Abstract
Translational control is a key level in regulating gene expression in oocytes and eggs because many mRNAs are synthesized and stored during oogenesis for latter use at various stages of oocyte maturation and embryonic development. Understanding the molecular mechanisms that underlie this translational control is therefore crucial. Another important issue is the evolutionary conservation of these mechanisms—in other words the determination of their universal and specific aspects. We report here a comparative analysis of a translational repression mechanism that depends on the EDEN (embryo deadenylation element) element. This small cis-acting element, localized in the 3′ untranslated region of c-mos and Eg mRNAs, was shown to be involved in a deadenylation process. We demonstrate here that in Xenopus embryos, mRNAs that contain an EDEN are translationally repressed. Next, transgenic flies were used to study the effect of the EDEN motif on translation in Drosophila oocytes. We show that this element also causes the translational repression of a reporter gene in Drosophila demonstrating that the EDEN-dependent translational repression is functionally conserved between Xenopus and Drosophila.
Keywords: yemanuclein-alpha‖oogenesis
During many crucial steps in the development of an organism, such as the specification of cell fate and pattern formation, the nucleus is transcriptionally silent. As a result, the regulation of gene expression at these stages of development relies on cytoplasmic events. This fact is particularly true for oocyte maturation because the oocyte nucleus is involved in the lengthy and complex step of meiosis, and also for early embryogenesis during which the nuclei of the zygote are rapidly dividing, which excludes any important transcriptional activity. In these cases, specific mRNAs are stored as inactive mRNA–protein complexes before they are activated in response to a biological stimulus. It has been clearly established that these processes of mRNA storage and translational control are important in the regulation of gene activity during development (1, 2). This control is exerted through the binding of trans-acting factors to specific cis sequences on the mRNA.
Two main mechanisms have been described that control mRNA translation. The first one is independent of the poly(A) tail. In Xenopus, this situation is exemplified by the FGF receptor mRNA (3). This mRNA is translationally repressed during oogenesis because of the presence of a sequence element named a TIE (translational inhibitory element). It is translated during oocyte maturation. However, no change in the length of the poly(A) tail of this mRNA was detected before or after maturation, ruling out an effect of the TIE on the poly(A) tail. In Drosophila, a similar situation has been described for oskar (4) and sex-lethal (5) mRNAs.
The second mechanism that controls mRNA translation is associated with regulated changes of the poly(A) tail. In Xenopus, mRNAs that have various adenylation/deadenylation [lengthening or shortening of the poly(A) tail] behaviors during early development have been studied (for review, see ref. 6). For instance, cytoplasmic polyadenylation during oocyte maturation relies on a specific sequence element localized within the 3′ untranslated region (3′UTR) and named CPE (cytoplasmic polyadenylation element) (6). mRNAs that are polyadenylated during oocyte maturation are generally recruited to the translation apparatus during the same period. A causal relationship has been demonstrated between the presence of a CPE, the poly(A) extension, and the translational activation of both reporter and natural maternal mRNAs (7, 8). A subclass of these mRNAs that are polyadenylated during oocyte maturation are deadenylated after fertilization. These are the Eg family/c-mos mRNAs (7, 9). The deadenylation of these maternal mRNAs, which is temporally correlated with translational repression (10), relies on another sequence element named EDEN (embryo deadenylation element) (11).
Examples of changes in translation associated with changes in poly(A) tail length have been reported in other organisms. This is the case for bicoid, Toll, Torso in Drosophila (12), or tPA in mouse (13).
In this study, we bring a direct demonstration that the EDEN sequence, which provokes mRNA deadenylation, is able to repress translation of a reporter mRNA in Xenopus embryos. Next, we show that an EDEN inserted in the 3′UTR of a maternally expressed lacZ reporter gene represses its translation in Drosophila oocytes, without affecting its localization or its stability. These results demonstrate the functional conservation of the EDEN-mediated translational repression between Xenopus and Drosophila.
Materials and Methods
Cloning Procedures.
