (200 μg ml−1; Thermo Fisher Scientific, catalogue no. 10687010). HEK-293T cells for viral vector production were cultured in high-glucose DMEM (4.5 g l−1; Thermo Fisher Scientific, catalogue no. 11995065) with 10% heat-inactivated FBS, l-glutamine (2 mM), and penicillin–streptomycin (100 U l−1).
MEFs with different genotypes were isolated from E13.5 mouse embryos and propagated in high-glucose DMEM supplemented with 10% heat-inactivated FBS, l-glutamine (2 mM) and penicillin–streptomycin (100 U l−1).
aNSCs were isolated and cultured following the protocol described by Ahmed et al.44. Upon isolation, aNSC were maintained as neurospheres and then allowed to attach to laminin-coated (Sigma-Aldrich, L2020) dishes in NeuroCult Stem Cell Basal Media with NeuroCult Proliferation Supplement (Mouse & Rat) (Stem Cells Technologies, catalogue no. 05702), 20 ng ml−1 EGF (Stem Cells Technologies, catalogue no. 78006), 10 ng ml−1 bFGF (Stem Cells Technologies, catalogue no. 78003), and 2 μg ml−1 heparin (Stem Cell Technologies, 07980).
Cerebellar stem cells were isolated from the cerebella of 5-day-old mice. Cerebella were digested using a Papain Dissociation system (Worthington, LK003150) to obtain a single-cell suspension. Cells were washed and suspended in flow buffer (phosphate-buffered saline (PBS) supplemented with 2% heat-inactivated FBS (Sigma-Aldrich, F2442), 2 mM EDTA and 25 mM HEPES (Thermo Scientific, catalogue no. 15630080)) and passed through a 40 µm cell strainer. Cells were stained for prominin-1 and lineage markers and sorted. Cerebellar stem cells (prominin-1-positive, lineage-negative) were allowed to attach to laminin-coated (Sigma-Aldrich, L2020) dishes in NeuroCult Stem Cell Basal Media with NeuroCult Proliferation Supplement (Mouse & Rat) (Stem Cells Technologies, catalogue no. 05702), 20 ng ml−1 EGF (Stem Cells Technologies, catalogue no. 78006), 10 ng ml−1 bFGF (Stem Cells Technologies, catalogue no. 78003) and 2 μg ml−1 heparin (Stem Cell Technologies, catalogue no. 07980).
Primary hepatocytes were isolated from the livers of adult mice. Following euthanasia, the liver was rapidly perfused with PBS, chopped into small pieces and digested in 3.42 mg ml−1 dispase II (Roche, catalogue no. 04942078001) and 1 mg ml−1 collagenase IV (Sigma-Aldrich, C5138) in DMEM. The hepatocyte fraction was isolated by centrifugation at 200g and plated on collagen-coated plates. Culture media for hepatocytes consisted of William’s E media (Gibco, A1217601) with supplements: 10% heat-inactivated FBS (Sigma-Aldrich, F2442), 10 mM nicotinamide, 2 mM glutamine, 0.1 mM dexamethasone, 1× ITS+ (Gibco, catalogue no. 41-400-045), 0.2 mM ascorbic acid, 20 mM HEPES (Thermo Scientific, catalogue no. 15630080), 1 mM sodium pyruvate, 14 mM glucose (Gibco, A2494001), penicillin–streptomycin (100 U l−1). Hepatocytes were subsequently immortalized by infection with Ad-Tbg-Cre (Addgene, 107787-AAV8).
All cells were negative for mycoplasma contamination. Cells were maintained in a humidified, 5% CO2 atmosphere at 37 °C.
In vivo tumorigenesis
For in vivo tumour formation assays, nude mice were injected in the flank with 400,000–1,000,000 pBABE-Puro-HRASG12V transduced MEFs of the indicated genotype. Mice were monitored every 2–3 days and euthanized when the tumour volume reached 2 cc.
ATAC-seq analysis
ATAC-seq libraries were generated and sequenced by the Memorial Sloan Kettering Cancer Center (MSKCC) genomic core. Fastq files from ATAC-seq and WGS were aligned to the mouse genome (mm10) using Bowtie2. Then, CNVkit (v.0.9.10) was applied to WGS bam files to identify copy number variations and ecDNA amplicon regions. To compare ATAC-seq signals between ecDNA amplicons and corresponding chromosomal regions, bamCoverage in deeptools (v.3.5.3) was used to calculate read counts with 10 kb bin size, and MACS (v.3.0.0b1) was used for peak calling. Read counts and peak numbers were normalized by segment length (10 kb), sequencing depth and copy number.
Homologous recombination in HCT116 cells
HCT116 cells were obtained directly from ATCC (ATCC, CCL-247). Cas9 protein, CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) were purchased from Integrated DNA Technologies and preassembled by incubation according to the manufacturer’s instructions.
