Abstract
TAP/hNXF1 is a key factor that mediates general cellular mRNA export from the nucleus, and its orthologs are structurally and functionally conserved from yeast to humans. Metazoans encode additional proteins that share homology and domain organization with TAP/hNXF1, suggesting their participation in mRNA metabolism; however, the precise role(s) of these proteins is not well understood. Here, we found that the human mRNA export factor hNXF2 is specifically expressed in the brain, suggesting a brain-specific role in mRNA metabolism. To address the roles of additional NXF factors, we have identified and characterized the two Nxf genes, Nxf2 and Nxf7, which together with the TAP/hNXF1's ortholog Nxf1 comprise the murine Nxf family. Both mNXF2 and mNXF7 have a domain structure typical of the NXF family. We found that mNXF2 protein is expressed during mouse brain development. Similar to TAP/hNXF1, the mNXF2 protein is found in the nucleus, the nuclear envelope and cytoplasm, and is an active mRNA export receptor. In contrast, mNXF7 localizes exclusively to cytoplasmic granules and, despite its overall conserved sequence, lacks mRNA export activity. We concluded that mNXF2 is an active mRNA export receptor similar to the prototype TAP/hNXF1, whereas mNXF7 may have a more specialized role in the cytoplasm.
INTRODUCTION
TAP/hNXF1 is the key factor mediating the nuclear export of mRNAs (1–3), and its orthologs in Saccharomyces cerevisiae, Drosophila melanogaster and Caenorhabditis elegans were shown to be essential for general cellular mRNA export (4–6). Metazoans encode additional NXF-like proteins, which together with the TAP/hNXF1 orthologs comprise a family of proteins termed nuclear export factors (NXFs) that are evolutionarily conserved from yeast to humans. Besides sequence homology, the NXFs share the domain architecture, and therefore are thought, by analogy to TAP/hNXF1, to participate in mRNA metabolism (7). For the human hNXF2 and hNXF3 proteins, the mRNA export activity has been confirmed by mRNA export assays (6,8). In Drosophila, while the NXF1 ortholog dmNXF1 is essential (9), additional NXF proteins were shown to be nonessential for general mRNA export in cell culture, suggesting roles that are more specialized than that of dmNXF1 (10). We previously found that the C.elegans NXF1 homolog Ce-NXF1 is essential for mRNA export (4). Although the RNAi depletion of Ce-NXF2 was not lethal (5), this protein was recently implicated in the post-transcriptional regulation of tra-2 mRNA, which is required for female development (11). A human hNXF5 nullisomy was linked to mental retardation (12). Taken together, these observations suggest that while the TAP/hNXF1 orthologs are essential for general mRNA export in metazoan species, additional NXF family members have more specialized roles. To understand these roles, we studied the family of the mouse NXF proteins. Here, we describe the isolation and characterization of two additional mouse Nxf genes, Nxf2 and Nxf7.
MATERIALS AND METHODS
Isolation of cDNAs encoding mNXF2
Gene-specific primers were designed using a short region of identity between the mouse BAC clone RP23 65A22 and human TAPX2/hNXF2 cDNA (13). 5′ and 3′ rapid amplification of cDNA ends (RACEs) were performed on mouse brain Marathon-ready cDNA (Clontech) using outward primers complementary to this identity region. The RACE products were cloned into TOPO vector (Invitrogen), and the cDNAs were sequenced by BigDye™ Terminator Sequencing Kit (AB Applied Biosystems). The mouse Nxf2 and Nxf7 genes were identified and sequenced from the mouse BAC clones RP23 65A22 and BAC441N13, respectively.
Recombinant DNA
Expression plasmids for the green fluorescent protein (GFP)-tagged mNXF2 or mNXF7 proteins or their deletion mutants were generated by PCR amplification of corresponding cDNAs, and subsequent insertion of PCR products into the SacII and NheI sites in plasmid pCMV-GFPsg25 (14) in-frame with GFP. GFP-βGal-tagged mNXF2 and its mutants were generated by PCR amplification, followed by insertion of SacII- and XbaI-digested PCR fragment into SacII and NheI sites in plasmid pGFP-βgal (13), respectively. Histidine-tagged mNXF2 was constructed by PCR amplification of full-length mNXF2 coding sequence, then cloned into BssHII and XhoI sites in pCMV37M1-10D (15) replacing gag. Single or multiple point mutations, using 40–80Gβgal as a template, were generated by the QuickChange XL Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer's protocol. The nucleotide sequences of each mutant plasmid were confirmed by automated BigDye fluorescence sequencing. To generate glutathione S-transferase (GST)-tagged mNXF2, the coding region for amino acids 1–400 of mNXF2 was PCR amplified and subcloned into the pGEX-2T vector (Promega) for expression in Escherichia coli. Mouse p15/NXT1 expression plasmid was generated based on GenBank accession no. NM_019761, and the human p15-1 expression plasmid was obtained from E. Izaurralde.
Cell culture, transfection and microscopy
Human HeLa and 293 cells and mouse NIH3T3 and PA317 cells were transfected with FuGene 6 reagent (Roche) according to the manufacturer's protocol. All transfections for subcellular localization analysis were performed in 35 mm glass-bottom plates. Approximately 24 h post-transfection cells were fixed with 3.7% formaldehyde in phosphate-buffered saline. For some experiments, 20 h post-transfection, fresh media containing 30 nM leptomycin B, actinomycin D (2 μg/ml) or 5,6-dichlororibofuranosylbenzimidazole (DRB, 30 μg/ml) were added, and the cells were incubated for 4 h prior to fixation. As control for the drug-induced relocalization experiments, the same treatments were performed on cells expressing GFP-tagged HIV Rev protein. Microscopic analysis of GFP fluorescence was performed as described previously (13). Three-dimensional reconstruction of fluorescent images from confocal Z-stacks was performed using the maximum intensity projection renderer implemented in Imaris software (Bitplane).