The pCAT-Eg2Delta1, Delta2, and Delta3 plasmids were constructed by inserting the chloramphenicol acetyltransferase (CAT) coding sequence into the XbaI and KpnI sites of pGbEg2–410Delta1, pGbEg2–410Delta2, and pGbEg2–410Delta3 (14), respectively. In these constructions, the Beta-globin 5′UTR is substituted by the CAT coding sequence.
In all of the Drosophila transformation constructs, we used the pCaspeR vector, which contains the white gene as a selectable marker. The lacZ gene from pPMC plasmid (Amersham Pharmacia) was fused in frame to a yem-alpha 5′ genomic fragment that spans the promoter region, the 5′UTR, and part of the coding sequence (up to the StyI site, nucleotide 2354), giving the pYZED plasmid (15). The 3′UTR of Eg5 mRNA, deleted of the endogenous EDEN (plasmid p3′Eg5ΔKN, ref. 11), and the SV40 polyadenylation signal were amplified by PCR and cloned into pGEMT-easy (Promega). Before this PCR amplification, the KpnI site in p3′Eg5ΔKN was removed by digestion with KpnI, blunt-ending, and ligation. The primers used were: Eg5 NC FOR, 5′-GCAGAGTCGACGCACTTGCTTAACC-3′; Eg5 NC REV2, 5′-GGGAATATTTATTCTAGAATAAAATTAAATACAA0–3′; SV40FOR2, 5′-GTAACCATTATATCTAGAAATAAACAAGTTAAC-3′; SV40REV2, 5′-GATCATAATCAGCGGTACCACATTTGTAG-3′. These primers introduce a XbaI site immediately 5′ to the AAUAAA motif in both plasmids, a KpnI site 3′ to the SV40 sequence, and a SalI site 5′ to the Eg5 sequence. Both plasmids were digested by XbaI and SacI (SacI site is in pGEMT-easy). The orientation of the cloned fragments was such that only the Eg5-derived fragment was excised. This fragment was purified and cloned into the digested SV40-containing plasmid, thereby producing pEg5ΔSV40 in which the Eg5 3′UTR is situated immediately 5′ of the SV40 nuclear polyadenylation signal, without a duplication of the AAUAAA motifs. To produce a similar hybrid 3′UTR (pEg5Δyema) in which the nuclear polyadenylation signal was derived from the yem-α gene, the appropriate region of the pyg1 plasmid (16) was amplified using the primers yemapaf3 (5′-TAGAGAACCATTTTCTAGAAATAAAGGCGAATAAGTTATG-3′) and yemapar2 (5′-GATGCTGGTACCTTTCCAGACGAAAAC-3′). After restriction with XbaI and KpnI the PCR product was cloned into pEg5ΔSV40 also restricted with these two enzymes.
Next, the plasmids pYZED, pEg5ΔSV40, and pEg5Δyema were digested with SalI and KpnI restriction enzymes. The fragments derived from pEg5ΔSV40 and pEg5Δyema were purified and cloned into the restricted pYZED. The resultant hybrid genes were excised by digestion with PstI and Acc65I and purified. These fragments were cloned into pCaspeR4 also restricted by these enzymes. The synthetic EDEN motifs in sense and antisense orientations were introduced by cloning the complementary oligonucleotides (TGTA12S, 5′-CTAGGTACCACTAGTGTATGTATGTATGTATGTATGTATGTATGTATGTATGTATGTATGTATGCTAAGCTT 3′, and TGTA12AS, 5′-CTAGAAGCTTAGATCTACATACATACATACATACATACATACATACATACATACATACATACATACACTAGTGGTAC-3′) into the NheI site localized at the position of the endogenous EDEN originally present in the Eg5 3′UTR. The following clones were used in the present study: CO6 pCaspeR4/5′yema/lacZ/Eg5ΔoligoS/3′SV40; CO10 pCaspeR4/5′yema/lacZ/Eg5ΔoligoAS/3′SV40; CO16 pCaspeR4/5′yema/lacZ/Eg5ΔoligoS/3′yema; CO1 pCaspeR4/5′yema/lacZ/Eg5ΔoligoAS/3′yema.