The two circularization cassettes were introduced sequentially. The donor cassettes were amplified using primers containing an 80 nucleotide 5′ homology sequence to the desired targeting site. Then, 100–500 ng of gel-purified PCR products were transfected into 500,000 HCT116 cells plated the day before in a well of a six-well plate using Lipofectamine 3000 (Thermo Fisher, L3000008). Two hours later, the preassembled Cas9–crRNA–tracrRNA were introduced using Lipofectamine CRISPR-Max (Thermo Fisher, CMAX00008) following the manufacturer’s instructions. Selection with either puromycin (5′ cassette, 2 µg ml−1) or hygromycin (3′ cassette, 200 µg ml−1) was started 48 h after transfection. Surviving clones were isolated and screened by PCR, followed by Sanger sequencing to detect correct targeting.
Animals
All animal experiments were approved by the Institutional Animal Care and Use Committee of MSKCC. To generate the Mycec (JAX strain: 039221) and Mdm2ec (JAX strain: 039222) mouse strains in a C57BL/6J background, zygote electroporation based on published protocols45 was carried out by the MSKCC Mouse Genetics Core Facility using crRNAs experimentally validated in mouse embryonic stem cells. Briefly, multiple zygotes placed in an electrode chamber were subjected to electroporation at one time. Each electroporation mixture contained the 5′ (AGATGCGCACAGAAAAGTGG) and 3′ (ATCATGAGTTGAGTTCACTC) breakpoint targeting crRNAs (25 ng μl−1), S.p. Cas9 V3 protein (100 ng μl−1; IDT), and two single-stranded oligodeoxynucleotides of 159 bp with asymmetric homology arms (200 ng μl−1) in a solution of 0.1% polyvinyl alcohol. Electroporated zygotes were cultured in KSOM medium at 37 °C and 5% CO2 until the two-cell stage; after that, they were transferred to the oviducts of pseudopregnant females on the day of the vaginal plug. N0 animals generated from the zygotes were genotyped and Sanger sequenced to confirm insertion of both loxP sites. Double-targeted N0 mice were mated to C57BL/6J wild-type mice to generate Mycec/+ F1 progeny. Primers for genotyping are listed in Supplementary Table 4. The Mdm2ec mouse was also generated as described above, except that guide RNAs were used to target chr. 10: 116711442 (TCTTACAGCATACTACGGTC TGG) and chr. 10: 118002454 (TTCTGCGATTCGTTATGCGT AGG) for the 5′ and 3′ loxP insertion sites, respectively.
Liver tumours were generated by hydrodynamic tail-vein injection of a solution of sterile saline containing 25 µg of pT3-EF1a-MYC-IRES-Luc and 5 µg of transposase-encoding vector (SB13). In brief, a volume equivalent to 10% of the mouse’s body weight was injected through the tail vein using a 3 ml syringe with 26-gauge × 5/8-inch needle. A mix of male and female mice were used for these experiments. Mice were monitored for tumour development and euthanized at the humane end point. The CMV-SB13 and pT3-loxP-EF1a-MYC-IRES-Luc-loxP were gifts from A. Lujambio (Icahn School of Medicine at Mount Sinai). pT3-EF1a-MYC-IRES-Luc was generated by removing the loxP sites from pT3-loxP-EF1a-MYC-IRES-Luc-loxP. p53fl (B6.129P2-Trp53tm1Brn/J) (strain: 008462)46, Actin–Cre (B6N.FVB-Tmem163Tg(ACTB-Cre)2Mrt/CjDswJ (strain: 019099)32, Vil1–Cre (B6.Cg-Tg(Vil1-cre)1000Gum/J; strain: 021504)47, Atoh1–Cre (B6.Cg-Tg(Atoh1-cre)1Bfri/J; strain: 011104)48, Alb–Cre (B6.Cg-Speer6-ps1Tg(Alb-cre)21Mgn/J; strain: 003574)49, Mx–Cre (B6.Cg-Tg(Mx1-cre)1Cgn/J; strain: 003556)20 and Nestin–Cre (B6.Cg-Tg(Nes-cre)1Kln/J)50,51 mouse strains were obtained from the Jackson Laboratory. Outbred athymic nude mice (stock: 007850) were purchased from the Jackson Laboratory.
Antibodies and immunoblots
Cells were lysed in Laemmli buffer, supplemented with protease (cOmplete; Roche, COEDTAF-RO) and phosphatase (EDTA-free Protease Inhibitor Cocktail; Roche, PHOSS-RO) inhibitors. Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and analysed by western blotting using standard procedures. After protein transfer, the nitrocellulose membranes (Bio-Rad, catalogue no. 1704271) were blocked by incubation with LICOR Intercept (TBS) blocking buffer. The following primary antibodies were used: anti-vinculin (1:5,000; Millipore, MAB3574), anti-c-MYC (1:1,000; Cell Signaling, D84C12), anti-MDM2 (1:1000; Cell Signaling, 51541), anti-α-tubulin (1:5000; Cell Signaling, 3873), anti-p53 (1:1000; Leica Biosystems, P53-PROTEIN-CM5) and anti-p21 (1:1000; Cell Signaling, 64016). The following secondary antibodies were used: IRDye 800 anti-rabbit (LICOR, 926-32213) and IRDye 680 anti-mouse (LICOR, 926-68072). Images were acquired using an Odyssey Imaging System (LICOR).
