Abstract
Cancer cells exhibit altered metabolism, often relying on glutamine (Gln) for growth. Breast cancer (BC) is a heterogeneous disease with varying clinical outcomes. We investigated the role of the amino acid transporter SLC1A5 (ASCT2) and its association with BC subtypes and patient outcomes. In large BC cohorts, SLC1A5 mRNA (n = 9488) and SLC1A5 protein (n = 1274) levels were assessed and correlated their expression with clinicopathological features, molecular subtypes, and patient outcomes. In vitro SLC1A5 knockdown and inhibition studies in luminal BC cell lines (ZR-75-1 and HCC1500) were used to further explore the role of SLC1A5 in Gln metabolism. Statistical analysis was performed using chi-squared tests, ANOVA, Spearman’s correlation, Kaplan–Meier analysis, and Cox regression. SLC1A5 mRNA and SLC1A5 protein expression were strongly correlated in luminal B, HER2 + and triple-negative BC (TNBC). Both high SLC1A5 mRNA and SLC1A5 protein expression were associated with larger tumour size, higher grade, and positive axillary lymph node metastases (P < 0.01). Importantly, high SLC1A5 expression correlated with poor BC-specific survival specifically in the highly proliferative luminal subtype (P < 0.001). Furthermore, SLC1A5 knockdown by siRNA or GPNA inhibition significantly reduced cell proliferation and glutamine uptake in ZR-75-1 cells. Our findings suggest SLC1A5 plays a key role in the aggressive luminal BC subtype and represents a potential therapeutic target. Further research is needed to explore SLC1A5 function in luminal BC and its association with Gln metabolism pathways.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-025-87292-1.
Keywords: SLC1A5, Breast cancer, Prognosis, Tumour marker
Subject terms: Cancer, Breast cancer, Cancer metabolism
Introduction
Deregulation of metabolic pathways has been readily accepted as one of the revised hallmarks of cancer, where cancer cells are able to regulate their metabolism to provide the energy and cellular building blocks required for growth1. Many cancer cells are highly reliant on amino acids for their growth, where endogenous synthesis may not provide rapidly proliferating cells with sufficient nutrients for nuclear biosynthesis. There is also increasing evidence that oncogenes and/or tumour suppressor genes can reprogramme tumour cell metabolism, including through direct regulation of the proline–glutamine regulatory axis by MYC and p532–4. This axis is the most important metabolic pathway in tumours, after which glucose, primarily glutamine, is used to replenish the tricarboxylic acid (TCA) cycle and supplies carbon and nitrogen for the synthesis of nucleotides, amino acids and glutathione. Indeed, some solid tumours exhibit glutamine-dependent cell growth or “glutamine addiction”5.
L-Glutamine (Gln) is a nonessential amino acid synthesised from glutamate and ammonia by glutamine synthetase (GS). Its utilisation, via reductive carboxylation, is necessary for sustained proliferation/survival and is linked with resistance to certain drugs6. A further role for Gln in cancer cell protein translation stems from observations that a master regulator of protein translation, rapamycin complex 1 (mTORC1), which regulates cell growth and protein translation, is also responsive to glutamine levels7,8. In BC, highly proliferative high-grade tumors, such as those in the triple negative (TN) class, have higher levels of glutamate and glutaminase (GLS) together with lower levels of Gln than low-grade tumours and normal breast epithelium9–13. The metabolic profiles of BC show that glutaminolysis metabolism is a key pathway for discriminating between TN and luminal/oestrogen receptor-positive (ER +) tumours14.
Solute carrier family 1 member 5 (SLC1A5)/ASC amino acid transporter 2 (ASCT2) is a cell surface sodium-dependent transporter that regulates the uptake of neutral amino acids, including Gln15,16. Inhibiting SLC1A5 resulted in reduced cellular proliferation in several cancer types, including non-small cell lung cancer17,18, renal cell carcinoma19, pancreatic carcinoma20, prostate carcinoma21 and melanoma22. In BC, it is reported to be highly expressed in HER2-positive but not in luminal A tumours10, although the uptake of Gln is only required for basal-like TNBC to sustain mTORC1 signalling8. SLC1A5 is also upregulated by MYC and downregulated by retinoblastoma (Rb)5,23.
With renewed interest in oncometabolism, metabolic enzymes are increasingly being targeted to improve therapeutic efficacy and reduce resistance. We therefore hypothesised that the proline-Gln axis is a key metabolic pathway regulated by MYC in BC, particularly because we have shown that proline dehydrogenase (PRODH) is downregulated2. This pathway could be used as a potential therapeutic target, particularly because the pleotropic MYC has thus far proven ineffective as a target. Therefore, the aim of this study was to assess SLC1A5 gene copy number and its expression at both the mRNA and protein levels in large and well-characterised annotated cohorts of BC patients combined with in vitro approaches to determine its biological, clinicopathological and prognostic value in different molecular classes, with particular interest in highly proliferative aggressive subgroups.
Materials and methods
SLC1A5 genomic and transcriptomic analysis
SLC1A5 gene expression was evaluated in a cohort of 1,980 BC samples using the Molecular Taxonomy of BC International Consortium (METABRIC) cohort24. RNA from fresh frozen tumours was subjected to transcriptional profiling using the Illumina HT-12 v3 platform, and the data were preprocessed and normalised as described previously24. In this cohort, patients with ER + and/or lymph node-negative tumours did not receive adjuvant chemotherapy, while those with ER-negative and/or lymph node-positive tumours received adjuvant chemotherapy. The relationships between copy number (CN) aberrations, both gains and losses, of SLC1A5 and TP53 mutations and SLC1A5 mRNA expression and patient outcome were investigated. The clinicopathological parameters for this dataset are summarised in Supplementary Table 1.
For the external validation of SLC1A5 mRNA expression, the BC Gene-Expression Miner v5.0 (n = 4712) (http://bcgenex.centregauducheau.fr) and Kaplan‒Meier plotter (n = 2,796) (http://kmplot.com) datasets were used.
SLC1A5 proteomic analysis
Patient cohort
Immunohistochemistry (IHC) was conducted using a large cohort of patients comprising a well-characterised consecutive series of early-stage (TNM stage I-III excluding T3 and T4 tumours) sporadic primary operable invasive BC. Patients (aged ≤ 70 years) who presented at Nottingham City Hospital between 1989 and 1998 (n = 1274) and were managed in accordance with a uniform protocol were enrolled in the Nottingham Tenovus Primary Breast Carcinoma Series. Patients’ clinical history, tumour characteristics, and information on therapy and outcomes were prospectively collected. Outcome data were collected on a prospective basis and included development and time to distant metastasis (DM) and BC-specific survival (BCSS). The BCSS was defined as the time (in months) from the date of primary surgery to the date of BC-related death. DM-free survival (DMFS) was defined as the time (in months) from the date of primary surgery to the appearance of DM. The clinicopathological parameters of this cohort of patients are summarised in Supplementary Table 1.
Tissue microarrays (TMAs) and IHC
Tumour samples (0.6 mm cores) were arrayed as previously described25. Immunohistochemical staining was performed on 4 μm sections using a Novolink polymer detection system (RE7150-K, Leica Biosystems, UK). Briefly, tissue slides were deparaffinised with xylene and rehydrated through 3 changes of alcohol. Heat-induced antigen epitope retrieval was performed in citrate buffer (pH 6.0) for 20 min using a microwave oven (Whirpool JT359 Jet Chef 1000 W). Endogenous peroxidase activity was blocked with a peroxidase block for 5 min. The slides were washed with Tris-buffered saline (TBS, pH 7.6), followed by the application of a protein block for 5 min. Following another TBS wash, a mouse monoclonal primary antibody against SLC1A5 (HPA035240, Sigma‒Aldrich, UK) diluted at 1:100 in Leica antibody diluent (RE7133 Leica, Biosysytems, UK) was applied, and the membrane was incubated for 30 min. The slides were washed with TBS, incubated with postprimary block for 30 min, and then washed with TBS. Novolink polymer was applied for 30 min. DAB working solution composed of 1:20 DAB chromogen in DAB substrate buffer was prepared and applied for 5 min. Slides were counterstained with Novolink haematoxylin for 6 min, dehydrated and coverslipped. Negative (omission of the primary antibody) and positive controls were included according to the manufacturer’s datasheet for each antibody.