Xenopus Embryos Methods.
Capped, radiolabeled CAT-Eg2Delta1, CAT-Eg2Delta2, and CAT-Eg2Delta3 transcripts were obtained by in vitro transcription using BamHI or EcoRV linearized plasmids to obtain, respectively, poly(A)− and poly(A)+ transcripts, T7 RNA polymerase (Promega), and Cap analog (Biolabs, Northbrook, IL). Microinjections of 20–30 nl of in vitro transcripts (1 fmol) into Xenopus two-cell embryos were done following standard procedures (14). Adenylation activities were analyzed by incubating the injected embryos at 22°C for the indicated times. After incubation, RNAs were extracted, electrophorized on a 4% acrylamide-urea gel, and autoradiographed as described (14). CAT activities were measured in extracts made from injected embryos following standard procedures (17).
P-Element-Mediated Transformation.
A yw fly stock was used as a recipient to construct all of the transgenic flies. The test DNA was injected at a concentration of 100 ng/μl together with the pUC hs (P Delta 2–3) helper DNA (300 ng/μl) as a source of transposase (18). An Eppendorf device was used for the embryo microinjection.
β-Galactosidase Histochemical Staining.
Newly hatched flies were reared for 3 days on fresh yeast. The ovaries were hand dissected and placed in ice cold PBS. They were fixed for 15 min at 4°C in a 0.25% glutaraldehyde/3.7% formaldehyde solution in PBS. After rinsing twice in PBS, the X-Gal staining was carried out at room temperature in a classical staining solution containing X-Gal at a 0.5 mg/ml final concentration (19). To account for effects of chromosomal location on gene activity, at least two transformed lines were systematically examined for each construct.
Whole-Mount in Situ Hybridization.
The ovaries were hand dissected as described above. The in situ hybridization to whole-mount ovaries was carried out essentially as described (20). The probes were made from gel-purified PCR fragments by using the pPMC recombinant plasmid (Amersham Pharmacia) as a template. The digoxigenin labeling of the probes was achieved by nick translation using the Roche nick translation kit.
RNA Extraction from Drosophila Ovaries and Northern Blot Analysis.
Total RNA was extracted from hand-dissected ovaries by using the RNAwiz reagent (Ambion, Austin, TX). The Northern blot experiments were performed using the glyoxal method as described (21). The lacZ and yem-alpha PCR amplified fragments were labeled using a Promega labeling kit. The PCR reactions were carried out with Taq Polymerase from Promega. The lacZ fragment is the same as used for the in situ hybridization experiments, and the sequence of the yem-alpha OA31 and OA10 primers is, respectively: forward, ATGTCAAAGGGGGGCGAGCACAAGCGAGTC; reverse, TCCAGTGCGCCACAGTTG.
Results
An EDEN Containing 3′ UTR Is Sufficient to Repress Translation of Reporter mRNAs in Xenopus Embryos.
Translation of the Eg family and c-mos mRNAs is repressed after fertilization (7, 10). This repression is temporally correlated with the deadenylation of these mRNAs that depends on the EDEN sequence (11). Therefore, we hypothesized that an EDEN sequence may confer translational repression to a reporter mRNA.
To test this hypothesis, three constructs (pCAT-Eg2Delta1 to pCAT-Eg2Delta3) were made. They are based on deletion mutants that have been characterized previously (14). These constructs were used as templates to synthesize RNAs containing the CAT coding sequence and deletion mutants of Eg2 3′UTR (Fig. 1A). Depending on the enzyme used to linearize the templates, the transcripts were polyadenylated (A65) or not. The Delta2 RNA is deleted of the EDEN sequence, and Delta1 and Delta3 RNAs, which are of identical size, have deletions in the regions flanking the EDEN.