Flow cytometry and cell sorting
Flow cytometry analysis of ecMDM2 and invMDM2 HCT116 cells was performed following collection of cells in a single-cell suspension. Cells were resuspended in flow buffer (PBS supplemented with 2% heat-inactivated FBS (Sigma-Aldrich, F2442), 2 mM EDTA, and 25 mM HEPES (Thermo Scientific, catalogue no. 15630080)) and passed through a 40 µm cell strainer to remove clumps. DAPI (Thermo Scientific, D1306) staining was used to discriminate between live and dead cells. Flow cytometry analyses were performed on LSR Fortessa (BD Biosciences) instruments. Cells were first gated on the basis of size (forward scatter, FSC) and density and/or granularity (side scatter, SSC), excluding debris and doublets; DAPI staining was used to gate the live fraction. Then, polygonal or quadrant gates were applied to isolate the population of interest on the basis of GFP and mScarlet expression. The same protocol was used to sort ecMDM2 and invMDM2 HCT116 double-positive cells using FACSAria (BD Biosciences) and FACSymphony (BD Biosciences) instruments.
For isolation of cerebellar stem cells, cells were incubated for 1 h with Cd133 (prominin-1) monoclonal antibody (13A4)-FITC (1:100; Thermo Scientific 311-1331-80), Cd81 monoclonal antibody (Eat2)-PE (1:50; Thermo Scientific, MA517941), anti-O4-PE (1:50; Miltenyi Biotec., 130-117-507) and anti-PSA-NCAM-PE (Miltenyi Biotec. 130-117-394, 1:50). After staining, cells were washed and sorted using a BD FACSAria III instrument (BD Biosciences) according to prominin-1-positive and lineage-negative (Cd81−; O4−; PSA/NCAM−) marker expression. The gating strategy is provided in Supplementary Fig. 1, which contains the uncropped gel images.
Tissue processing and immunohistochemistry
Subcutaneous tumours and livers were fixed for 24 h in 10% formalin and then transferred to 70% ethanol for at least 24 h before paraffin embedding and sectioning. Unstained 5 μm tissue sections were deparaffinized in Histo-Clear II (Electron Microscopy Sciences, catalogue no. 64110-04) and rehydrated with graded alcohols to distilled water. Endogenous peroxidase activity was inactivated using 3% H2O2 (MP Biomedicals, catalogue no. 194057) for 15 min. Tissue slides were pretreated for antigen retrieval by heating in Antigen Unmasking Solution, Citrate-Based (Vector Laboratories H-3300) using a pressure cooker. Non-specific protein interactions were blocked in 10% normal serum for 30 min. Slides were incubated in primary antibody overnight at 4 °C at the following dilutions: HNF4α (1:500; CST, 3113T); CK19 (1:500; Abcam, AB52625). VectaStain ABC-HRP Rabbit IgG (Vector Laboratories, PK-6101) was used as the secondary antibody, with incubation according to the manufacturer’s instructions. ImmPACT DAB (Vector Laboratories SK-4105) was used as a substrate. Haematoxylin (Vector Laboratories, H-3401) was used as the counterstain. Cover slides were mounted with VectaMount Express Mounting Medium (Vector Laboratories, H-5700). Bright-field images were obtained using a Zeiss Axiocam microscope. Haematoxylin and eosin slides were examined by C.A. and C.S., board-certified pathologists with extensive expertise in human sarcomas and liposarcomas (C.A.) and human liver cancers (C.S.).
RNA extraction and qPCR with reverse transcription
Total RNA was isolated using an RNeasy Mini Kit (QIAGEN, catalogue no. 74106) according to the manufacturer’s instructions. After treatment with DNAse I (Ambion, AM2222), 1 μg of purified RNA was retrotranscribed with oligos d(T)18 using a SuperScript IV First-strand System (Invitrogen, catalogue no. 18091050). For qPCR with reverse transcription (RT–qPCR), an aliquot of the RT reaction was analysed with PowerUp SYBR Green (Applied Biosystems, A25777) and a QuantStudio 6 Flex real-time PCR system (Applied Biosystems). Target transcript levels were normalized to those of the indicated reference genes. The expression of each gene was measured in at least three independent experiments.
Milademetan dose–response curve
A total of 1,500 cells were seeded in each well of a 96-well plate, with 100 µl of complete medium per well. Cells were allowed to adhere for 12 h in the incubator before the treatment was initiated. Milademetan (MedChemExpress, HY-101266) doses ranging from 20 to 10,240 nM were prepared, along with a vehicle control at the highest treatment dose. The doses were added to the respective wells in a volume of 100 µl, ensuring that the cells received milademetan doses ranging from 10 to 5,120 nM. After a 72 h incubation period, cell viability was determined using the Cell-TiterGlo assay (Promega, G7570), following manufacturer’s instructions. Briefly, the assay reagent was added to the wells, and the plates were gently agitated to ensure thorough mixing. The luminescence signal was then measured using a Synergy 2 plate reader (Biotek) according to the manufacturer’s guidelines.