The stained TMA sections were scored using high-resolution digital images (NanoZoomer; Hamamatsu Photonics, Welwyn Garden City, UK) at × 20 magnification. Assessment of SLC1A5 staining was based on a semiquantitative assessment of core digital images using a modified histochemical score (H-score), which includes an assessment of both the intensity of staining and the percentage of stained cells [29]. TMA cores were only assessed if the tumour burden was > 15%26,27.
Immunohistochemical staining and dichotomisation of the other biomarkers included in this study were performed according to previous publications2,25,28–38. ER and progesterone receptor (PR) positivity was defined as ≥ 1% staining. The immunoreactivity of HER2 in the TMA cores was scored using standard HercepTest guidelines (Dako). Chromogenic in situ hybridisation (CISH) was used to quantify HER2 gene amplification in borderline cases using the HER2 FISH pharmDx™ plus HER2 CISH pharmDx™ kit (Dako) and was assessed according to the American Society of Clinical Oncology guidelines. BC molecular subtypes were defined based on the IHC profile as follows: ER + /HER2- low proliferation (Ki67 < 10%); ER + /HER2- high proliferation (Ki67 ≥ 10%); and the HER2 + class, HER2 + regardless of the ER status and triple negative (TN) subtype, ER-, PR- and HER2-. The basal phenotype was defined as those tumours expressing cytokeratin (Ck) 5/6 and/or Ck14 and/or Ck17.
Cell culture
The luminal BC cell lines MDA-MB-175, ZR-75-1 (ER + /PR-/HER2-), T47D, MCF7, and HCC1500 (ER + /PR + /HER2-) were obtained from the American Type Culture Collection (Rockville, MD, USA). The cells were cultured in Roswell Park Memorial Institute (RPMI-1640) medium (Sigma‒Aldrich, UK) supplemented with 10% foetal bovine serum (Sigma‒Aldrich, UK). Mycoplasma testing was carried out on a routine basis using the MycoAlert Detection Kit (R&D Systems).
SLC1A5 inhibition and knockdown
SLC1A5 inhibition was achieved in ZR-75-1 and HCC1500 cells cultured in 96-well plates at a density of 1 × 103 cells per well. Cultures were treated with the SLC1A5 inhibitor gamma-l-glutamyl-p-nitroanilide (GPNA) (1 mM; Sigma‒Aldrich, UK).
SLC1A5 knockdown in ZR-75-1 and HCC1500 cells was performed via transfection of 2 × 105 cells/well in 6-well plates via the reverse transfection method with 100 pmol of siRNA (Thermo Fisher Scientific, UK) and 5 µl of Lipofectamine RNAiMAX (Thermo Fisher Scientific, UK) according to the manufacturer’s protocol. The sequence of the antisense siRNA used was 5’-AAAGAGUAAACCCACAUCCtc-3’. Scrambled siRNA was used alongside the experiment as a negative control. SLC1A5 knockdown was confirmed by Western blotting. Functional assays were carried out in the transfected and control cells 24 h after transfection and in cultured cells with or without GPNA.
Cell proliferation assay
Cells were seeded at a density of 1 × 103 cells per well in a 96-well plate in triplicate. MTS assays were conducted every day for 4 days to assess cell growth according to the manufacturer’s instructions (MTS CellTiter 96 Aqueous One Solution) (Promega, UK). The absorbance was recorded at 490 nm using a microplate reader (TECAN Infinite F50). The background absorbance from the empty wells was subtracted from that of the sample wells.
Glutamine uptake assay
A total of 5 × 104 cells were incubated with 3H-L-glutamine (250 µci; Perkin Elmer) in glutamine-free media (Sigma‒Aldrich, UK) for 30 min at 37 °C in the presence/absence of transfection or inhibitor. The cells were washed twice with Dulbecco’s phosphate-buffered saline (Sigma‒Aldrich, UK). The cell pellets were resuspended in glutamine-free media and loaded onto Luma plates (Perkin Elmer). Radioactivity was measured using a scintillation counter (PerkinElmer, USA).
Western blotting
Cells were harvested and lysed in lysis buffer. The samples were subjected to SDS‒polyacrylamide gel electrophoresis (PAGE) and transferred to PVDF membranes (Immobilon-FL). A 1:250 dilution of the primary SLC1A5 antibody and a 1:5000 dilution of the mouse monoclonal anti-β-actin primary antibody (A5441, Sigma‒Aldrich, UK) were used as loading controls. IRDye 800CW donkey anti-rabbit fluorescent secondary antibody (1:15,000 dilution) and IRDye 600RD donkey anti-mouse fluorescent secondary antibody (926–32,213 and 926–68,072, LI-COR Biosciences, UK) were used. A PageRuler Plus Prestained Protein Ladder (26,619, Thermo Fisher Scientific, UK) was used. An Odyssey Fc with Image Studio 4.0 was used to visualise the bands (LI-COR Biosciences).
Statistical analysis
Statistical analysis was performed using SPSS software (SPSS Inc., Chicago, IL, USA). The chi-squared test was used to evaluate the significance of associations with clinicopathological parameters. One-way ANOVA with the post hoc Tukey multiple comparison test and Spearman’s correlation coefficient were used for continuous data. The Kaplan–Meier method and Cox proportional hazards model were used to evaluate the prognostic results. Dichotomisation of SLC1A5 mRNA and SLC1A5 protein expression was performed using X-tile software (version 3.6.1, Yale University, USA) based on outcome prediction. A P value < 0.05 was considered to indicate statistical significance.
Results
SLC1A5 copy number and mRNA expression in breast cancer
SLC1A5 CN gains were observed in 3% and CN loss in 2% of invasive BC patients, whereas high SLC1A5 mRNA expression was observed in 61.4% of the tumours. Those with a CN loss of the SLC1A5 gene exhibited significantly lower SLC1A5 mRNA expression (P = 0.001; Fig. 1A). Conversely, tumours with CN gain of SLC1A5 did not show a corresponding increase in SLC1A5 mRNA levels.
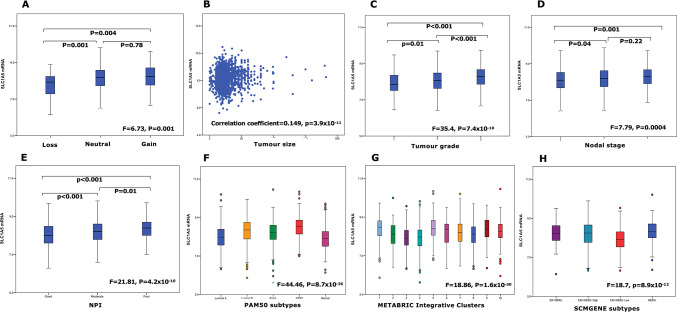
Fig. 1.
SLC1A5 mRNA expression and its association with invasive breast cancer clinicopathological parameters and molecular subtypes in the METABRIC cohort: (A) SLC1A5 copy number variation, (B) tumour size, (C) tumour grade, (D) lymph node stage, (E) Nottingham Prognostic Index (NPI), F) PAM50 subtypes (G) METABRIC Integrative Clusters, (H) SMCGENE subtypes.
Clinicopathological parameters and molecular subtypes
High SLC1A5 mRNA expression was significantly associated with larger tumour size, higher grade and nodal stage together with poor NPI (all P < 0.001, Fig. 1B–E). These associations were confirmed using bc-GenExMiner (Supplementary Fig. 1).
High SLC1A5 mRNA expression was significantly associated with ER-negative, PR-negative, and HER2-positive tumours together with TN tumours (all P < 0.01, Table 1). When comparing the levels of SLC1A5 mRNA expression in the intrinsic (PAM50) subtypes [39], high expression was observed in basal, HER2-enriched and luminal B tumours (P < 0.001, Fig. 1F). Similarly, within the METABRIC Integrative Clusters, high SLC1A5 mRNA expression was associated with clusters 1 (luminal B subgroup), 5 (ERBB2 amplified), 9 (luminal B subgroup) and 10 (triple negative/basal-like) (P < 0.001, Fig. 1G). In the SCMGENE subtypes, there was greater expression of SLC1A5 mRNA in the ER + /HER2- high proliferation class (luminal B) than in the ER + /HER2- low proliferation class (luminal A) (P < 0.001, Fig. 1H). The associations of SLC1A5 mRNA with ER- and PR- tumours, as well as with HER2 + basal and luminal B tumours (PAM50), were confirmed using bc-GenExMiner (Supplementary Fig. 1). High SLC1A5 mRNA expression was detected in the luminal androgen receptor (LAR) subtype compared with the basal-like immunosuppressed (BLIS), basal-like immune-activated (BLIA), and mesenchymal (MES) TN subtypes (Supplementary Fig. 1H).