Protein analysis
Chloramphenicol acetyltransferase (CAT) assays and luciferase measurements were performed as described previously (13). For western blot analyses, transfected cells were extracted in 500 μl of lysis buffer (0.5% Triton X-100 and 100 mM Tris–HCl, pH 7.4), separated on denaturing polyacrylamide gels and blotted onto nitrocellulose membrane. Polyclonal antisera against human TAPX2/hNXF2 and mouse mNXF2 were raised in rabbits using purified cellulose-binding domain (CBD) fusion protein containing amino acids 102–372 of TAPX2, or GST fusion containing full-length mNXF2 as immunogens. The TAPX2/hNXF2 antibodies were affinity purified on immobilized CBD-TAPX2 immunogen. For western blots, after probing with rabbit anti-TAPX2/hNXF2 or anti-mNXF2 antibody (1:1000) in 5% nonfat dry milk, and subsequent incubation with donkey anti-rabbit horseradish peroxidase-conjugated secondary antibody, immunoreactive proteins were visualized by enhanced chemiluminescence (ECL plus Western Blotting Detection System, Amersham) and autography. Pre-made western blots of multiple human tissues (GenoTech) and ‘mouse brain aging’ blots (RNAWAY) were quality controlled by the manufacturers to ensure equal loading and transfer efficiency.
In vitro protein binding assays
Reticulocyte-produced proteins were synthesized and metabolically labeled in coupled transcription/translation system (TNT T7 Coupled Reticulocyte Lysate System, Promega), using T7 promoter-containing PCR fragments as templates, and were adjusted with unprogrammed extract to equal molar concentrations. These stocks were used in the binding reactions that contained equimolar amounts of reticulocyte-produced proteins and 1–2 μg of E.coli-produced GST-tagged proteins that were immobilized on glutathione–Sepharose beads (Amersham). The binding was performed in 200 μl RBB buffer (15 mM HEPES, pH 7.9, 50 mM KCl, 0.1 mM EDTA and 0.2% Triton X-100) supplemented with 200 mM NaCl. Following incubation for 15 min at room temperature, the beads were pelleted and washed three times with binding buffer. Bound proteins were eluted by boiling in SDS–PAGE sample buffer, separated by SDS–PAGE and detected using Phosphoimager.
Biocomputing
Database similarity searches, multiple sequence alignments and phylogenetic analyses were performed using the standard programs of Genetics Computer Group package, with default parameters.
Nucleotide sequence accession
The sequences of mouse Nxf genes and cDNAs were submitted previously to GenBank under the accession numbers AY017476, AF490577 for Nxf2, and AY260550, AY266683 for Nxf7.
RESULTS
Identification of mouse NXF-related genes
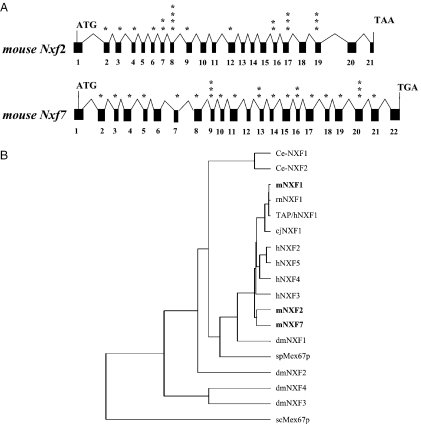
As a first step to study the role of the NXF family of proteins, we have identified the complete cDNAs as well as the exonic structures of the mouse Nxf2 (GenBank accession numbers AY017476 and AF490577) and Nxf7 genes (GenBank accession numbers AY260550 and AY266683). To achieve the identification of the mouse homologs of human TAP/hNXF1-related proteins, database searches for NXF-related mouse expressed sequence tags (ESTs) or cDNAs were employed, which revealed a homology in the mouse BAC clone RP23 65A22 to the human TAPX2/hNXF2 cDNA (13), a hNXF2 isoform. A full-length cDNA clone, termed Nxf2, was obtained using mouse brain cDNA library as template and the intron sequences were identified from the BAC clone. By comparison of the cDNA and genomic sequences, the exon and intron structure of the mouse Nxf2 gene was determined (Figure 1A). We found that the mouse Nxf2 gene is >17 kb in length and consists of 21 coding exons with 20 in-frame AUGs present in 11 exons. Comparison of our mNXF2 protein sequence with those published by Wang et al. (16) (GenBank accession no. NM_031259) and Jun et al. (12) (GenBank accession no. NP_112549) shows 99% identity with a single E347D amino acid change. Database searches further revealed a partial cDNA sequence of an additional Nxf-related gene on another BAC clone. By PCR amplification based on EST homology, a full-length cDNA corresponding to mouse Nxf7 was obtained and its exonic structure was determined (Figure 1A). We found that the mouse Nxf7 gene contains 22 exons, spans >14 kb (GenBank accession no. AY266683) and encodes a predicted protein of 620 amino acids. Our cDNA clone has a perfect sequence match with all the exonic sequences in the mouse Nxf7 genomic DNA. The proteins encoded by Nxf-a1 (GenBank accession no. AJ305317) and Nxf-a2 (GenBank accession no. AJ305318) have a high sequence homology to mNXF7, although both protein isoforms lack 110 amino acids at the N-terminus as compared with mNXF7. Comparison of the mNXF7 protein with NXF-a1 and NXF-a2 reveals the following changes: L354P and L361P (NXF-a2); the replacement of 121-STF-123 with two valine residues (NXF-a1 and NXF-a2); NXF-a1 is a splice variant (lacking exon 10), which results in an internal deletion of 36 amino acids (nt 271–306) and removes the sequence between L271 and L306.
Figure 1.