Gene copy number assay
Myc, MDM2 and Mdm2 gene amplification was evaluated using TaqMan Copy Number Assays (probe Mm00734221_cn, probe Hs02873318_cn and probe Mm00312030_cn) using the QuantStudio 6 Flex real-time PCR system. Briefly, genomic DNA was amplified using the TaqPath ProAmp Master Mix (Applied Biosystems, A30865) kit and following the supplier’s instructions. TaqMan Copy Number Reference Assay (Tfrc, 4458367 and TERT, 4403316) were used in duplex as references for mouse and human genomes, respectively. Relative copy number variations were calculated using the ddCt method.
Digital droplet PCR
Assays specific for the detection of Cre-mediated circularization and excision of the mouse Mycec or Mdm2ec allele were designed and ordered through Bio-Rad (Supplementary Table 4). Cycling conditions were tested to ensure optimal annealing and extension temperatures, as well as optimal separation of positive and empty droplets. Optimization was done with a known positive control. After PicoGreen quantification, 0.4–9.0 ng genomic DNA was combined with locus-specific primers, FAM- and HEX-labelled probes, BamHI and digital PCR Supermix for probes (no dUTP). All reactions were performed on a QX200 digital droplet PCR system (Bio-Rad, catalogue no.: 1864001), and each sample was evaluated in technical duplicates using Ptger2 as a reference. Reactions were partitioned into a mean of approximately 21,000 droplets per well using a QX200 droplet generator. Emulsified PCRs were run on a 96-well thermal cycler, using cycling conditions identified during the optimization step (95 °C 10 min; 40 cycles of 94 °C 30 s and 56 °C 1 min; 98 °C 10 min; 4 °C hold). Plates were read and analysed with QuantaSoft software (Bio-Rad; v.1.7) to assess the number of droplets positive for the gene of interest, reference gene, both or neither.
RNA-seq and gene set enrichment analysis
Paired-end RNA libraries were sequenced by the MSKCC integrated genomics core. Reads were mapped to mm10 using the STAR aligner52, and differential gene expression was calculated using the DESeq2 R package53. For gene set enrichment analysis, genes were ranked on the basis of their moderated P value [−log10(adjusted P) × sign(log2(fold change))], and gene sets were obtained from the HALLMARK msigdb pathways database. Enrichment was calculated using the fgsea R package.
Shallow whole-genome sequencing
The sWGS was carried out by the Integrated Genomics Operation core at MSKCC. Briefly, after PicoGreen quantification and quality control by Agilent BioAnalyzer, 100 ng of genomic DNA was sheared using a LE220-plus Focused-ultrasonicator (Covaris, catalogue no. 500560), and sequencing libraries were prepared using a KAPA Hyper Prep Kit (Kapa Biosystems, KK8504) with eight cycles of PCR. Samples were run on a NovaSeq 6000 in a PE100 run, using a NovaSeq 6000 S4 Reagent Kit v.1.5 (200 cycles) (Illumina). Paired-end genomic DNA libraries were sequenced by the MSKCC integrated genomics core. Reads were mapped to mm10 or hg19 using the Bowtie2 aligner54. To calculate genome coverage and copy number changes, we used the QDNAseq R package55 with 15 kb bins. Plots were generated using the Gviz package56.
Amplicon Architect analysis
We used the AmpliconSuite-pipeline (v.0.1555.1, https://github.com/AmpliconSuite/AmpliconSuite-pipeline) to invoke AmpliconArchitect57 (v.1.3.r5) on a collection of copy-number seed regions generated using CNVkit58 (v.0.9.9), with default settings.
Metaphase chromosome spread analysis
Cells were incubated with KaryoMAX (catalogue no. 15212012; Thermo Fisher Scientific) treatment at 0.05 μg ml−1 for 1 h (mouse cells) or at 0.1 μg ml−1 for 1 h and 30 min (human cells). A single-cell suspension was then collected, washed in PBS, and treated with 75 mM KCl for 10 min at 37 °C. Samples were then fixed in ice-cold 3:1 methanol/glacial acetic acid (Carnoy’s fixative) for 20 min and washed a further three times with Carnoy’s fixative. Fixed cells were dropped on to a glass slide in a humidified chamber and counterstained with DAPI (Vector Laboratories, H-1800). Images were acquired with an AX10 Imager.Z1 Zeiss microscope through a ×63 objective lens. Zeiss Zen 2.3 Pro software was used for image acquisition. Fiji (v.2.0.0-rc-65/1.15w) was used for image analysis and for brightness and contrast adjustments. Fiji’s ‘Invert LUT’ and ‘Shadows’ postprocessing commands were sequentially applied to better visualize ecDNAs.