Table 1.
Association of SLC1A5 expression at mRNA and protein levels and other molecular biomarkers in breast cancer.
| SLC1A5 | ||||||||
|---|---|---|---|---|---|---|---|---|
| mRNA | Protein | |||||||
| Low n (%) |
High n (%) |
χ2 (p-value) |
Adjusted p-value | Low n (%) |
High n (%) |
χ2 (p-value) |
Adjusted p-value | |
| ER | ||||||||
| Negative | 123 (26.1) | 348 (73.9) |
40.2 (2.3 × 10–10) |
< 0.0001 | 107 (18.0) | 488 (82.0) |
120.0 (6.2 × 10–28) |
< 0.0001 |
| Positive | 639 (42.4) | 868 (57.6) | 918 (42.5) | 1242 (57.5) | ||||
| PR | ||||||||
| Negative | 298 (31.8) | 640 (68.2) |
34.4 (4.6 × 10–9) |
< 0.0001 | 306 (28.0) | 785 (72.0) |
61.37 (4.7 × 10–15) |
< 0.0001 |
| Positive | 464 (44.6) | 576 (55.4) | 691 (42.9) | 921 (57.1) | ||||
| HER2 | ||||||||
| Negative | 710 (41.0) | 1023 (59.0) |
35.4 (2.8 × 10–9) |
< 0.0001 | 929 (39.5) | 1424 (60.5) |
45.3 (1.6 × 10–11) |
< 0.0001 |
| Positive | 52 (21.2) | 193 (78.8) | 73 (20.9) | 277 (79.1) | ||||
| Triple negative | ||||||||
| No | 661 (39.8) | 999 (60.2) |
7.3 (0.007) |
0.008 | 942 (40.8) | 1367 (59.2) |
83.1 (7.7 × 10–20) |
< 0.0001 |
| Yes | 101 (31.8) | 217 (68.2) | 74 (17.5) | 349 (82.5) | ||||
| TP53 mutation | ||||||||
| Wild-type | 316 (44.1) | 401 (55.9) |
22.3 (0.000002) |
< 0.0001 | N/A | |||
| Mutation | 19 (19.2) | 80 (80.8) | ||||||
| P53 protein | ||||||||
| Negative | N/A | 344 (40.6) | 503 (59.4) |
32.7 (1.1 × 10–8 ) |
< 0.0001 | |||
| Positive | 86 (23.5) | 280 (76.5) | ||||||
Significant values are in [bold].
High SLC1A5 mRNA expression in breast tumors correlated with a high frequency of TP53 mutations (P < 0.001, Table 1). Gene CN analysis revealed that luminal B tumours had the greatest proportion of SLC1A5 CN loss (P = 0.02, Supplementary Table 2), whereas no significant association was detected between SLC1A5 gain and the intrinsic BC subtypes (P > 0.05, Supplementary Table 2).
Regulatory and amino acid transporter genes
The correlation between SLC1A5 mRNA and other related genes was investigated (Table 2). The genes were selected based on previous publications, as they are either regulatory genes or others that share the biological functions of amino acid transporters, which are primarily focused on glutamine transport and glutamine metabolism.
Table 2.
Correlation of SLC1A5 mRNA expression with the expression of other related genes in primary invasive breast cancer using the METABRIC cohort.
| SLC1A5 mRNA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| All cases (n = 1,980) | Luminal A (n = 368) | Luminal B (n = 367) | HER2 + (n = 110) | Triple negative (n = 150) | ||||||
| Correlation Coefficient (p-value) | Adjusted p-value | |||||||||
| Regulatory and other associated genes | ||||||||||
| MYC | − 0.126 (1.6 × 10–8) | < 0.0001 | − 0.179 (0.001) | 0.01 | − 0.041 (0.438) | 5.1 | − 0.263 (0.006) | 0.13 | − 0.031 (0.709) | 3.5 |
| Rb | − 0.23 (2.4 × 10–25) | < 0.0001 | − 0.154 (0.00003) | 0.0007 | − 0.168 (0.0001) | 0.002 | − 0.285 (0.000007) | 0.0002 | − 0.305 (1.7 × 10–8) | < 0.0001 |
| PIK3CA | 0.196 (1.5 × 10–18) | < 0.0001 | 0.208 (1.7 × 10–8) | < 0.0001 | 0.134 (0.003) | 0.007 | 0.126 (0.05) | 0.56 | 0.236 (0.00001) | 0.0003 |
| AKT1 | 0.148 (3.5 × 10–11) | < 0.0001 | 0.230 (0.000008) | 0.0002 | 0.006 (0.903) | 2.7 | 0.189 (0.048) | 0.48 | 0.204 (0.012) | 0.16 |
| RAF1 | 0.136 (1.1 × 10–9) | < 0.0001 | 0.103 (0.006) | 0.11 | 0.098 (0.03) | 0.63 | 0.134 (0.03) | 0.45 | 0.112 (0.04) | 0.60 |
| BRAF | 0.108 (0.000001) | < 0.0001 | 0.08 (0.01) | 0.12 | − 0.042 (0.357) | 4.9 | 0.09 (0.162) | 1.41 | 0.215 (0.00008) | 0.001 |
| KRAS | 0.127 (1.4 × 10–8) | < 0.0001 | 0.07 (0.03) | 0.27 | 0.119 (0.009) | 0.19 | − 0.04 (0.49) | 1.98 | 0.149 (0.007) | 0.13 |
| EPHA2 | 0.068 (0.002) | 0.02 | 0.109 (0.003) | 0.30 | 0.001 (0.987) | 0.98 | 0.114 (0.07) | 0.70 | 0.08 (0.13) | 1.56 |
| ATF4 | 0.273 (4.3 × 10–35) | < 0.0001 | 0.106 (0.042) | 0.33 | 0.215 (0.00004) | 0.001 | 0.445 (0.000001) | < 0.0001 | 0.370 (0.000003) | 0.0001 |
| Enzymes involved in glutamine metabolism | ||||||||||
| GLS | − 0.087 (0.0001) | 0.001 | − 0.111 (0.033) | 0.30 | − 0.064 (0.219) | 3.7 | − 0.183 (0.056) | 0.60 | 0.076 (0.357) | 1.58 |
| ALDH4A1 | 0.042 (0.064) | 0.30 | − 0.144 (0.006) | 0.11 | − 0.037 (0.482) | 5.2 | 0.220 (0.021) | 0.34 | 0.055 (0.505) | 3.04 |