Mouse Nxf2 and Nxf7. (A) Exon–intron structure of mouse Nxf2 and Nxf7 genes. The cDNAs were isolated and the sequences and the genomic structure were determined. Exons are shown in black boxes. ATG, translational initiation codon; TAA and TGA, translation termination codons. Asterisks indicate location of in-frame ATG codons. (B) Dendrogram illustrating the NXF family of proteins. Abbrevations: rn, Rattus norvegicus; cj, Coturnix japonica; dm, Drosophila melanogaster; sp, Schizosaccharomyces pombe; sc, Saccharomyces cerevisiae. The mouse family members are shown in bold. (C) Multiple sequence alignment of human TAP/hNXF1 and mouse NXF proteins. The protein domains based on Herold et al. (7) are indicated. The identified domains mediating nuclear localization of human TAP/hNXF1 and mNXF2, localization to cytoplasmic granules of mNXF7 and the rim association of mNXF2 are underlined. RBD, RNA-binding domain; LRR, leucine-rich repeat; NTF2, nuclear transport factor (NTF2)-like domain; UBA-like, ubiquitin associated-like domain.
Taken together, we identified and isolated two genes termed mouse Nxf-2 and Nxf-7, whose sequences we have previously submitted to GenBank. Both genes are located on X chromosome, suggesting that they arose from a gene amplification, whereas Nxf1 is located on chromosome 19. Thus, the mouse NXF family consists of three members, whereas the human NXF family comprises five (7). Figure 1B shows a dendrogram of NXF family proteins from yeast to humans, illustrating significant homology both across and within species. On this tree, the mouse mNXF1 clusters with the other TAP/hNXF1 orthologs, from birds to humans, as expected (7). Interestingly, the ‘additional’ mouse factors mNXF2 and mNXF7 are close together and further cluster with the ‘additional’ human factors. We then performed a more detailed comparison of the mouse NXF family with TAP/hNXF1 protein, a prototype NXF factor (Figure 1C). While the mouse mNXF1 and the human TAP/hNXF1 share 90% identity, the comparison of mNXF1 to mNXF2 and mNXF7 shows reduced homologies of 47 and 49% amino acid identity, respectively. Despite this, the mNXF2 and mNXF7 proteins are predicted to share the domain organization with the human TAP/hNXF1 protein (7), including a non-canonical RNP-type RNA-binding domain (RBD), the leucine-rich repeats (LRRs), the NTF2-like domain (mediating interactions with p15/NXT1) and the ubiquitin associated-like domain (UBA-like) that is part of nucleoporin-binding region. mNXF2 also has an insertion of 5 tandem 12 amino acid imperfect repeats located at the C-terminus of its LRR domain, as previously noted (12), but its role was not further investigated.
Expression of mNXF2 protein in the brain
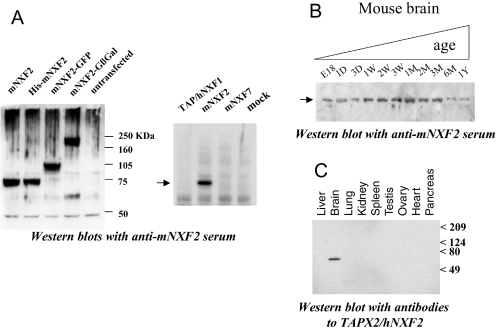
To study the mNXF2 protein expression, we generated an antiserum specific against GST-tagged mNXF2 in rabbits. Western blot analysis showed that the antiserum recognized untagged, His-tagged, GFP-tagged or GFP-βGal-tagged mNXF2 protein (Figure 2A, left panel), whereas it did not react with proteins from untransfected human 293 cells. In addition, it did not cross-react with the human TAP/hNXF1 or mouse mNXF7 proteins (Figure 2A, right panel). The untagged mNXF2 protein migrated with an apparent molecular mass of ∼75 kDa.
Figure 2.
Specific expression of NXF-related proteins in brain tissue. (A) Western immunoblot analysis of 293 cells transfected with untagged and tagged (HIS, GFP and Gβgal) mNXF2 (left panel) and different NXF expression plasmids (right panel) using a rabbit anti-mNXF2 antiserum. (B) Western blot analysis of mouse brain tissue using monospecific rabbit anti-mNXF2 antiserum. Pre-made ‘mouse brain aging’ blots (RNAWAY) contained normalized amounts of whole brain lysates from fetus to 1-year-old as indicated. (C) Western blot analysis of human tissues using the monospecific antiserum to human TAPX2/hNXF2. Pre-made blots (GenoTech) contained normalized amounts of proteins from the indicated tissues.
It has been previously reported that the mouse NXF-related mRNAs can be detected using RT–PCR in the brain (12). Using western immunoblot analysis, we found that the mNXF2 protein can also be detected in the mouse brain at different stages of development (Figure 2B). While the mNXF2 protein could be detected in a brain sample from an 18-week mouse embryo (E18), its level of expression is slightly increased at 1 week after birth and kept constant for ∼3 months and is still detectable, although at lower levels, after the age of 6 months. We were unable to address the expression of mNXF7 protein because of the lack of specific antibodies.
Similarly, we used a monospecific rabbit antiserum generated to human TAPX2 that represents an isoform of hNXF2 (13), to probe a human multiple tissue blot. This analysis revealed that TAPX2/hNXF2 was specifically expressed in the brain (Figure 2C), but could not be detected in other tissues examined, such as testis, where mRNA was readily detectable (data not shown). Taken together, these results confirmed that NXF-related proteins, such as the human and mouse NXF2, are expressed in the brain, suggesting the roles in brain-specific mRNA metabolism.