FISH analysis
DNA FISH was performed on fixed cells using a two-colour probe. MDM2 (green dUTP) and centromeric control (orange dUTP) FISH probes were purchased from Empire Genomics (SKU MDM2-CHR12). BAC clones containing the murine Myc locus (RP23-130M7, RP23-342F3 RP23-454G15) were labelled with red dUTP, and RP23-333G9 (15qA1) labelled with green dUTP served as the control. Myc (RP23-307D14-ORANGE) and centromeric control (RP23-333G9-GREEN) FISH probes were also purchased from Empire Genomics. All RP23 clones were purchased from the Roswell Park Cancer Institute Genomics Shared Resource.
Probe labelling, hybridization, posthybridization washing and fluorescence detection were performed according to procedures established at the Molecular Cytogenetics Core Facility. Slides were scanned using a Zeiss Axioplan 2i epifluorescence microscope (Carl Zeiss Microscopy) equipped with Isis imaging software (MetaSystems Group Inc.) or Leica SP5 confocal microscope (Leica) with a ×63 objective. The entire hybridized area was scanned through a ×63 objective lens to assess the quality of hybridization and signal pattern. To the extent possible, apoptotic cells and bodies were excluded.
The BAC clone RP23-428D5 (BACPAC) was used for detection of the murine Mdm2 locus, and the BAC clone RP23-309H16 was used for the detection of murine 10qA1 (Cen10). Following inoculation, bacterial cells were pelleted, and BAC DNA was extracted using a NucleoBond Xtra BAC kit (Takara, catalogue no. 740436) as per the manufacturer’s instructions. The probe for Mdm2 was labelled with ChromaTide Alexa Fluor 568-5-dUTP (Thermo, C11399), and the Cen10 probe was labelled with ChromaTide Alexa Fluor 488-5-dUTP (Thermo, C11397) as per manufacturer’s instructions. Before hybridization, the labelled probe was preannealed with mouse COT-1 DNA in hybridization buffer (2× SSC, 50% formamide, 10% dextran sulfate) for 90 min at 37 °C. Hybridization of slides using preannealed probes was performed at 72 °C for 2 min, followed by 37 °C overnight. Posthybridization washes were conducted in 0.4× SSC/0.3% Igepal at 72 °C for 2 min, followed by 2× SSC at room temperature for 5 min. Slides were then rinsed briefly in water, air-dried, counterstained with DAPI (Thermo, D1306) and mounted with Prolong Diamond Antifade Mountant (Thermo, P36965). Images were acquired in the MSKCC Molecular Cytology Core using a Zeiss Imager equipped with a Zeiss AxioCam Mrm camera and a ×100 oil objective. Specimens for DNA FISH were embedded in optimal cutting temperature compound and stored at −80 °C before analysis.
RNA in situ hybridization
Mdm2 RNA FISH was performed in collaboration with the MSKCC Molecular Cytology Core. Briefly, paraffin-embedded tissue sections were cut at 5 μm and kept at 4 °C. Samples were loaded into a Leica Bond RX autostainer, baked for 30 min at 60 °C, dewaxed with Bond Dewax Solution (Leica, AR9222) and pretreated with EDTA-based epitope retrieval ER2 solution (Leica, AR9640) for 15 min at 95 °C. The mouse Mdm2 probe (Advanced Cell Diagnostics (ACD), catalogue no. 447648) was hybridized for 2 h at 42 °C. Mouse PPIB (ACD, catalogue no. 313918) and dapB (ACD, catalogue no. 312038) probes were used as positive and negative controls, respectively. The hybridized probes were detected using an RNAscope 2.5 LS Reagent Kit – Brown (ACD, catalogue no. 322100) according to the manufacturer’s instructions with the following modifications: DAB application was omitted and replaced with fluorescent CF594/tyramide (Biotium, B40953) for 20 min at room temperature. Images were acquired using a Zeiss Imager equipped with a Zeiss AxioCam Mrm camera, a ×20 air objective and a ×100 oil objective.
Statistics and reproducibility
Paired or unpaired Student’s two-tailed t-test and one-way ANOVA (corrected for multiple comparisons, Tukey test), were used to compare two or more groups, respectively, and to determine statistical significance (GraphPad Prism 9 and R software). Welch’s correction was used for populations with unequal variances. Unless otherwise indicated, the mean value and the standard deviation of each condition are shown. Differences were considered significant at P < 0.05.
Materials availability
Materials are available from the corresponding authors upon request. Plasmids containing the ‘circularization cassettes’ described in this paper are available through Addgene (Plasmids #219563, #219564, #219565). Mycec (strain: 039221) and Mdm2ec (strain: 039222) are available through the JAX repository (https://www.jax.org/).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-024-08318-8.
Supplementary information
Contains all uncropped gels images for the main figures and the extended data figures. It also includes gating strategies for all flow cytometry experiments.