| PRODH | 0.052 (0.021) | 0.18 | 0.179 (0.001) | 0.01 | − 0.014 (0.795) | 4.7 | 0.222 (0.020) | 0.36 | 0.071 (0.385) | 2.10 |
| PYCR1 | 0.335 (4.7 × 10–53) | < 0.0001 | 0.267 (1.9 × 10–7) | < 0.0001 | 0.245 (0.000002) | 0.0001 | 0.445 (0.000001) | < 0.0001 | 0.362 (0.000005) | 0.0001 |
| ALDH18A1 | 0.164 (2.4 × 10–13) | < 0.0001 | 0.227 (0.00001) | 0.0002 | 0.057 (0.278) | 4.3 | 0.278 (0.003) | 0.07 | 0.217 (0.008) | 0.14 |
| GLUL | 0.016 (0.478) | 1.40 | 0.158 (0.002) | 0.02 | − 0.065 (0.214) | 3.5 | − 0.072 (0.454) | 1.62 | − 0.067 (0.415) | 2.52 |
| GLUD1 | 0.134 (2.5 × 10–9) | < 0.0001 | 0.278 (5.8 × 10–8) | < 0.0001 | 0.087 (0.096) | 0.80 | 0.116 (0.228) | 1.45 | 0.076 (0.358) | 2.00 |
| Amino acid transporters | ||||||||||
| SLC7A5 | 0.29 (4.5 × 10–41) | < 0.0001 | 0.170 (0.000005) | 0.0001 | 0.150 (0.001) | 0.02 | 0.208 (0.001) | 0.02 | 0.25 (0.000002) | < 0.0001 |
| SLC3A2 | − 0.098 (0.00001) | 0.0001 | − 0.188 (0.0003) | 0.006 | − 0.077 (0.142) | 2.6 | − 0.209 (0.028) | 0.38 | − 0.129 (0.115) | 1.43 |
| SLC6A19 | 0.01 (0.47) | 0.94 | 0.02 (0.44) | 1.3 | 0.01 (0.74) | 5.9 | − 0.11 (0.03) | 0.46 | 0.04 (0.50) | 3.42 |
| SLC7A6 | 0.129 (8.8 × 10–9) | < 0.0001 | 0.03 (0.29) | 1.4 | 0.01 (0.76) | 5.3 | 0.137 (0.01) | 0.20 | 0.18 (0.004) | 0.08 |
| SLC7A7 | − 0.01 (0.40) | 1.60 | − 0.104 (0.005) | 0.10 | − 0.04 (0.28) | 4.2 | − 0.04 (0.39) | 1.60 | − 0.21 (0.001) | 0.02 |
| SLC7A8 | − 0.07 (0.001) | 0.01 | 0.06 (0.09) | 0.54 | 0.007 (0.88) | 3.2 | − 0.03 (0.62) | 1.80 | − 0.20 (0.0002) | 0.003 |
| SLC7A9 | 0.057 (0.01) | 0.08 | 0.06 (0.101) | 0.70 | − 0.01 (0.72) | 6.4 | 0.15 (0.006) | 0.13 | 0.05 (0.38) | 2.50 |
| SLC38A1 | 0.066 (0.003) | 0.03 | -0.028 (0.596) | 1.1 | 0.042 (0.427) | 5.4 | 0.105 (0.275) | 1.54 | 0.227 (0.005) | 0.10 |
| SLC38A2 | 0.18 (1.5 × 10–16) | < 0.0001 | 0.17 (0.000004) | 0.0001 | 0.14 (0.002) | 0.02 | 0.19 (0.002) | 0.02 | 0.17 (0.002) | 0.04 |
| SLC38A3 | 0.069 (0.002) | 0.02 | 0.053 (0.307) | 1.2 | − 0.020 (0.704) | 6.0 | 0.121 (0.206) | 1.44 | 0.128 (0.119) | 1.45 |
| SLC38A5 | 0.04 (0.03) | 0.18 | − 0.05 (0.11) | 0.6 | 0.01 (0.79) | 4.7 | 0.10 (0.06) | 0.65 | 0.05 (0.42) | 2.87 |
| SLC38A7 | 0.241 (1.2 × 10–27) | < 0.0001 | 0.203 (4.0 × 10–8) | < 0.0001 | 0.145 (0.001) | 0.03 | 0.168 (0.002) | 0.05 | 0.31 (0.000001) | < 0.0001 |
| SLC38A8 | 0.01 (0.48) | 0.48 | 0.01 (0.79) | 0.79 | 0.005 (0.91) | 1.8 | − 0.03 (0.47) | 1.80 | − 0.01 (0.79) | 3.85 |
There was a significant correlation between SLC1A5 mRNA expression and the expression of all regulatory genes that were previously identified in the literature, including MYC, Rb, ATF4, PIK3CA, EphA2 and genes involved in the MAPK pathway (RAF1, BRAF and KRAS) (P < 0.05, Table 2). While many regulatory genes displayed a positive correlation with SLC1A5, an inverse relationship was observed between MYC and Rb, with the latter showing a consistent negative correlation across all BC subtypes (P < 0.01). However, the negative correlation between MYC and SLC1A5 expression was significant only for luminal A tumours (P = 0.01) and not for the other subtypes (P > 0.05). PIK3CA was the only gene that showed a positive relationship in all BC subtypes, excluding HER2 + tumours (P < 0.01 and P > 0.05).
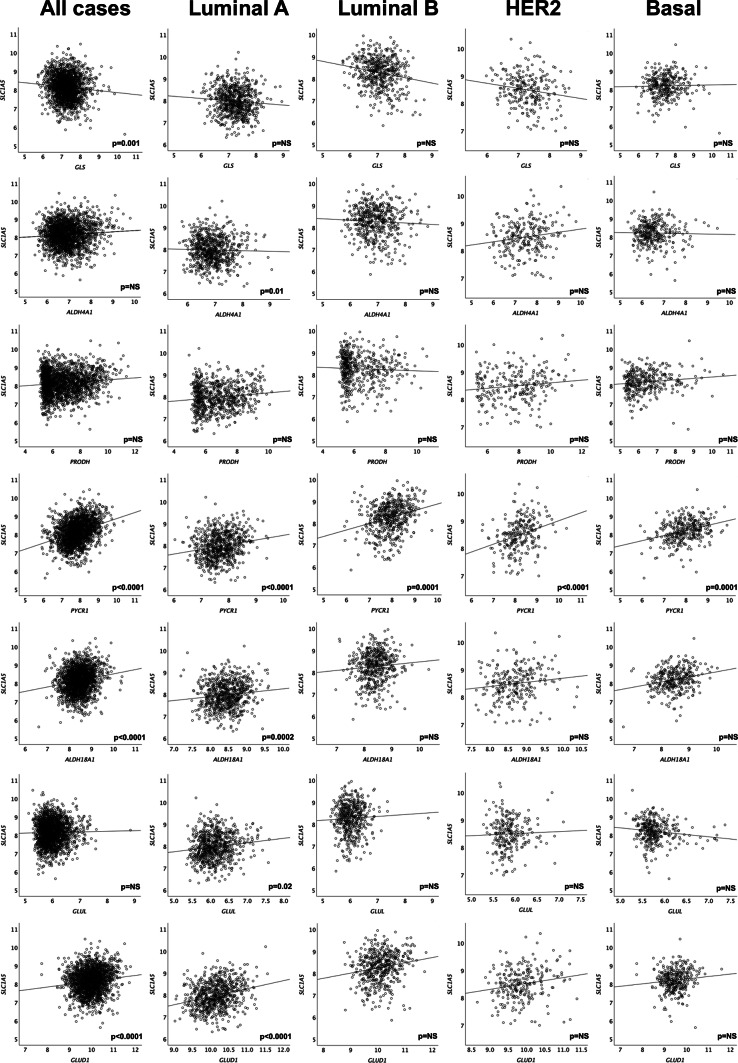
Regarding associations with glutamine metabolic enzymes, SLC1A5 expression was positively correlated with enzymes involved in the conversion of glutamine to proline (GLS, PYCR1 and ALDH18A1) (P < 0.01, Fig. 2 and Table 2). A positive relationship was also observed with the enzyme GLUD1, which catalyses the formation of α-KG from glutamate (P < 0.01). Most enzymes were significantly associated with luminal A tumours (P < 0.05), the only subtype that was positively associated with glutamine synthetase enzyme (GLUL) (P < 0.05). Many amino acid transporters were significantly associated with SLC1A5 expression, primarily in TN tumours and, to a lesser extent, in luminal tumours (P < 0.05). SLC7A5 and SLC38A2 were significantly differentially expressed from SLC1A5 in all subtypes (P < 0.05).
Fig. 2.
Correlation of SLC1A5 mRNA expression in invasive breast cancer molecular subtypes with the expression of genes associated with the proline-gluatmine regulatory axis (GLS, ALDH4A1, PRODH, PYCR1, ALDH18A1, GLUL, GLUD1) in the METABRIC cohort. NS = not significant.