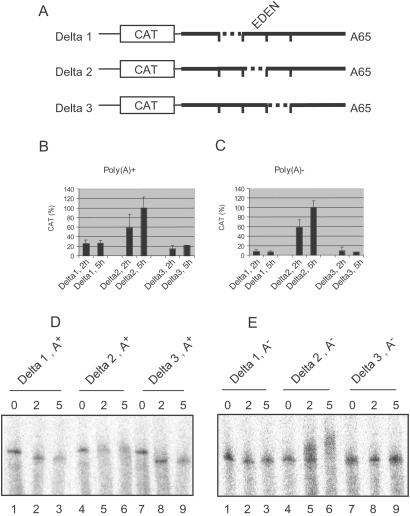
Figure 1.
The EDEN represses translation of a reporter RNA in Xenopus embryos. (A) Schematic drawing (not to scale) of the transcripts used. The CAT coding sequence is boxed. Polylinker sequences are represented by a thin line; Eg2 3′UTR is represented by a thick line. The positions of the 40-nt deletions (Delta1 to Delta3) are indicated by dotted lines. The EDEN is enclosed within the Delta2 deletion. (B) Batches of three embryos injected at the two-cell stage with the indicated poly(A)+ RNAs were pooled 2 and 5 h after injection, and the CAT activities from these pools were measured. For each time point and transcript, the mean activities (±SD) of five pools are represented and normalized to 100% for the maximal activity. (C) Same as in 1B with poly(A)− RNAs. (D) Two-cell embryos were injected with the indicated capped, radiolabeled poly(A)+ RNAs. RNAs were extracted either immediately, after a 2-h or a 5-h incubation from pools of five embryos. The extracted RNAs were resolved by denaturing electrophoresis, and the dried gel was analyzed using a PhosphorImager (Molecular Dynamics). (E) Same as in D with poly(A)− RNAs.
Capped, radiolabeled CAT-Eg2Delta1, CAT-Eg2Delta2, and CAT-Eg2Delta3 RNAs were injected into two-cell Xenopus embryos either in a polyadenylated or in a nonadenylated form. The CAT activities present in the injected Xenopus embryos were measured 2 and 5 h after injection of the chimeric transcripts. When injected as a polyadenylated RNA, the Delta2 transcript that contains no EDEN produced a CAT activity that was readily detected after 2 h of incubation and that has further increased after 5 h of incubation (Fig. 1B Center). Therefore, this transcript is translated between 0 and 2 h, and this translation persists during all or part of the next 3 h of incubation. In contrast, the embryos injected with the EDEN-containing Delta1 and Delta3 transcripts displayed only weak, background levels of CAT activity that were not significantly different between the 2-h and 5-h time points (Left and Right, respectively).
The CAT activity was also determined for the nonadenylated chimeric CAT-Eg2Delta1, CAT-Eg2Delta2, and CAT-Eg2Delta3 transcripts. As observed for the polyadenylated molecules, the CAT activity was significantly stronger in embryos injected with the nonadenylated Delta2 transcript (Fig. 1C). Again, the CAT activity from the Delta2 transcript increased between the 2-h and 5-h time points, indicating a persistence of translation after 2 h. The basal activity (as observed in Delta1 and Delta3) is greater when using initially polyadenylated transcripts (compare Fig. 1 B and C). This result may be attributed to the translation of the injected transcripts before deadenylation has occurred.
To analyze the polyadenylation status of the injected chimeric transcripts, RNAs were extracted immediately after injection or after a 2-h or 5-h incubation, and analyzed by PAGE and autoradiography.