Excel file containing RNA-seq differential gene expression in Mycec/+p53fl/fl versus p53fl/fl aNSCs 5 weeks after AdCre infection.
Excel file containing RNA-seq differential gene expression in AdCre-treated Mdm2ec/+HRASG12V versus p53fl/flHRASG12V MEFs.
Word document containing a summary of mouse strains generated to test the oncogenic potential of the Mycec allele in vivo. For the Mx1–Cre experiments, Cre expression was induced by intraperitoneal administration of pIpC. The numbers of mice in each cohort and the duration of follow-up are also indicated.
Excel file containing oligonucleotides, guide RNAs and PCR primers used in this study.
Acknowledgements
We thank the group of G. Lozano for sharing the Mdm2−/−p53−/− double-knockout MEFs used to validate antibodies for western blotting. This work was supported by grants from NIH-NCI (P30 CA008748 to the MSK Cancer Center, and R01 CA282913 to A.V.), the American Cancer Society Discovery Boost grant (A.V.), The Mark Foundation for Cancer Research ASPIRE grant (A.V.) and a rapid response grant from the Functional Genomics Initiative (A.V.). The research was also supported in part by grants U24CA264379 and R01GM114362 to V.B. D.P. was supported by an AIRC fellowship for Abroad, M.Z. is a recipient of the Paul Calabresi Career Development Award for Clinical Oncology from the Memorial Sloan Kettering K12 Clinical Translational Cancer Research Training Program, M.W. is supported by the Erwin Schrodinger Fellowship from the Austrian Science Fund (FWF, J4723), and M.A.Y. is supported by the Harold E. Varmus Fellowship in Cancer Biology and the Marie-Josée Kravis Women in Science Endeavor (WiSE) fellowship. K.M.G. was supported by an ETIUDA scholarship (no. 2018/28/T/NZ5/00467) funded by the National Science Center, Poland. This work was delivered as part of the eDyNAmiC team supported by the Cancer Grand Challenges partnership funded by Cancer Research UK (CGCATF-2021/100012 and CGCATF-2021/100025) and the National Cancer Institute (OT2CA278688 and OT2CA278635) (H.Y.C., P.S.M., V.B. and A.V.). We thank members of the Cancer Research Emphasis Think Tank, G. Lozano, T. Kelly, B. Stillman, P. Jallepalli, D. Remus and members of the Benezra, Joyner and Lowe laboratories for helpful discussion and advice. Figures 7a, 10a and 12a of the first response in the Peer Review reports were created using BioRender. We thank A. Drilon and M. Repetto for sharing unpublished data on ecDNA in human cancers.
Extended data figures and tables
Author contributions
A.V. conceived the project and designed the general strategy to generate ecDNAs in vitro and in vivo. R.G., M.A.Y., F.G. and A.V. generated and tested the circularization cassettes; K.M.G., T.M., D.P., M.L., M.A.Y. and A.V. generated and characterized the HCT116 invMDM2 and ecMDM2 cell lines. R.G. generated the Mycec mouse; D.P. characterized the Mycec mouse and performed the experiments in aNSCs, with assistance from H.M.H.N., A.A.Z. and M.L.; M.Z. generated the Mdm2ec mouse and, together with A.V., M.A.Y. and M.W., characterized it and studied the effects of ecDNA formation in MEFs. M.A.Y. and M.Z. generated and characterized the liver cancer model. D.P. and M.Z. generated the RNA-seq data and, together with A.V., analysed them. A.V. analysed the sWGS datasets; Y.C.F. tested the effects of milademetan on Mdm2ec/+-transformed MEFs and performed the PCR with reverse transcription; and G.L.R. and Z.J. performed the western blots. C.R.A. analysed the tumours generated from Mdm2ec/+ cells; J.L., B.D. and V.B. performed the Amplicon Architect analysis, Y.P., Z.Z., S.Z., P.S.M., E.G.S. and H.Y.C. analysed the chromatin accessibility datasets. C.M., K.C. and M.O. genotyped and maintained the mouse strains. C.R.A. and C.S. analysed the histology of sarcomas and liver cancers, respectively. D.P., M.Z. and A.V. wrote the manuscript with assistance and comments from all coauthors.
Peer review
Peer review information
Nature thanks Eric Holland and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
All datasets have been deposited and made publicly available: sWGS data (GSE264005), bulk RNA-seq data (GSE264003) and ATAC-seq data (GSE264240). AmpliconArchitect outputs for murine Mdm2-containing ecDNA (https://ampliconrepository.org/project/649b3c097dc54138a9d391b3) and murine Myc-containing ecDNA (https://ampliconrepository.org/project/64839f137dc54138a9d39122) are available through the AmpliconRepository online repository (https://ampliconrepository.org/). Materials are available from the corresponding authors upon request.