Patient outcomes
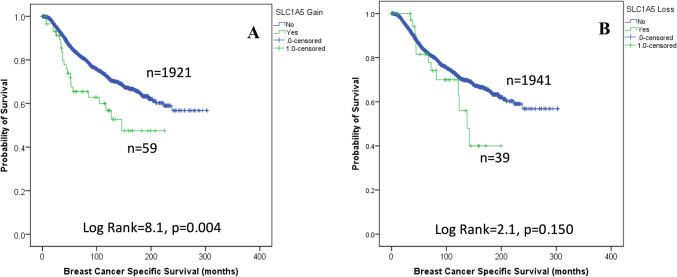
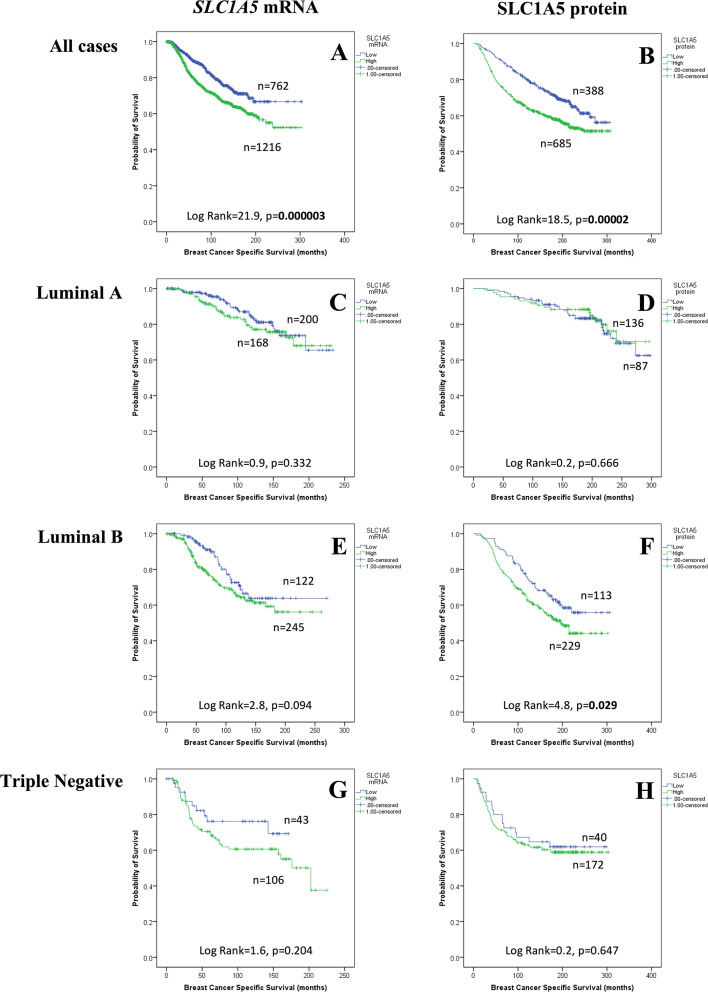
The results demonstrated that CN gain of SLC1A5, but not CN loss, was associated with poor patient survival (P = 0.004, Fig. 3). High expression of SLC1A5 mRNA was associated with shorter BCSS in all patients (P < 0.001, Fig. 4A), and shorter overall survival (OS) was also observed in the bc-GenExMiner (P < 0.001) but not KM plotter (P > 0.05, Supplementary Fig. 2A-B) datasets. When investigating the expression of SLC1A5 mRNA in molecular subtypes, high SLC1A5 mRNA expression tended to be associated with high BCSS in luminal B tumours (P = 0.094, Fig. 4E). There was no association between SLC1A5 mRNA and outcomes in patients with other molecular subtypes (P > 0.100, Fig. 4). In the bc-GenExMiner validation dataset, high SLC1A5 mRNA was predictive of poor OS in patients with luminal A tumours only (P = 0.025, Supplementary Fig. 2C). There was no association between SLC1A5 mRNA and any of the molecular subtypes according to the KM Plotter dataset (Supplementary Fig. 2).
Fig. 3.
SLC1A5 copy number aberrations in invasive breast cancer and their relationship with breast cancer-specific survival: (A) copy number gain (B) copy number loss.
Fig. 4.
SLC1A5 and breast cancer patient outcome: SLC1A5 mRNA vs breast cancer specific survival in (A) all cases, (C) ER + /HER2- Low Proliferation, (E) ER + /HER2- High Proliferation, (G) Triple Negative tumours and I) HER2 + tumours; SLC1A5 protein vs BCSS in (B) all cases, (D) ER + /HER2- Low Proliferation, (F) ER + /HER2- High Proliferation, (J) Triple Negative tumours; (K) SLC1A5 protein vs distant metastases free survival in all cases. Green = high; blue = low.
According to multivariable Cox regression analysis, SLC1A5 mRNA remained an independent predictor of BCSS in all patients (P = 0.005, Table 3) but not in any specific subtype (data not shown).
Table 3.
Multivariate analysis of prognostic variables and SLC1A5 mRNA and protein expressions in primary invasive breast cancer.
| Parameter | SLC1A5 mRNA | SLC1A5 protein | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) | p-value | Hazard ratio (95% CI) | p-value | |
| SLC1A5 | 1.55 (1.14–2.12) | 0.005 | 1.09 (0.89–1.34) | 0.36 |
| Nodal stage | 1.72 (1.43–2.05) | 2.7 × 10–9 | 1.89 (1.68–2.12) | 1.4 × 10–27 |
| Tumour size | 1.45 (1.02–2.06) | 0.03 | 1.34 (1.12–1.61) | 0.001 |
| Tumour grade | 1.41 (1.10–1.82) | 0.007 | 2.41 (2.03–2.85) | 2.5 × 10–24 |
Significant values are in [bold].
SLC1A5 protein expression in breast cancers
There was variable expression of the SLC1A5 protein in the membrane of breast tumour cells, ranging from absent to high (Supplementary Fig. 3A–B). Compared to that in invasive breast tumour cells, SLC1A5 protein expression in normal breast epithelium was lower (Supplementary Fig. 3C). High SLC1A5 protein expression (> 40 H-score) was observed in 63% of the tumours. Many patients with high SLC1A5 mRNA expression also expressed high levels of SLC1A5 protein (correlation coefficient = 0.35, P = 3.3 × 10−7).
Clinicopathological parameters and biological subtypes
Similar associations with SLC1A5 mRNA were observed with high SLC1A5 protein expression, including larger tumour size, high tumour grade, high pleomorphism, high mitotic count, less tubular formation, poor NPI and the presence of lymphovascular invasion (P < 0.001, Table 4). A significant association was also observed between high SLC1A5 expression and medullary-like tumours (P < 0.001). Regarding BC metastatic sites, high SLC1A5 protein levels were associated with the development of distant metastasis to the bone (P = 0.0006) and liver (P = 0.005, Table 4).
Table 4.
Clinicopathological associations of SLC1A5 protein expression in primary invasive breast cancer.
| SLC1A5 protein | ||||
|---|---|---|---|---|
| Low n (%) | High n (%) | χ2 (p-value) | Adjusted P value | |
| Tumour size | ||||
| < 2.0 cm | 670 (43.7) | 864 (56.3) | 57.82 (2.8 × 10–14) | < 0.0001 |
| ≥ 2.0 cm | 365 (29.6) | 868 (70.4) | ||
| Tumour Grade | ||||
| 1 | 304 (68.0) | 143 (32.0) | 353.77 (1.5 × 10–77) | < 0.0001 |
| 2 | 459 (44.8) | 565 (55.2) | ||
| 3 | 270 (20.9) | 1023 (79.1) | ||
| Mitosis | ||||
| 1 | 609 (53.8) | 524 (46.2) | 280.62 (1.1 × 10–61) | < 0.0001 |
| 2 | 190 (36.2) | 355 (63.8) | ||
| 3 | 200 (19.1) | 847 (80.9) | ||
| Pleomorphism | ||||
| 1 | 33 (71.7) | 13 (28.3) | 252.04 (1.8 × 10–55) | < 0.0001 |
| 2 | 507 (56.0) | 399 (44.0) | ||
| 3 | 459 (26.2) | 1295 (73.8) | ||
| Tubular formation | ||||
| 1 | 124 (70.1) | 53 (29.9) | 123.69 (1.3 × 10–27) | < 0.0001 |
| 2 | 369 (42.4) | 501 (57.6) | ||
| 3 | 507 (30.5) | 1153 (69.5) | ||
| Vascular Invasion | ||||
| Negative | 780 (41.3) | 1110 (58.7) | 40.77 (1.7 × 10–10) | < 0.0001 |
| Positive | 248 (28.6) | 619 (71.4) | ||
| Lymph Node Stage | ||||
| 1 | 696 (40.8) | 1010 (59.2) | 28.73 (5.7 × 10–7) | < 0.0001 |
| 2 | 275 (34.1) | 531 (65.9) | ||
| 3 | 62 (24.9) | 187 (75.1) | ||
| Nottingham Prognostic Index | ||||
| Good | 505 (56.4) | 390 (43.6) | 219.55 (2.1 × 10–48) | < 0.0001 |
| Moderate | 438 (30.8) | 985 (69.2) | ||
| Poor | 91 (20.4) | 354 (79.6) | ||
| Biological Subtypes | ||||
| ER + /HER2- Low Proliferation | 704 (49.8) | 710 (50.2) | 231.2 (7.5 × 10–50) | < 0.0001 |
| ER + /HER2- High Proliferation | 99 (22.3) | 344 (77.7) | ||
| Triple Negative | 77 (18.0) | 351 (82.0) | ||
| HER2 + | 61 (22.5) | 210 (77.5) | ||
| Histological type | ||||
| Ductal (including mixed) | 836 (35.2) | 1536 (64.8) | 87.18 (2.6 × 10–17) | < 0.0001 |
| Lobular | 103 (47.7) | 113 (52.3) | ||
| Medullary | 3 (7.5) | 37 (92.5) | ||
| Miscellaneous | 9 (50.0) | 9 (50.0) | ||
| Special type | 83 (70.9) | 34 (29.1) | ||
| Tubular | 1 (33.3) | 2 (66.7) | ||
| Site of distant metastasis | ||||
| Brain | ||||
| No | 973 (55.9) | 767 (44.1) | 1.73 (0.18) | 0.54 |
| Yes | 45 (48.9) | 47 (51.1) | ||
| Lung | ||||
| No | 929 (55.8) | 737 (44.2) | 0.282 (0.59) | 1.18 |
| Yes | 89 (53.6) | 77 (46.4) | ||
| Bone | ||||
| No | 848 (57.8) | 620 (42.2) | 14.45 (0.0001) | 0.0006 |
| Yes | 170 (46.7) | 194 (53.3) | ||
| Liver | ||||
| No | 903 (57.1) | 679 (42.9) | 10.73 (0.001) | 0.005 |
| Yes | 115 (46.0) | 135 (54.0) | ||
Significant values are in [bold].