Distinct subcellular localization of mNXF2 and mNXF7
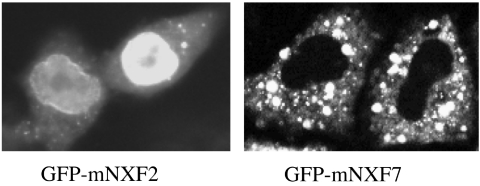
We next studied the subcellular localization of mNXF2 and mNXF7 upon transfection of human HeLa cells with plasmids expressing GFP-tagged fusion proteins. Figure 3 shows that mNXF2 localizes to the nucleus, but is excluded from the nucleolus, accumulates at the nuclear rim and is present in the cytoplasm. Its localization is similar to that observed for the human TAP/hNXF1 (13,17,18). In contrast, mNXF7 is localized exclusively to the cytoplasm (Figure 3), where it accumulates in granules. Similar localization patterns of mNXF2 and mNXF7 were found in human 293 cells as well as in mouse NIH3T3 and PA317 cells (data not shown); thus, the distinct subcellular localization is independent of the tested cell lines. The localization of both proteins was not affected in the presence of Leptomycin B, which excludes a role of CRM1 in protein export. Also, the presence of Actinomycin D or DRB did not alter the localization of mNXF2 and mNXF7 (data not shown), indicating that their nucleocytoplasmic trafficking is transcription-independent, while it affected the localization of HIV-1 Rev as expected (19).
Figure 3.
Distinct subcellular localization of mNXF2 and mNXF7 proteins. HeLa cells were transfected with GFP-tagged mNXF2 and mNXF7 expression plasmids, as indicated and the proteins were visualized in living cells. The images were obtained by fluorescent microscopy (Axiovert135TV, Zeiss) and by the use of a CCD camera and processed using IPLab Spectrum software (13). Similar results were obtained in numerous experiments and are typical of the vast majority of expressing cells in each individual experiment. Representative cells are shown.
Identification of the active nuclear localization signal of mNXF2
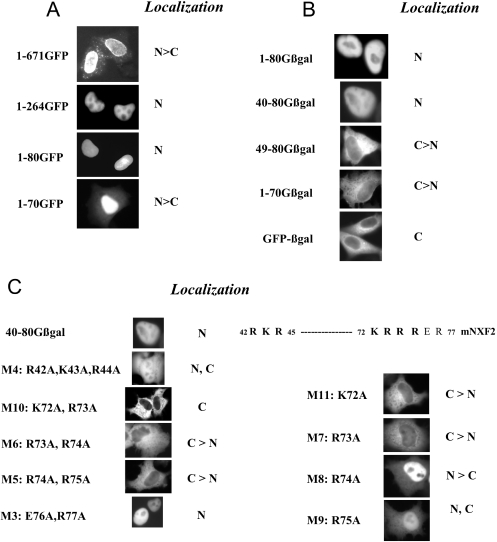
We next determined the nuclear localization signal (NLS) of mNXF2 using GFP-tagged deletion mutants (Figure 4A). We found that mutant 1–264 which lacks 407 amino acids from the C-terminus (Figure 4A) lost the nuclear rim association as expected, because it lacks the conserved UBA-like domain (see Figure 1C). Mutant 1–80GFP still localized to the nucleus (Figure 4A, 1–80GFP), whereas further deletion to amino acid 70 resulted in cytoplasmic accumulation of the mutant protein (Figure 4A, 1–70GFP). This suggests a NLS located between amino acid 1 and 80 at the N-terminus.
Figure 4.
Identification of NLS of mNXF2. (A and B) HeLa cells were transfected with plasmids producing mNXF2 and N- and C-terminal deletions fused to GFP (A) or GFP-βgal (B) and the proteins were visualized as described in Figure 2. The localization of the proteins is indicated. N, nucleus; C, cytoplasm. (C) Fine mapping of the NLS. Alanine substitution in 40–80Gβgal generated a series of mutants and the mutant proteins are visualized. The protein sequence containing the NLS regions is shown and the critical residues are indicated in bold.
Since GFP is a small protein, a GFP-tagged small polypeptide may localize to the nucleus due to passive diffusion. To distinguish passive diffusion from active import, we used GFP-β-galactosidase (Gβgal) fusion protein as a tag (13). The Gβgal fusion protein is localized to the cytoplasm because the fusion protein has a higher molecular mass, and neither GFP nor βgal contains an active nuclear import signal (Figure 4B). In contrast, insertion of the N-terminal 80 residues of mNXF2 (1–80Gβgal) conferred nuclear localization on the otherwise cytoplasmic GβGal, demonstrating that mNXF2 contains an active nuclear import signal. We also tested this nuclear import signal using GST–GFP (13) as a tag (data not shown). The GST–GFP fusion protein is localized to the cytoplasm because GST can form a dimer, and neither GST nor GFP contains an active nuclear import signal. Fusion of GST–GFP to amino acids 2–80 or to amino acids 40–80 of mNXF2 resulted in nuclear localization (data not shown).
We further mapped the minimal NLS within amino acids 1–80 of mNXF2 and examined the localization of several deletion mutants (Figure 4B). For the N-terminal deletion mutants, we found that deletion of amino acids 1–39 did not alter nuclear localization of the mutant protein (40–80Gβgal), whereas further deletion to residue 49 (49–80Gβgal) resulted in cytoplasmic accumulation of the fusion protein. Deletion of 10 amino acids from the C-terminus (1–70Gβgal) also abolished nuclear localization of the protein. These results demonstrate that amino acids 40–49 and amino acids 71–80 define the N- and C-terminal boundaries of the mNXF2 NLS. Taken together, these results demonstrate that amino acids 40–80 of mNXF2 are necessary and sufficient to promote nuclear import and this peptide constitutes the minimal NLS.
Fine mapping of the mNXF2 NLS
Inspection of the N-terminal region (40-QGRKRGVNY-48) and the C-terminal region (71-MKRRRERCSY-80) of mNXF2 revealed stretches of basic amino acids. To further study their role in NLS function, alanine substitutions were introduced within 40–80Gβgal (Figure 4C). In mutant M4, changes of three basic amino acids in the N-terminal part of the NLS domain (R42A, K43A, R44A) led to an impairment of the NLS and significant cytoplasmic accumulation of the mutant protein. Double alanine substitutions in the C-terminal part of the NLS as in mutants M10 (K72A, R73A), M6 (R73A, R74A), and M5 (R74A, R75A) resulted in loss of NLS function, whereas mutant M3 (E76A, R77A) has a fully functional NLS. Using single alanine substitutions, we further found that the NLS function is abrogated in M11 (K72A) and M7 (R73A) or significantly affect in M9 (R75A), but the NLS function is only slightly affected in M8 (R74A). Taken together, our results demonstrate that the mNXF2 core NLS consists of two short stretches of basic amino acids (amino acids 42–44 and amino acids 72–75), which are essential for maintaining full NLS activity. Similarly, we previously found that the NLS of the human TAP/NXF1 is bipartite consisting of a C-terminal region contributing (Figure 1C, dotted line) and an N-terminal region (solid line) being essential for NLS activity (13). In both the human TAP/hNXF1 and mNXF2, the core NLS is composed of few basic residues located toward the N-terminus of the proteins.