Competing interests
H.Y.C. is a cofounder of Accent Therapeutics, Boundless Bio, Cartography Biosciences and Orbital Therapeutics and an advisor for 10x Genomics, Arsenal Biosciences, Chroma Medicine, Exai Bio and Spring Discovery. P.S.M. is a cofounder of, chairs the scientific advisory board of and has equity interest in Boundless Bio. V.B. is a cofounder of, consultant for and SAB member of and has equity interest in Boundless Bio, Inc. and Abterra, Inc. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Davide Pradella, Minsi Zhang, Rui Gao, Melissa A. Yao
Extended data
is available for this paper at 10.1038/s41586-024-08318-8.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-024-08318-8.
References
- 1.Beroukhim, R. et al. The landscape of somatic copy-number alteration across human cancers. Nature463, 899–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yi, E., Chamorro Gonzalez, R., Henssen, A. G. & Verhaak, R. G. W. Extrachromosomal DNA amplifications in cancer. Nat. Rev. Genet.23, 760–771 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosswog, C. et al. Chromothripsis followed by circular recombination drives oncogene amplification in human cancer. Nat. Genet.53, 1673–1685 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Shoshani, O. et al. Chromothripsis drives the evolution of gene amplification in cancer. Nature591, 137–141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox, D., Yuncken, C. & Spriggs, A. I. Minute chromatin bodies in malignant tumours of childhood. Lancet1, 55–58 (1965). [DOI] [PubMed] [Google Scholar]
- 6.Wu, S., Bafna, V., Chang, H. Y. & Mischel, P. S. Extrachromosomal DNA: an emerging hallmark in human cancer. Annu. Rev. Pathol.17, 367–386 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noer, J. B., Horsdal, O. K., Xiang, X., Luo, Y. & Regenberg, B. Extrachromosomal circular DNA in cancer: history, current knowledge, and methods. Trends Genet.38, 766–781 (2022). [DOI] [PubMed] [Google Scholar]
- 8.Kim, H. et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat. Genet.52, 891–897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner, K. M. et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature543, 122–125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.deCarvalho, A. C. et al. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat. Genet.50, 708–717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu, Y. et al. Oncogenic extrachromosomal DNA functions as mobile enhancers to globally amplify chromosomal transcription. Cancer Cell39, 694–707.e697 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi, E. et al. Live-cell imaging shows uneven segregation of extrachromosomal DNA elements and transcriptionally active extrachromosomal DNA hubs in cancer. Cancer Discov.12, 468–483 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nathanson, D. A. et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science343, 72–76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange, J. T. et al. The evolutionary dynamics of extrachromosomal DNA in human cancers. Nat. Genet.54, 1527–1533 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu, S. et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature575, 699–703 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu, S., Bafna, V. & Mischel, P. S. Extrachromosomal DNA (ecDNA) in cancer pathogenesis. Curr. Opin. Genet. Dev.66, 78–82 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Morton, A. R. et al. Functional enhancers shape extrachromosomal oncogene amplifications. Cell179, 1330–1341.e1313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung, K. L. et al. ecDNA hubs drive cooperative intermolecular oncogene expression. Nature600, 731–736 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luebeck, J. et al. Extrachromosomal DNA in the cancerous transformation of Barrett’s oesophagus. Nature616, 798–805 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn, R., Schwenk, F., Aguet, M. & Rajewsky, K. Inducible gene targeting in mice. Science269, 1427–1429 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Abremski, K. & Hoess, R. Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J. Biol. Chem.259, 1509–1514 (1984). [PubMed] [Google Scholar]
- 22.Lengauer, C., Kinzler, K. W. & Vogelstein, B. Genetic instability in colorectal cancers. Nature386, 623–627 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Haupt, Y., Maya, R., Kazaz, A. & Oren, M. Mdm2 promotes the rapid degradation of p53. Nature387, 296–299 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Kubbutat, M. H., Jones, S. N. & Vousden, K. H. Regulation of p53 stability by Mdm2. Nature387, 299–303 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Oliner, J. D., Saiki, A. Y. & Caenepeel, S. The role of MDM2 amplification and overexpression in tumorigenesis. Cold Spring Harb. Perspect. Med.6, a026336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, S. N., Roe, A. E., Donehower, L. A. & Bradley, A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature378, 206–208 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Montes de Oca Luna, R., Wagner, D. S. & Lozano, G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature378, 203–206 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Song, K. et al. Plasticity of extrachromosomal and intrachromosomal BRAF amplifications in overcoming targeted therapy dosage challenges. Cancer Discov.12, 1046–1069 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell88, 593–602 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Binh, M. B. et al. MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well-differentiated and dedifferentiated liposarcoma subtypes: a comparative analysis of 559 soft tissue neoplasms with genetic data. Am. J. Surg. Pathol.29, 1340–1347 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Gounder, M. M. et al. A first-in-human phase I study of milademetan, an MDM2 inhibitor, in patients with advanced liposarcoma, solid tumors, or lymphomas. J. Clin. Oncol.41, 1714–1724 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewandoski, M., Meyers, E. N. & Martin, G. R. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb. Symp. Quant. Biol.62, 159–168 (1997). [PubMed] [Google Scholar]