High SLC1A5 protein expression was significantly associated with ER-negative, PR-negative and HER2-positive tumours (P < 0.001, Table 1). Additionally, SLC1A5 protein expression was associated with TN tumours (P < 0.001, Table 1). SLC1A5 protein expression in the IHC-defined molecular subtypes was significantly lower in the luminal A tumours than in the other subtypes (P < 0.001, Table 4).
Regulatory and amino acid transporter proteins
The associations of SLC1A5 protein expression with other proteins were also examined (Table 5). SLC1A5 protein was significantly expressed in breast tumours with high Ki67 (P < 0.001) and MYC (P = 0.02) expression. PIK3CA was also significantly expressed in breast tumours with high SLC1A5 expression (P = 0.005). Interestingly, high SLC1A5 protein expression was significantly associated with all enzymes involved in the glutamine-proline regulatory axis, including enzymes that convert glutamine to proline (GLS, PYCR1 and ALDH18A1) (P < 0.001) and enzymes that catalyse proline to glutamine (PRODH and ALDH4A1) (P < 0.05). Apart from SLC7A8, all the following amino acid transporters, SLC7A5, SLC3A2, SLC7A11 and SLC38A2, were significantly expressed in breast tumours with high SLC1A5 expression (P < 0.01).
Table 5.
Association of SLC1A5 protein and other biological markers in primary invasive breast cancer.
| SLC1A5 protein | ||||
|---|---|---|---|---|
| Low, n (%) | High, n (%) | χ2 (p-value) |
Adjustedp-value | |
| c-MYC | ||||
| Negative | 292 (35.4) | 534 (64.6) |
7.27 (0.007) |
0.02 |
| Positive | 41 (24.6) | 126 (75.4) | ||
| Ki67 | ||||
| Negative | 184 (55.1) | 150 (44.9) |
89.8 (2.5 × 10–21) |
< 0.0001 |
| Positive | 184 (25.3) | 544 (74.7) | ||
| PIK3CA | ||||
| Negative | 97 (43.7) | 125 (56.3) |
10.68 (0.001) |
0.005 |
| Positive | 227 (31.7) | 488 (68.3) | ||
| GLUD1 | ||||
| Negative | 292 (33.7) | 575 (66.3) |
1.78 (0.18) |
0.36 |
| Positive | 128 (37.8) | 211 (62.2) | ||
| GLS | ||||
| Negative | 216 (39.2) | 335 (60.8) |
28.79 (8.0 × 10–8) |
< 0.0001 |
| Positive | 92 (22.8) | 312 (77.2) | ||
| PYCR1 | ||||
| Negative | 171 (39.7) | 260 (60.3) |
29.89 (4.5 × 10–8) |
< 0.0001 |
| Positive | 80 (21.7) | 289 (78.3) | ||
| ALDH4A1 | ||||
| Negative | 166 (38.2) | 268 (61.8) |
10.39 (0.001) |
0.006 |
| Positive | 121 (27.9) | 312 (72.1) | ||
| ALDH18A1 | ||||
| Negative | 183 (40.1) | 273 (59.9) |
20.59 (0.000006) |
< 0.0001 |
| Positive | 112 (25.8) | 322 (74.2) | ||
| PRODH | ||||
| Negative | 240 (33.8) | 471 (66.2) |
6.30 (0.01) |
0.03 |
| Positive | 46 (24.2) | 144 (75.8) | ||
| SLC7A5 | ||||
| Negative | 825 (41.3) | 1174 (58.7) |
161.7 (4.7 × 10–37) |
< 0.0001 |
| Positive | 39 (9.0) | 394 (91.0) | ||
| SLC3A2 | ||||
| Negative | 423 (41.3) | 601 (58.7) |
32.40 (1.2 × 10–8) |
< 0.0001 |
| Positive | 308 (29.4) | 741 (70.6) | ||
| SLC38A2 | ||||
| Negative | 487 (34.7) | 917 (65.3) |
12.92 (0.0003) |
0.002 |
| Positive | 27 (19.6) | 111 (80.4) | ||
| SLC7A8 | ||||
| Negative | 430 (33.4) | 856 (66.6) |
1.50 (0.22) |
0.44 |
| Positive | 47 (28.7) | 117 (71.3) | ||
| SLC7A11 | ||||
| Negative | 430 (38.5) | 686 (61.5) |
27.8 (1.3 × 10–7) |
< 0.0001 |
| Positive | 197 (26.7) | 541 (73.3) | ||
Significant values are in [bold].
Patient outcomes
The results demonstrated that high SLC1A5 protein expression was associated with shorter BCSS in all patients (P < 0.001; Fig. 4B). When investigating the expression of SLC1A5 protein in biological subtypes, high expression was only predictive of shorter BCSS in luminal B tumours (P < 0.05, Fig. 4F). There was no association between SLC1A5 protein and outcome in luminal A (Fig. 4D), TN (Fig. 4H) or HER2 + (Fig. 4J) tumours. According to multivariable analysis, SLC1A5 protein and other clinicopathological parameters were not significantly associated with BCSS (P > 0.05, Supplementary Table 3).
SLC1A5 is required for cell proliferation and glutamine uptake
SLC1A5 protein expression in a normal basal mammary cell line (MCF10) and a panel of luminal BC cell lines was greater in ZR-75-1 and HCC1500 cells than in the other cells analysed (Supplementary Fig. 4A). SLC1A5 knockdown and SLC1A5 inhibition were confirmed in ZR-75-1 and HCC1500 cells by western blotting (Supplementary Fig. 4B-C). Cell proliferation was significantly decreased by GPNA inhibition in ZR-75-1 cells but not in HCC1500 cells (Fig. 5A). However, targeted knockdown of SLC1A5 did not significantly impair the proliferation of ZR-75-1 cells or HCC1500 cells (Fig. 5B). GPNA reduced glutamine uptake in ZR-75-1 cells but not in HCC1500 cells (Fig. 5C). Glutamine uptake was lower in both ZR-75-1 and HCC1500 cells transfected with siRNA targeting SLC1A5 than in control cells (Fig. 5D).
Fig. 5.
The effect of SLC1A5 inhibition on the growth (A–B) and glutamine uptake (C-D) of luminal breast cancer cell lines using the GPNA inhibitor (A, C) and SLC1A5 mRNA knockdown (B, D) using siRNA. NS = not significant, *p < 0.05, **p < 0.01.
Discussion
ER + /luminal tumours, which constitute approximately 75% of BCs39,40, remain a heterogeneous group in terms of molecular biology and patient outcomes. Therefore, there is a clear need for improved understanding of the biology of the luminal class of BC, with subsequent translation into more effective methods for the diagnosis and management of this most common form of BC.
Metabolic reprogramming in cancer plays a vital role in the provision of supplementary elements, including nutrients and energy, which are essential for cellular growth. It has been reported that tumour cells rely on glutamine metabolism and become “addicted” to this amino acid for sustained proliferation/survival. Studies that address the prognostic significance of the key Gln transporter SLC1A5 in BC and its potential influence on Gln metabolism in different subtypes remain limited, particularly in luminal BC. We therefore investigated SLC1A5 mRNA and SLC1A5 protein expression in a large number of breast tumours to better understand the potential role of this important transporter of Gln in BC and its molecular subtypes, particularly in luminal ER + disease.