Association of mNXF2 and mNXF7 with nuclear envelope
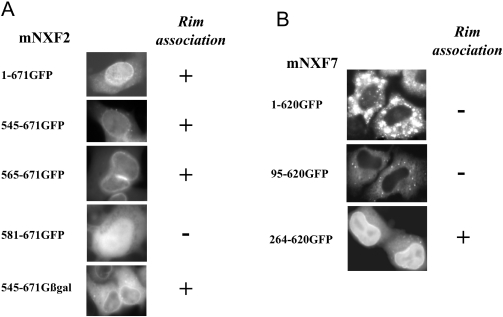
As previously shown for human TAP/hNXF1, mNXF2-GFP protein also localized to the nuclear envelope (Figures 3 and 5A). Fusion of the C-terminal portion 545–671 to GFP-βgal led to the accumulation of the mutant protein in the cytoplasm and the nuclear periphery (Figure 5A, 545–671Gβgal), whereas fusion to GFP resulted in diffusion of the protein to the nucleus and accumulation at the nuclear rim. Deletion mutant 565–671GFP still accumulated at the nuclear rim, whereas further deletion to amino acids 581 abolished rim association (581–671GFP). Rim association of the mutants 545–671GFP and 565–671GFP was not affected by digitonin treatment prior to fixation (data not shown), indicating strong interactions with the nuclear envelope. Therefore, mNXF2 contains a signal for rim association at the C-terminus, which shows conservation in location and sequence with other human NXF proteins (7) (Figure 1C).
Figure 5.
Association of mNXF2 and mNXF7 with nuclear envelope. HeLa cells were transfected with plasmids producing the indicated GFP-tagged mNXF2 and mNXF7 deletion mutants and the images were analyzed as described in Figure 2.
Interestingly, we found that although GFP-tagged full-length mNXF7 or the N-terminal deletion 95–620GFP did not show apparent nuclear rim association, the deletion of the N-terminal 263 residues led to the accumulation of the protein in the nucleus and to its association with the nuclear rim (Figure 5B, 264–620GFP). Inspection of mNXF7 protein sequence confirmed the conservation of the C-terminal portion containing the nuclear rim association determinants (Figure 1C). By analogy, we previously reported that deletion of the N-terminal NLS of TAP/hNXF1 resulted in a mutant protein, which was found to accumulate at the nuclear rim and the nucleoplasm, suggesting that the ability of NXF to associate with mobile factors of the nuclear pore complex (NPC) facilitates its nuclear import even in the absence of the active N-terminal NLS (13,17,18). We conclude that although mNXF7 protein shares the key signals with the prototype TAP/hNXF1, other regions within the protein are responsible for its distinct localization.
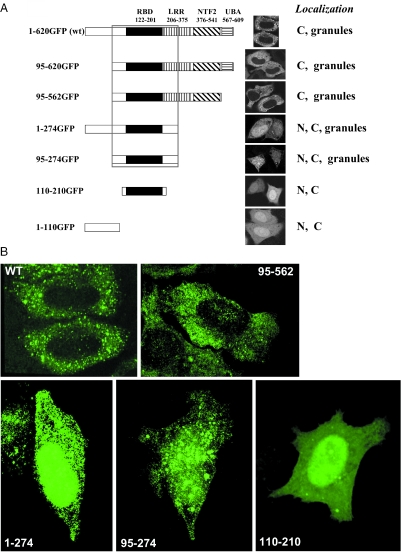
Mapping of mNXF7 domain responsible for granule formation
A key property distinguishing mNXF7 from the other members of the NXF family is its localization to cytoplasmic granules. Figure 6A shows a series of N- and C-terminal deletion mutants of mNXF7 designed to identify the domains responsible for granule formation. Removal of part of the LRR, the NTF2 and the UBA-like domains as well as the N-terminal 95 residues did not abolish the localization to granules. The minimal region necessary and sufficient for granule formation spans amino acids 95–274, and further truncation to amino acids 110–210 resulted in uniform localization of the mutant (Figure 6). The localization of some mutants was further verified using 3D rendering, an approach that allows to better distinguish the granular and homogeneous localization patterns. As shown in Figure 6B, this analysis indeed led to better visualization of the granular pattern of the wild-type mNXF7 and its mutants encompassing amino acids 95–562, 1–274 and 95–274, whereas the mutant spanning amino acids 110–210 showed mostly homogeneous localization, confirming the granule phenotypes presented in Figure 6A. We note that the granule localization determinant in mNXF7 corresponds to part of TAP/hNXF1's ‘substrate-binding domain’ through which it interacts with nuclear export cofactors and assembles with mRNP complexes (2,7,20–22).
Figure 6.