When the chimeric transcripts were injected in a polyadenylated form (Fig. 1D), after a 2-h incubation, the EDEN-containing transcripts (Delta1 and Delta3) migrated at the position of the nonadenylated transcripts (lanes 2 and 8), indicative of a deadenylation. Although some degradation appears to have occurred after 5 h of incubation, it is clear that between the 2-h and 5-h time points these RNAs were maintained in a deadenylated form (lanes 3 and 9). In contrast, the Delta2 transcript displayed a continuous smearing around the size of the injected polyadenylated transcript (lanes 4–6). When the transcripts were injected in a nonadenylated form (Fig. 1E), those containing an EDEN (Delta1 and Delta3) remained poly(A)− (lanes 2 and 3, and 8 and 9), whereas the Delta2 transcript showed a marked decrease in electrophoretic mobility indicative of a polyadenylation (lanes 5 and 6). We have previously shown that the Delta2 deletion disrupts the Eg2 EDEN, thereby permitting the CPE (cytoplasmic polyadenylation element) in the 3′UTR of this mRNA to direct polyadenylation (14).
In conclusion, these results show that the EDEN-containing transcripts are translationally repressed. This translational repression is correlated with the resultant poly(A)− status of the RNA in the embryo; it is independent of the presence of a poly(A) tail on the transcript at the time of injection.
An EDEN Is Sufficient to Suppress Translation in Drosophila Ovaries.
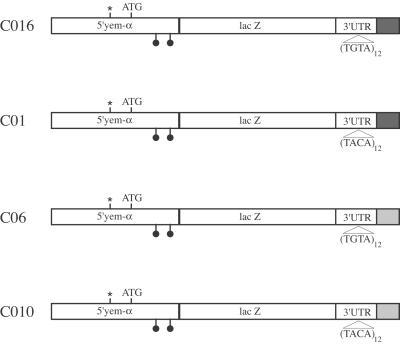
The results described above show a clear role of the EDEN in translational repression in Xenopus eggs. To test the role of EDEN in Drosophila translation, we used transgenic flies that carry different constructs with EDEN-bearing 3′UTRs. We have previously shown that multiple (UGUA) motifs constitute a synthetic EDEN whose efficiency increases with the number of repetitions (22). In the experiments described in this section, a functional EDEN containing twelve contiguous (UGUA) motifs was used. To assess its specificity, this EDEN element was inserted in both orientations (Fig. 2). The sense or antisense EDEN was inserted in place of the natural EDEN in Eg5 3′UTR. The 3′UTR contains no Drosophila sequence. Two different contexts were used downstream to the 3′UTR as pre-mRNA maturation signals, either Drosophila sequences [yem-alpha genomic sequence (16)] or a SV40 sequence.
Figure 2.
Structure of the various constructs used in the transformation experiments. The constructs are under the control of the yem-alpha Drosophila germ line promoter. All of the constructs bear the Eg5 3′UTR in which the natural EDEN was replaced either by a synthetic sense EDEN (TGTA)12 or antisense EDEN (TACA)12. Either yem-alpha (black box) or SV40 (gray box) genomic sequences located downstream to the polyadenylation signal were fused to the 3′end of the constructs. The symbols below the sequence represent the introns present on the yem-alpha genomic fragment used in these constructs. The star indicates yem-alpha transcription start. The ATG is the yem-alpha translation start.
The engineered 3′UTR was fused downstream of a lacZ gene placed under the control of the yem-alpha gene 5′ sequences. In addition to the promoter that is active exclusively in the female germ line, these sequences bear the 5′UTR and the first kilobase of the yem-alpha ORF. The latter sequences are known to be necessary and sufficient for a proper transport of the yem-alpha RNA from the nurse cells to the oocyte (15, 16, 23). The chimeric transcripts produced from these constructs, after transformation of Drosophila embryos, are therefore expressed as a maternal mRNA and efficiently transported into the oocyte (see Fig. 3B). The level of the β-galactosidase protein was determined with an indirect test for its activity, using the X-Gal chromogenic substrate. We assumed that the β-galactosidase activity reflects the protein level.