In this study, we have shown that SLC1A5, which is a primary transporter for Gln uptake, is highly expressed in a subset of ER + tumours that have high proliferation, i.e., luminal B tumours, and is related to poor patient outcome in this group. The high expression of SLC1A5 in luminal B tumours is perhaps not surprising because these tumours have greater demands for nutrients and energy, which are essential for cell survival and proliferation. SLC1A5 has previously been shown to be a poor prognostic factor in BC41. Similarly, Jeon et al. showed that high SLC1A5 was associated with shorter disease-free survival in patients with ER + BC, but neither investigated the molecular subtypes15. We further demonstrated that the association of SLC1A5 with patient outcome occurs only in luminal B and not luminal A tumours.
We confirmed that the SLC1A5 protein is expressed in TNBC and HER2 + cells, which is in accordance with the findings of Van Geldermalsen et al.8, and we additionally showed that SLC1A5 mRNA is also highly expressed in these subtypes10, confirming the possible role of the transcription and translation of this amino acid transporter in driving the uptake of Gln, which is required for proliferation in these highly proliferative subtypes of BC. However, in both subtypes, there was no association between SLC1A5 mRNA or protein expression and patient outcome.
Previous studies have raised awareness and revealed the importance of the proline-glutamine (Pro-Gln) regulatory axis in BC as part of tumour metabolism in addition to glycolysis according to different BC subtypes42, mainly focusing on either Gln or Pro metabolism only. The acquisition of glutamine via SLC1A5 is undoubtedly important, but proline metabolism may be an alternative source of glutamine. The coexpression of genes encoding Pro-Gln enzymes with SLC1A5 suggested that luminal A tumours might be partly glutamine independent rather than relying on uptake via SLC1A5, as in the basal HCC1806 cell line8. We therefore compared the gene expression of Pro-Gln enzymes with that of SLC1A5 and showed highly variable expression of this regulatory axis across molecular subtypes. Luminal A tumours had the highest number of correlates focused on a positive association with enzymes involved in the conversion of Pro to Gln, whereas those genes involved in the conversion of Gln to Pro were primarily downregulated. The correlation between SLC1A5 and GLUD1, which is involved in the conversion of Gln to alpha-ketoglutarate for the TCA cycle and subsequent gluconeogenesis, in luminal A tumours suggests that Gln is utilised for this process. Only PCYR1 was associated with SLC1A5 in luminal B tumours, suggesting that the primary source of Gln in these tumours is via uptake rather than neosynthesis. This might explain why MCF-7 luminal A cells are not affected by Gln deprivation when SLC1A5 is blocked with GPNA8.
Previous studies have shown that SLC1A5 is regulated by other proteins, including the tumour oncogene MYC, which induces SLC1A5. In the current study, we sought to understand the relationship between SLC1A5 and other regulatory proteins in terms of both mRNA and protein expression. We observed a positive correlation between SLC1A5 and MYC at the protein level but not at the mRNA level. MYC also induces apoptosis via ATF4 upon glutamine deprivation, and we observed a positive correlation between ATF4 and SLC1A5 gene expression, in line with expectations. We have recently investigated ATF4 protein expression in invasive BC and its coexpression with SLC1A5 protein is associated with poor patient outcome in ER + tumours42. Chemotherapy treatment in BC promotes the degradation of SLC1A5 via RNF5 ubiquitination, leading to mTOR inactivation15, although we did not observe any association between the gene expression of SLC1A5 and RNF5 or its pseudogene RNFP1.
There are more than 24 amino acid transporters, and we further investigated whether SLC1A5 expression was associated with any of the key transporters. SLC7A5 functionally couples with SLC1A5 to allow cellular influx and efflux of Gln. The coexpression of SLC1A5 and SLC7A5 in all BC subtypes suggested that they play a key role in Gln transport. Indeed, SLC7A5 has previously been incorporated into the Mammostrat® risk test used to stratify BC patients treated with tamoxifen43. SLC1A5 also requires SLC7A11 for functional coupling of glutamine efflux and cystine entry, which in turn is converted to cysteine, which rules SLC1A5-mediated glutamine entry, although in our study, SLC7A11 was associated with luminal A tumours44. The coexpression of SLC1A5, SLC7A11 and SLC7A5 proteins has yet to be determined in BC and is therefore important for understanding the potential transport of Gln in and out of tumour cells.
With the increasing number of treatment strategies available for BC patients, it is important that effective strategies that can support the personalisation of care and allow tailored treatment planning appropriate for patients’ tumour biology, both to maximise treatment benefit and to avoid the adverse effects associated with over- or undertreatment, emerge. This would minimise harmful side effects to patients and reduce treatment costs by focusing expensive and valuable resources on those who would optimally benefit from the new generation of targeted molecular therapies. For instance, it has been proposed that one of the mechanistic actions of tamoxifen involves the suppression of glutamine uptake and the induction of apoptosis45. SLC1A5 has also been linked to endocrine therapy resistance in luminal BC. Inhibiting or depleting SLC1A5 in these cells has been shown to increase sensitivity to tamoxifen and decrease proliferation in aromatase inhibitor-resistant cells46,47. Additionally, high SLC1A5 expression in clinical samples correlates with endocrine therapy resistance and worse patient outcomes in luminal BC46.
Blocking SLC1A5 using the small molecule inhibitor GPNA inhibited Gln uptake and subsequent tumor growth in basal-like TNBC but not in luminal A tumours using MCF-7 cells as an in vitro model8. Although the consequences of blocking SLC1A5 in luminal B tumours remain undetermined, our data suggest that SLC1A5 could be used as a target in luminal B tumours to reduce Gln uptake and thus cell proliferation and growth. In addition to GPNA, 2-amino-4-bis (aryloxybenzyl) aminobutanoic acids have recently been identified as novel inhibitors of Gln uptake via SLC1A5 48. Evaluation of these and other inhibitors is therefore warranted in luminal B BC.
Therefore, we believe that continued refinement of the understanding of the biological diversity of BC, particularly the luminal B subtype, with linked development of classification strategies suitable for routine clinical use is essential to achieve a personalised approach to BC management. Further assessment of the metabolic pathways associated with glutamine and its uptake is therefore essential in the luminal B subtype and other subtypes, including HER2 + tumours.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the Nottingham Health Science Biobank and Breast Cancer Now Tissue Bank for the provision of tissue samples.
Author contributions
Conceptualisation, A.R.G., R.E.A. and M.L.C; Methodology and Formal Analysis, L.H.A., R.E.A, B.E., A.F., M.L.C., M.A.A., K.W.C, and A.R.G. Data Curation, I.O.E., E.A.R, and A.R.G.; Writing—Original Draft Preparation, L.H.A., R.E.A., M.L.C., and A.R.G.; Writing—Review & Editing, L.H.A., R.E.A, B.E., A.F., M.L.C., M.A.A., K.W.C, I.O.E., E.A.E. and A.R.G.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed This work was performed according to REMARK guidelines or tumour prognostic study and obtained ethics approval by the North West–Greater Manchester Central Research Ethics Committee under the title: Nottingham Health Science Biobank (NHSB), reference number 15/NW/0685. We can declare that this study is complying with Helsinki declaration.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lutfi H. Alfarsi and Rokaya El Ansari contributed equally to this work.