Identification of signal mediating granule localization of mNXF7. (A) A panel of N- and C-terminal deletion mutants of GFP-tagged mNXF7 deletion mutants was analyzed upon transfection of HeLa cells. N, nucleus; C, cytoplasm. Confocal images of GFP fluorescence in the mid-sections through the nuclei. (B) Three-dimensional rendering of GFP images from confocal Z-stacks.
mNXF2, but not mNXF7, can promote mRNA export
To test whether mNXF2 or mNXF7 play a role in mRNA export, we used the CAT reporter plasmid pDM128. Plasmid pDM128 contains CAT coding sequence and the Rev response element located within the env intron of HIV-1, flanked by a splice donor and acceptor sites (23). In 293 cells, such transcripts are retained in the nucleus and, therefore, produce only background levels of CAT enzyme, whereas coexpression of export activators, such as HIV-1 Rev or TAP/hNXF1, leads to the stimulation of CAT production. We have previously demonstrated that TAP/hNXF1 indeed enhances the nuclear export of CAT transcripts (22); therefore, CAT production directly reflects the nuclear export of this reporter mRNA.
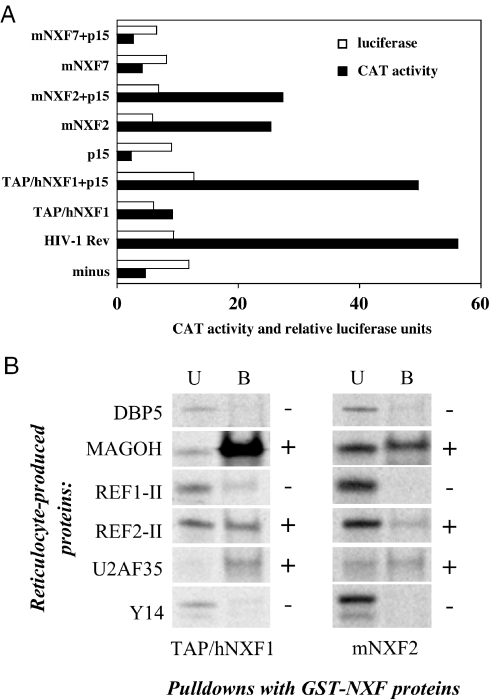
As a positive control, we coexpressed pDM128 with HIV-1 Rev and found a strong (∼10-fold) activation of CAT production (Figure 7A), as expected (23). We further showed that the presence of TAP/hNXF1 resulted in an ∼2-fold activation, whereas cotransfection with its cofactor, the human p15/NXT1, led to ∼10-fold activation, in agreement with previous observations (7,24), whereas the presence of p15/NXT1 alone had no effect. Using this assay, we found that cotransfection of mNXF2 alone yielded a significant ∼5-fold activation of CAT expression, demonstrating that mNXF2 is a bonafide mRNA export factor. Cotransfection with human or mouse p15/NXT1 did not significantly affect mNXF2 activity. This is in contrast to the human TAP/hNXF1, which depends on the exogenous p15/NXT1 for maximal activity in this assay. We hypothesize that the mNXF2 has a higher affinity to the NPC similar to a TAP/hNXF1 mutant having a duplication of its nucleoporin-binding sites (25) or to endogenous p15/NXT1; therefore, it can act independently of exogenous p15/NXT1. In contrast, mNXF7 failed to activate CAT production both in the absence and presence of p15/NXT1 (Figure 7A), consistent with the lack of mRNA export function of this cytoplasmic protein. This finding suggests that the mouse NXF7 protein functions at post-export steps in the mRNA metabolism. Similar results were obtained in mouse PA317 cells (data not shown).
Figure 7.
mNXF2 is an active mRNA export factor. (A) Human 293 cells were transfected with the cat reporter pDM128 either alone, in the presence of HIV-1 rev expression plasmid or in the presence of the indicated untagged NXF producing plasmids. As indicated, the transfection mixtures contained NXF expression plasmids together with human p15/NXT1 or p15/NXT1 alone. Two days later, the cells were harvested and the CAT production, measured as percentage of chloramphenicol acetylation, is shown by filled bars. Expression of the cotransfected luciferase expression plasmid was analyzed for each plate and the relative luciferase values are shown in open bars. A typical experiment is shown. (B) mNXF2 binds to TAP/hNXF1 export cofactors. Bacterially produced GST-tagged NXF proteins were immobilized on glutathione–Sepharose beads and used in pull-down assays with reticulocyte-produced, metabolically labeled factors (shown to the left). The bound (B) and 1:100 aliquots of the unbound (U) fractions were separated on SDS–PAGE and visualized on Phosphoimager.
To understand the mechanism of mNXF2's nuclear export activity, we studied its interactions with Y14/MAGOH, the REF proteins and U2AF35, which are known to bind to TAP/hNXF1 directly and are thought to facilitate the addition of TAP/hNXF1 to its export substrates (22,26–30). The bacterially produced recombinant proteins spanning the substrate-binding domains of mNXF2 and TAP/hNXF1 (amino acids 1–400) were immobilized and used in pull-down assays together with metabolically labeled binding factors. As a negative control, we used the mRNA export factor DBP5 (31), which does not bind TAP/hNXF1 directly. Figure 7B shows that MAGOH, REF2-II and U2AF35 bound readily to both TAP/hNXF1 and mNXF2, whereas the binding of Y14 and DBP5 was low or undetectable. The observed binding of MAGOH, REF2-II and U2AF35 to TAP/hNXF1 is consistent with previous findings (22,26–30). Since the MAGOH subunit binds to TAP/hNXF1 much more avidly than Y14 (28), the lack of REF1-II and Y14 binding confirmed that only strong interactions were revealed under our assay conditions. However, we cannot exclude that the reticulocyte-produced REF1-II and/or Y14 were misfolded or lacked proper post-translational modifications, and this possibility was not further addressed. Although TAP/hNXF1 and mNXF2 proteins were used at similar amounts, the overall binding efficiency of the tested proteins to TAP/hNXF1 was higher. This effect could be attributed to a better folding of this recombinant protein, and it was not further examined. In conclusion, this study shows that mNXF2 is an mRNA export factor sharing the localization and functional properties with TAP/hNXF1.
DISCUSSION
In this report, we present the identification and functional characterization of the mouse Nxf family genes and their corresponding proteins. Since some mammalian NXF factors are believed to be expressed specifically in the brain, and because human hNXF5 nullisomy is linked to mental retardation (12), we wished to understand the possible roles of such proteins. We have chosen to study the Nxf gene family in mouse, because this species is widely used as a model to study human neurological disease, yet possesses only two Nxf1-related genes.