- 33.Moon, H., Park, H. & Ro, S. W. c-Myc-driven hepatocarcinogenesis. Anticancer Res.41, 4937–4946 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Molina-Sanchez, P. et al. Cooperation between distinct cancer driver genes underlies intertumor heterogeneity in hepatocellular carcinoma. Gastroenterology159, 2203–2220.e2214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moller, H. D. et al. CRISPR-C: circularization of genes and chromosome by CRISPR in human cells. Nucleic Acids Res.46, e131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoff, Von et al. Hydroxyurea accelerates loss of extrachromosomally amplified genes from tumor cells. Cancer Res.51, 6273–6279 (1991). [PubMed] [Google Scholar]
- 37.Weng, W., Liu, X., Lui, K. O. & Zhou, B. Harnessing orthogonal recombinases to decipher cell fate with enhanced precision. Trends Cell Biol.32, 324–337 (2022). [DOI] [PubMed] [Google Scholar]
- 38.Penn, L. J., Brooks, M. W., Laufer, E. M. & Land, H. Negative autoregulation of c-myc transcription. EMBO J.9, 1113–1121 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grignani, F. et al. Negative autoregulation of c-myc gene expression is inactivated in transformed cells. EMBO J.9, 3913–3922 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sivak, L. E. et al. Autoregulation of the human N-myc oncogene is disrupted in amplified but not single-copy neuroblastoma cell lines. Oncogene15, 1937–1946 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Jones, S. N., Hancock, A. R., Vogel, H., Donehower, L. A. & Bradley, A. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc. Natl Acad. Sci. USA95, 15608–15612 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donehower, L. A. et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature356, 215–221 (1992). [DOI] [PubMed] [Google Scholar]
- 43.Jacks, T. et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol.4, 1–7 (1994). [DOI] [PubMed] [Google Scholar]
- 44.Ahmed, A., Isaksen, T. J. & Yamashita, T. Protocol for mouse adult neural stem cell isolation and culture. STAR Protoc.2, 100522 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashimoto, M., Yamashita, Y. & Takemoto, T. Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse. Dev. Biol.418, 1–9 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Marino, S., Vooijs, M., van Der Gulden, H., Jonkers, J. & Berns, A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev.14, 994–1004 (2000). [PMC free article] [PubMed] [Google Scholar]
- 47.Madison, B. B. et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem.277, 33275–33283 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Matei, V. et al. Smaller inner ear sensory epithelia in Neurog1 null mice are related to earlier hair cell cycle exit. Dev. Dyn.234, 633–650 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Postic, C. et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem.274, 305–315 (1999). [DOI] [PubMed] [Google Scholar]
- 50.Giusti, S. A. et al. Behavioral phenotyping of Nestin-Cre mice: implications for genetic mouse models of psychiatric disorders. J. Psychiatr. Res.55, 87–95 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Tronche, F. et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet.23, 99–103 (1999). [DOI] [PubMed] [Google Scholar]
- 52.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scheinin, I. et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res.24, 2022–2032 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hahne, F. & Ivanek, R. Visualizing genomic data using Gviz and Bioconductor. Methods Mol. Biol.1418, 335–351 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Deshpande, V. et al. Exploring the landscape of focal amplifications in cancer using AmpliconArchitect. Nat. Commun.10, 392 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Talevich, E., Shain, A. H., Botton, T. & Bastian, B. C. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput. Biol.12, e1004873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gorkin, D. U. et al. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature583, 744–751 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contains all uncropped gels images for the main figures and the extended data figures. It also includes gating strategies for all flow cytometry experiments.
Excel file containing RNA-seq differential gene expression in Mycec/+p53fl/fl versus p53fl/fl aNSCs 5 weeks after AdCre infection.
Excel file containing RNA-seq differential gene expression in AdCre-treated Mdm2ec/+HRASG12V versus p53fl/flHRASG12V MEFs.
Word document containing a summary of mouse strains generated to test the oncogenic potential of the Mycec allele in vivo. For the Mx1–Cre experiments, Cre expression was induced by intraperitoneal administration of pIpC. The numbers of mice in each cohort and the duration of follow-up are also indicated.
Excel file containing oligonucleotides, guide RNAs and PCR primers used in this study.
Data Availability Statement
All datasets have been deposited and made publicly available: sWGS data (GSE264005), bulk RNA-seq data (GSE264003) and ATAC-seq data (GSE264240). AmpliconArchitect outputs for murine Mdm2-containing ecDNA (https://ampliconrepository.org/project/649b3c097dc54138a9d391b3) and murine Myc-containing ecDNA (https://ampliconrepository.org/project/64839f137dc54138a9d39122) are available through the AmpliconRepository online repository (https://ampliconrepository.org/). Materials are available from the corresponding authors upon request.