References
- 1.Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: The next generation. Cell144, 646–674. 10.1016/j.cell.2011.02.013 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Green, A. R. et al. MYC functions are specific in biological subtypes of breast cancer and confers resistance to endocrine therapy in luminal tumours. Br. J. Cancer114, 917–928. 10.1038/bjc.2016.46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu, W. et al. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl. Acad. Sci. USA107, 7455–7460. 10.1073/pnas.1001006107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polyak, K., Xia, Y., Zweier, J. L., Kinzler, K. W. & Vogelstein, B. A model for p53-induced apoptosis. Nature389, 300–305. 10.1038/38525 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Wise, D. R. et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. USA105, 18782–18787. 10.1073/pnas.0810199105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alberghina, L. & Gaglio, D. Redox control of glutamine utilization in cancer. Cell Death Dis.5, e1561. 10.1038/cddis.2014.513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Csibi, A. et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell153, 840–854. 10.1016/j.cell.2013.04.023 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Geldermalsen, M. et al. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene35, 3201–3208. 10.1038/onc.2015.381 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross, M. I. et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther.13, 890–901. 10.1158/1535-7163.MCT-13-0870 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Kim, S., Kim, D. H., Jung, W. H. & Koo, J. S. Expression of glutamine metabolism-related proteins according to molecular subtype of breast cancer. Endocr. Relat. Cancer20, 339–348. 10.1530/ERC-12-0398 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Budczies, J. et al. Glutamate enrichment as new diagnostic opportunity in breast cancer. Int. J. Cancer136, 1619–1628. 10.1002/ijc.29152 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Cassago, A. et al. Mitochondrial localization and structure-based phosphate activation mechanism of Glutaminase C with implications for cancer metabolism. Proc. Natl. Acad. Sci. USA109, 1092–1097. 10.1073/pnas.1112495109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Timmerman, L. A. et al. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell24, 450–465. 10.1016/j.ccr.2013.08.020 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao, M. D. et al. Metabolic characterization of triple negative breast cancer. BMC Cancer14, 941. 10.1186/1471-2407-14-941 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeon, Y. J. et al. Regulation of glutamine carrier proteins by RNF5 determines breast cancer response to ER stress-inducing chemotherapies. Cancer Cell27, 354–369. 10.1016/j.ccell.2015.02.006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larriba, S. et al. ATB(0)/SLC1A5 gene. Fine localisation and exclusion of association with the intestinal phenotype of cystic fibrosis. Eur. J. Hum. Genet.9, 860–866. 10.1038/sj.ejhg.5200726 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Shimizu, K. et al. ASC amino-acid transporter 2 (ASCT2) as a novel prognostic marker in non-small cell lung cancer. Br. J. Cancer110, 2030–2039. 10.1038/bjc.2014.88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yazawa, T. et al. Clinical significance of coexpression of L-type amino acid transporter 1 (LAT1) and ASC amino acid transporter 2 (ASCT2) in lung adenocarcinoma. Am. J. Transl. Res.7, 1126–1139 (2015). [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, Y. et al. High expression of solute carrier family 1, member 5 (SLC1A5) is associated with poor prognosis in clear-cell renal cell carcinoma. Sci. Rep.5, 16954. 10.1038/srep16954 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaira, K. et al. Relationship between CD147 and expression of amino acid transporters (LAT1 and ASCT2) in patients with pancreatic cancer. Am. J. Transl. Res.7, 356–363 (2015). [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, Q. et al. Targeting ASCT2-mediated glutamine uptake blocks prostate cancer growth and tumour development. J. Pathol.236, 278–289. 10.1002/path.4518 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, Q. et al. Targeting glutamine transport to suppress melanoma cell growth. Int. J. Cancer135, 1060–1071. 10.1002/ijc.28749 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Reynolds, M. R. et al. Control of glutamine metabolism by the tumor suppressor Rb. Oncogene33, 556–566. 10.1038/onc.2012.635 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curtis, C. et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature486, 346–352. 10.1038/nature10983 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abd El-Rehim, D. M. et al. High-throughput protein expression analysis using tissue microarray technology of a large well-characterised series identifies biologically distinct classes of breast cancer confirming recent cDNA expression analyses. Int. J. Cancer116, 340–350. 10.1002/ijc.21004 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Nath, P. et al. The amino acid transporter SLC7A11 expression in breast cancer. Cancer Biol. Ther.25, 2291855. 10.1080/15384047.2023.2291855 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ilyas, M. et al. Guidelines and considerations for conducting experiments using tissue microarrays. Histopathology62, 827–839. 10.1111/his.12118 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Abdel-Fatah, T. M. et al. Proposal for a modified grading system based on mitotic index and Bcl2 provides objective determination of clinical outcome for patients with breast cancer. J. Pathol.222, 388–399. 10.1002/path.2775 (2010). [DOI] [PubMed] [Google Scholar]
- 29.Aleskandarany, M. A. et al. Clinicopathologic and molecular significance of phospho-Akt expression in early invasive breast cancer. Breast Cancer Res. Treat.127, 407–416. 10.1007/s10549-010-1012-y (2011). [DOI] [PubMed] [Google Scholar]
- 30.Aleskandarany, M. A. et al. PIK3CA expression in invasive breast cancer: a biomarker of poor prognosis. Breast Cancer Res. Treat122, 45–53. 10.1007/s10549-009-0508-9 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Aleskandarany, M. A. et al. MIB1/Ki-67 labelling index can classify grade 2 breast cancer into two clinically distinct subgroups. Breast Cancer Res Treat127, 591–599. 10.1007/s10549-010-1028-3 (2011). [DOI] [PubMed] [Google Scholar]
- 32.El Ansari, R. et al. The solute carrier SLC7A8 is a marker of favourable prognosis in ER-positive low proliferative invasive breast cancer. Breast Cancer Res Treat181, 1–12. 10.1007/s10549-020-05586-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El Ansari, R. et al. The multifunctional solute carrier 3A2 (SLC3A2) confers a poor prognosis in the highly proliferative breast cancer subtypes. Br. J. Cancer118, 1115–1122. 10.1038/s41416-018-0038-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El Ansari, R. et al. The amino acid transporter SLC7A5 confers a poor prognosis in the highly proliferative breast cancer subtypes and is a key therapeutic target in luminal B tumours. Breast Cancer Res.20, 21. 10.1186/s13058-018-0946-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elsheikh, S. et al. CCND1 amplification and cyclin D1 expression in breast cancer and their relation with proteomic subgroups and patient outcome. Breast Cancer Res. Treat109, 325–335. 10.1007/s10549-007-9659-8 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Green, A. R. et al. Biological and clinical significance of PARP1 protein expression in breast cancer. Breast Cancer Res. Treat149, 353–362. 10.1007/s10549-014-3230-1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jerjees, D. A. et al. The mammalian target of rapamycin complex 1 (mTORC1) in breast cancer: The impact of oestrogen receptor and HER2 pathways. Breast Cancer Res. Treat150, 91–103. 10.1007/s10549-015-3308-4 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Lancashire, L. J. et al. A validated gene expression profile for detecting clinical outcome in breast cancer using artificial neural networks. Breast Cancer Res. Treat120, 83–93. 10.1007/s10549-009-0378-1 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Allred, D. C. Commentary: Hormone receptor testing in breast cancer: A distress signal from Canada. Oncologist13, 1134–1136. 10.1634/theoncologist.2008-0184 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Rakha, E. A. et al. Biologic and clinical characteristics of breast cancer with single hormone receptor positive phenotype. J. Clin. Oncol.25, 4772–4778. 10.1200/JCO.2007.12.2747 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Zhao, X., Jin, L., Liu, Y., Liu, Z. & Liu, Q. Bioinformatic analysis of the role of solute carrier-glutamine transporters in breast cancer. Ann. Transl. Med.10, 777 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel, R. et al. ATF4 as a prognostic marker and modulator of glutamine metabolism in oestrogen receptor-positive breast cancer. Pathobiology10.1159/000539564 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartlett, J. M. et al. Mammostrat as a tool to stratify breast cancer patients at risk of recurrence during endocrine therapy. Breast Cancer Res.12, R47. 10.1186/bcr2604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhutia, Y. D., Babu, E., Ramachandran, S. & Ganapathy, V. Amino Acid transporters in cancer and their relevance to “glutamine addiction”: Novel targets for the design of a new class of anticancer drugs. Cancer Res.75, 1782–1788. 10.1158/0008-5472.CAN-14-3745 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Todorova, V. K., Kaufmann, Y., Luo, S. & Klimberg, V. S. Tamoxifen and raloxifene suppress the proliferation of estrogen receptor-negative cells through inhibition of glutamine uptake. Cancer Chemother. Pharmacol.67, 285–291. 10.1007/s00280-010-1316-y (2011). [DOI] [PubMed] [Google Scholar]
- 46.Alfarsi, L. H. et al. SLC1A5 co-expression with TALDO1 associates with endocrine therapy failure in estrogen receptor-positive breast cancer. Breast Cancer Res. Treat189, 317–331. 10.1007/s10549-021-06298-1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen, Z., Wang, Y., Warden, C. & Chen, S. Cross-talk between ER and HER2 regulates c-MYC-mediated glutamine metabolism in aromatase inhibitor resistant breast cancer cells. J. Steroid. Biochem. Mol. Biol.149, 118–127. 10.1016/j.jsbmb.2015.02.004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulte, M. L., Khodadadi, A. B., Cuthbertson, M. L., Smith, J. A. & Manning, H. C. 2-Amino-4-bis(aryloxybenzyl)aminobutanoic acids: A novel scaffold for inhibition of ASCT2-mediated glutamine transport. Bioorg. Med. Chem. Lett.26, 1044–1047. 10.1016/j.bmcl.2015.12.031 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.