The mouse mNXF2 and the human hNXF2 are expressed in the brain
Previously, mRNA analysis was used as evidence of brain-specific expression for the NXF factors, such as mNXF2, mNXF7, and hNXF5. Using hNXF2 as a model, we found that this approach can lead to gross overestimation of the predicted protein expression levels, because of high abundance of aberrant, non-coding splice products in certain tissues such as testis (A. Zolotukhin, S. Lindtner, S. Smulevitch and B.K. Felber, unpublished data). We therefore sought to verify the brain-specific expression of NXF family factors at the protein level and raised antibodies to mNXF2 and its human counterpart, TAPX2/hNXF2. By using these antibodies, we show here that the expression of the human TAPX2/hNXF2 protein is restricted to the brain (Figure 2C), and that the mouse mNXF2 expression levels in the brain are developmentally regulated (Figure 2B), suggesting brain-related function(s) of these proteins.
mNXF2 is an active mRNA export receptor, with properties similar to those of TAP/hNXF1
We show here that mNXF2 behaves like a typical NXF factor, since it shows mRNA export activity, localizes mostly to the nucleus and the nuclear envelope, and contains active nuclear localization and NPC-association signals that are positioned similar to those of TAP/hNXF1. We also show that mNXF2 and the human TAP/hNXF1 share the ability to interact with mRNA splicing/export cofactors, such as MAGOH, REF and U2AF35, strongly suggesting a shared nuclear export mechanism. While this manuscript was under review, another group (32) has reported that mNXF2, but not mNXF7, binds in vitro to the TAP/hNXF1 export cofactor p15, further supporting our conclusions. The few differences we observed between mNXF2 and the prototype export factor, TAP/hNXF1, include (i) a more pronounced cytoplasmic and nuclear envelope localization of mNXF2, whereas TAP/hNXF1 is mostly found in the nucleoplasm (ii) mNXF2 shows a more relaxed requirement for the cofactor p15/NXT1 in the CAT nuclear export assays used.
We believe that these small differences do not likely account for a brain-specific role of mNXF2 that is distinct from that of TAP/hNXF1. Rather, we propose that mNXF2 functions differently from TAP/hNXF1 due to its restricted substrate specificity. In this model, mNXF2, unlike the promiscuous TAP/hNXF1, only associates with a specific subset of transcripts in the brain, leading to their regulation during development. Further work is required to characterize such transcripts and the responsible determinants within mNXF2.
mNXF7 is atypical of NXF family
Unexpectedly, we found that mNXF7 is a cytoplasmic protein that is completely excluded from the nucleus. However, these localization data at steady-state do not rule out that mNXF7 can enter the nucleus, especially considering the presence of a cryptic NLS (see below). Independent of its fusion to detection tags (such as GFP and HA), species and cell lines, mNXF7 forms granules in the cytoplasm and is targeted to these granules via a region that spans amino acids 95–274. Interestingly, when devoid of this region, mNXF7 assumes the subcellular localization that is typical of a generic NXF factor, revealing nuclear import and nuclear envelope localization determinants. Apparently, these determinants are overridden in the wild-type mNXF7 protein by a potent granule localization signal. However, the conservation of active, NXF-like signals within mNXF7 points to their functional importance and suggests that the role of this protein may require nuclear entry and NPC binding. In agreement with its unusual localization, mNXF7 did not show nuclear export activity in CAT assays, probably due to its absence from or short dwelling time in the nucleus. We cannot exclude though, that mNXF7 may have an export activity that is strictly specific toward a subclass of transcripts and is not revealed using the cat mRNA reporter. In summary, we propose that mNXF7 plays a highly specialized role(s) in the cytoplasm that is distinct from that of other NXFs studied so far. Based on homology to TAP/hNXF1 and shared domain organization, it is likely that mNXF7 is engaged in mRNA metabolism. At present, we do not know whether such proposed activities include the mRNA nuclear export per se, or are restricted to post-export events. Understanding the precise biological role of mNXF7 and the underlying molecular mechanisms remains a goal for future studies.
Acknowledgments
We thank S. Lockett, J. Brenner, E. Cho and S. Costes for help with confocal microscopy, P. Aplan, G. Dreyfuss, E. Izaurralde and F. Stutz for their generous gifts of plasmids and T. Jones for editorial assistance. We are grateful to our summer students P. Sood and A. Witten, and our Werner H. Kirsten Student Intern program recipients D. Waddelow and C. Jodrie for their contributions. Funding to pay the Open Access publication charges for this article was provided by NCI.
Conflict of interest statement. None declared.