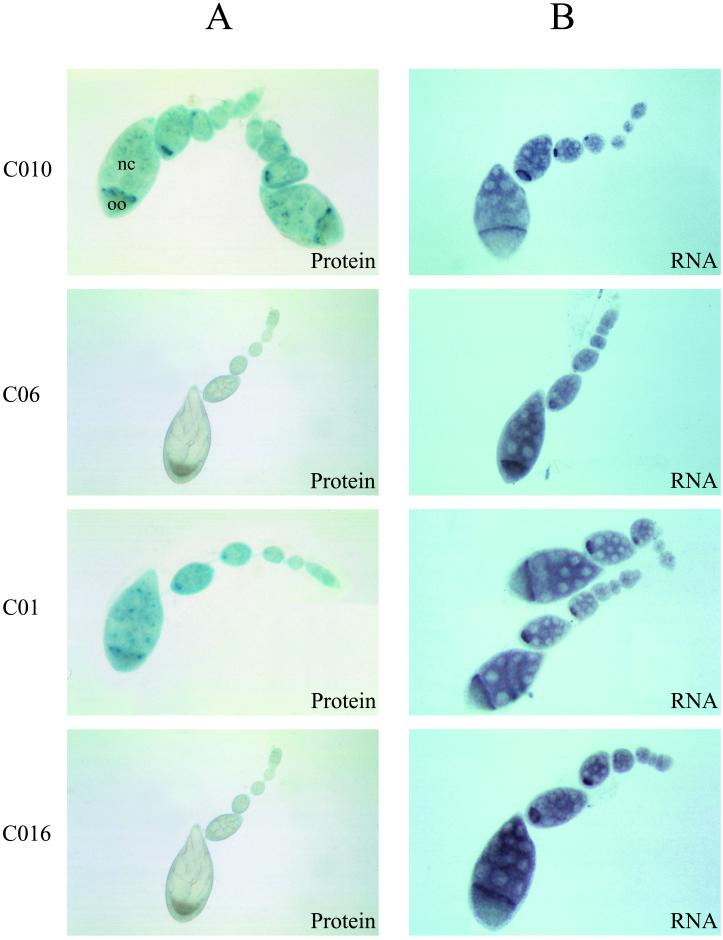
Figure 3.
Expression of β-galactosidase protein and lacZ RNA in Drosophila ovaries. In A are reported the histochemical staining of the various transgenic flies for β-galactosidase activity. In B are shown the in situ hybridization experiments of whole-mount ovaries with a digoxigenin-labeled lacZ probe. Note the concentration of both the β-galactosidase activity and the lacZ RNA in the oocyte (oo).
As shown in Fig. 3A, the β-galactosidase activity was not detectable in ovaries from transgenic flies that bear a sense EDEN, whatever the stage of the examined oocytes. Moreover, this effect was not dependent on the context of the 3′ genomic sequences because no difference was observed between the yem-alpha (CO16) or SV40 (CO6) nuclear polyadenylation signals. In contrast, the β-galactosidase activity was clearly detected in the ovaries from the antisense EDEN-bearing flies (C01, CO10). The EDEN element was therefore sufficient to induce a decrease in protein levels.
To verify whether the decrease in β-galactosidase was due to a genuine translational repression mechanism rather than to a failure of the chimeric RNA to be exported from the nucleus and/or an EDEN induced degradation, we carried out in situ hybridization experiments and Northern blots.
In situ hybridization experiments were performed using a digoxigenin-labeled lacZ probe and the results are shown in Fig. 3B. All of the chimeric RNAs displayed the same localization pattern in the ovaries of all of the transgenic flies—that is, regardless of the orientation of the EDEN element or the 3′ genomic sequences. Moreover, the RNA levels appeared to be similar in all of the ovaries. No staining by the lac Z probe was observed with ovaries from nontransgenic flies (data not shown).