REFERENCES
- 1.Grüter P., Tabernero C., von Kobbe C., Schmitt C., Saavedra C., Bachi A., Wilm M., Felber B.K., Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 2.Braun I., Rohrbach E., Schmitt C., Izaurralde E. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 1999;18:1953–1965. doi: 10.1093/emboj/18.7.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erkmann J.A., Kutay U. Nuclear export of mRNA: from the site of transcription to the cytoplasm. Exp. Cell Res. 2004;296:12–20. doi: 10.1016/j.yexcr.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Tan W., Zolotukhin A.S., Bear J., Patenaude D.J., Felber B.K. The mRNA export in Caenorhabditis elegans is mediated by Ce-NXF-1, an ortholog of human TAP and Saccharomyces cerevisiae Mex67p. RNA. 2000;6:1762–1772. doi: 10.1017/s1355838200000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segref A., Sharma K., Doye V., Hellwig A., Huber J., Luhrmann R., Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herold A., Teixeira L., Izaurralde E. Genome-wide analysis of nuclear mRNA export pathways in Drosophila. EMBO J. 2003;22:2472–2483. doi: 10.1093/emboj/cdg233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herold A., Suyama M., Rodrigues J.P., Braun I.C., Kutay U., Carmo-Fonseca M., Bork P., Izaurralde E. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol. Cell. Biol. 2000;20:8996–9008. doi: 10.1128/mcb.20.23.8996-9008.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J., Bogerd H.P., Wang P.J., Page D.C., Cullen B.R. Two closely related human nuclear export factors utilize entirely distinct export pathways. Mol. Cell. 2001;8:397–406. doi: 10.1016/s1097-2765(01)00303-3. [DOI] [PubMed] [Google Scholar]
- 9.Wilkie G.S., Zimyanin V., Kirby R., Korey C., Francis-Lang H., Van Vactor D., Davis I. Small bristles, the Drosophila ortholog of NXF-1, is essential for mRNA export throughout development. RNA. 2001;7:1781–1792. [PMC free article] [PubMed] [Google Scholar]
- 10.Herold A., Klymenko T., Izaurralde E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA. 2001;7:1768–1780. [PMC free article] [PubMed] [Google Scholar]
- 11.Kuersten S., Segal S.P., Verheyden J., LaMartina S.M., Goodwin E.B. NXF-2, REF-1, and REF-2 affect the choice of nuclear export pathway for tra-2 mRNA in C.elegans. Mol. Cell. 2004;14:599–610. doi: 10.1016/j.molcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Jun L., Frints S., Duhamel H., Herold A., Abad-Rodrigues J., Dotti C., Izaurralde E., Marynen P., Froyen G. NXF5, a novel member of the nuclear RNA export factor family, is lost in a male patient with a syndromic form of mental retardation. Curr. Biol. 2001;11:1381–1391. doi: 10.1016/s0960-9822(01)00419-5. [DOI] [PubMed] [Google Scholar]
- 13.Bear J., Tan W., Zolotukhin A.S., Tabernero C., Hudson E.A., Felber B.K. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol. Cell. Biol. 1999;19:6306–6317. doi: 10.1128/mcb.19.9.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stauber R., Gaitanaris A.S., Pavlakis G.N. Analysis of trafficking of Rev and transdominant Rev proteins in living cells using green fluorescent protein fusions: transdominant Rev blocks the export of Rev from the nucleus to the cytoplasm. Virology. 1995;213:439–449. doi: 10.1006/viro.1995.0016. [DOI] [PubMed] [Google Scholar]
- 15.Schneider R., Campbell M., Nasioulas G., Felber B.K., Pavlakis G.N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P.J., McCarrey J.R., Yang F., Page D.C. An abundance of X-linked genes expressed in spermatogonia. Nature Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 17.Truant R., Kang Y., Cullen B.R. The human tap nuclear RNA export factor contains a novel transportin- dependent nuclear localization signal that lacks nuclear export signal function. J. Biol. Chem. 1999;274:32167–32171. doi: 10.1074/jbc.274.45.32167. [DOI] [PubMed] [Google Scholar]
- 18.Bachi A., Braun I.C., Rodrigues J.P., Pante N., Ribbeck K., von Kobbe C., Kutay U., Wilm M., Gorlich D., Carmo-Fonseca M., Izaurralde E. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Agostino D.M., Ciminale V., Pavlakis G.P., Chieco-Bianchi L. Intracellular trafficking of the human immunodeficiency virus type 1 Rev protein: involvement of continued rRNA synthesis in nuclear retention. AIDS Res. Hum. Retroviruses. 1995;11:1063–1071. doi: 10.1089/aid.1995.11.1063. [DOI] [PubMed] [Google Scholar]
- 20.Liker E., Fernandez E., Izaurralde E., Conti E. The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J. 2000;19:5587–5598. doi: 10.1093/emboj/19.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stutz F., Bachi A., Doerks T., Braun I.C., Seraphin B., Wilm M., Bork P., Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zolotukhin A.S., Tan W., Bear J., Smulevitch S., Felber B.K. U2AF participates in the binding of TAP (NXF1) to mRNA. J. Biol. Chem. 2002;277:3935–3942. doi: 10.1074/jbc.M107598200. [DOI] [PubMed] [Google Scholar]
- 23.Hope T.J., Huang X., McDonald D., Parslow T.G. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc. Natl Acad. Sci. USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guzik B.W., Levesque L., Prasad S., Bor Y.C., Black B.E., Paschal B.M., Rekosh D., Hammarskjold M.L. NXT1 (p15) is a crucial cellular cofactor in TAP-dependent export of intron-containing RNA in mammalian cells. Mol. Cell. Biol. 2001;21:2545–2554. doi: 10.1128/MCB.21.7.2545-2554.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braun I.C., Herold A., Rode M., Izaurralde E. Nuclear export of mRNA by TAP/NXF1 requires two nucleoporin-binding sites but not p15. Mol. Cell. Biol. 2002;22:5405–5418. doi: 10.1128/MCB.22.15.5405-5418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strasser K., Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stutz F., Bachi A., Doerks T., Braun I., Seraphin B., Wilm M., Bork P., Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kataoka N., Diem M.D., Kim V.N., Yong J., Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon–exon junction complex. EMBO J. 2001;20:6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kataoka N., Yong J., Kim V.N., Velazquez F., Perkinson R.A., Wang F., Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 30.Blanchette M., Labourier E., Green R.E., Brenner S.E., Rio D.C. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol. Cell. 2004;14:775–786. doi: 10.1016/j.molcel.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 31.Schmitt C., von Kobbe C., Bachi A., Pante N., Rodrigues J.P., Boscheron C., Rigaut G., Wilm M., Seraphin B., Carmo-Fonseca M., Izaurralde E. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J. 1999;18:4332–4347. doi: 10.1093/emboj/18.15.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki M., Takeda E., Takano K., Yomogida K., Katahira J., Yoneda Y. Molecular cloning and functional characterization of mouse Nxf family gene products. Genomics. 2005;85:641–653. doi: 10.1016/j.ygeno.2005.01.003. [DOI] [PubMed] [Google Scholar]