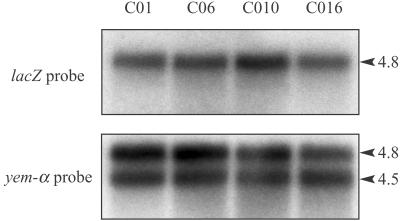
To confirm these data, more quantitative Northern blot analyses were performed. Total RNA from the transgenic ovaries were probed with a lacZ probe (Fig. 4). No significant difference in β-galactosidase RNA level could be detected between RNAs extracted from ovaries of the sense or antisense EDEN transgenic flies. The use of a yem-alpha probe, which revealed both the chimeric RNA (4.8 kb) and the endogenous yem-alpha RNA (4.5 kb), showed that there was no significant variation in RNA loading. This experiment, therefore, rules out RNA degradation as being the cause of the decreased protein levels in the EDEN-bearing transgenic ovaries. In addition, from the difference in electrophoretic mobility between the chimeric (4.8 kb) and endogenous (4.5 kb) RNAs, we calculated that differences in the sizes of the chimeric transcripts as small as 50 nucleotides would have been detected. Therefore, the lengths of the poly(A) tails of the sense and antisense EDEN containing transcripts do not differ by more than 50 nucleotides.
Figure 4.
Northern blot with total RNA from the ovaries of transgenic flies. 20 μg of RNA were subjected to electrophoresis on a 1.2% agarose gel after heat denaturation in the presence of glyoxal and DMSO. After transfer, the membrane was probed either with a lacZ or a yem-alpha probe. The 4.5-kb band corresponds to the yem-alpha endogenous transcript, whereas the 4.8-kb RNA is the chimeric lacZ transcript.
Altogether these data show that the EDEN motif can also repress translation in Drosophila.
Discussion
The EDEN Sequence Represses Translation in Xenopus Eggs and Drosophila Oocytes.
In this report, we show that an EDEN sequence represses the translation of a reporter RNA in Xenopus embryos. This mechanism is active even on mRNAs whose 5′UTR and coding sequences are not derived from natural mRNA targets for the EDEN-mediated repression. The EDEN-mediated translation regulation was also examined during Drosophila oogenesis in transgenic flies transformed with constructs that bear an EDEN in their 3′UTR. Because this mechanism was shown to act on maternally inherited mRNAs in Xenopus, the constructs used in the Drosophila experiments produce maternally expressed mRNA. The yem-alpha promoter, which is specifically active in the female germ line during oogenesis, was used to attain this goal. Visualizing the reporter protein (β-galactosidase) by its enzymatic activity furnished a first line of evidence indicating that the EDEN mediates a decrease in the amount of protein expressed from chimeric transcripts in the Drosophila egg chamber. This decrease depends on the proper orientation of the EDEN sequence in the 3′UTR. Northern blot analysis and in situ hybridization experiments showed that this effect is not due to transcript decay or incorrect export of the EDEN-containing transcripts from the nucleus.
In Xenopus, our data show an EDEN-mediated translational repression of a chimeric RNA after fertilization. This finding is in accordance with previous reports showing that endogenous, EDEN-containing RNAs are translationally repressed after but not before fertilization (7, 10). In Drosophila, the EDEN-mediated mechanism is already active from the very first steps of oogenesis. In fact, this translational repression persists in the preblastoderm Drosophila embryos. No β-galactosidase activity could be detected in embryos obtained from mothers that express the chimeric sense EDEN RNA, despite the maintenance of the chimeric RNA at this stage (data not shown).
In conclusion there is a functional conservation between Xenopus and Drosophila of the EDEN element as a translation regulator.
EDEN-Mediated Translational Repression and Deadenylation.
The mechanism by which the EDEN sequence represses translation in Xenopus embryos probably relies on deadenylation. Arguments for this come from the observation that the Delta2 deletion, which disrupts the EDEN and permits translation, also abrogates deadenylation. In addition, the Delta1 and Delta3 mutants that contain an active EDEN are deadenylated and translationally repressed. Furthermore, the difference in translational activity between EDEN-bearing and EDEN-deleted RNAs is less with initially polyadenylated than with initially deadenylated transcripts. This difference can be imputed to the translation of the polyadenylated Delta1 and Delta3 transcripts before they are deadenylated.
In Drosophila, we observed no difference in transcript size between the sense and the antisense EDEN-containing transcript indicating that the poly(A) tails do not differ by more than 50 nucleotides.
There are two mechanistic models that are compatible with these results. In the first model, in both Xenopus and Drosophila, the EDEN-dependent removal of the poly(A) tail causes a translational repression of the EDEN-containing RNAs by removing the binding site for poly(A)-binding proteins. For this model to be compatible with the data obtained with Drosophila, it is necessary to posit that in the Drosophila egg chamber the chimeric RNA containing the inactive antisense EDEN only has a short poly(A) tail (less than 50 nucleotides). Indeed short poly(A) tails have been documented for several Drosophila maternal mRNAs (12, 24). An EDEN-dependent removal of such a short adenosine tract would not have been detected in the analyses reported here.
In the second model, the mechanisms of EDEN-dependent translational repression are different in the two organisms studied. In Xenopus it depends on deadenylation, whereas in Drosophila it does not require deadenylation. Such cases have been reported in Drosophila. For example, the BRE (bruno responsive element)-mediated translational repression of oskar mRNA in oocyte extracts is active on both poly(A)+ and poly(A)− (24). Also, by using a RNA injection assay, Chagnovich and Lehmann (25) showed that the translational repression of hunchback mRNA in the posterior of Drosophila embryos does not require deadenylation.
Trans-Acting Factors for EDEN-Dependent Translational Repression.
Because EDEN-BP is required for EDEN-dependent deadenylation (11), it is a very likely candidate for mediating EDEN-dependent translational repression in Xenopus embryos. In Drosophila, a candidate for mediating EDEN-dependent translational repression would be Bruno. This factor represses the translation of oskar mRNA, by means of binding to a specific sequence element named a BRE (4). The BRE shows some similarity with EDEN and the Bruno protein shares 50% similarities with EDEN-BP (11, 26). Despite these similarities, our data and data published earlier argue against EDEN and BRE acting through the same pathway. The existence of the EDEN-dependent mechanism in the early Drosophila embryo is a strong argument against Bruno being the genuine EDEN-BP homologue because Bruno protein is restricted to the oocyte (26). Moreover the BRE is not sufficient to induce translational repression of reporter mRNAs even in an eight tandem copy if not used in the oskar 3′UTR context (24). We show here that EDEN is sufficient to repress translation in Drosophila. This repression is independent of the presence of Drosophila sequences in the 3′UTR.
Conservation of Translational Control Regulators in Animals.
Our data show a striking conservation of the EDEN-dependent translational repression between Xenopus and Drosophila. The TGE is another sequence element, different from EDEN, that confers translational repression in an evolutionary conserved manner between vertebrates and non-vertebrates. This sequence represses the translation of tra-2 mRNA in Caenorhabditis Elegans and also of a chimeric RNA in Xenopus embryos (27). A functional conservation of the sequence elements mediating cytoplasmic polyadenylation between Xenopus, mouse, and Drosophila has also been described (28). Although it has not been directly tested, it is highly probable that interspecies polyadenylation signals influence the translational efficiencies.
Together, these and the present results show a surprising functional conservation of the elements regulating translation over the animal kingdom.
Acknowledgments
Thanks are due to Patrick Atger (Institut de Génétique Humaine) for iconographic work. N.E. is a recipient of a fellowship from Lebanon. This work was supported essentially by the Centre National de la Recherche Scientifique and by Research Grants to O.A.-A. (Ministère de La Recherche, ACC-SV, ARC 9932, AFM) and H.B.O. (Ministère de La Recherche, ACC-SV, ARC 9529).
Abbreviations
- EDEN
embryo deadenylation element
- UTR
untranslated region
- CAT
chloramphenicol acetyltransferase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Curtis D, Lehmann R, Zamore P D. Cell. 1995;81:171–178. doi: 10.1016/0092-8674(95)90325-9. [DOI] [PubMed] [Google Scholar]
- 2.Wickens M, Kimble J, Strickland S. In: Translational Control. Hershey J, Mathews M, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Press Lab. Press; 1996. pp. 411–450. [Google Scholar]
- 3.Robbie E P, Peterson M, Amaya E, Musci T J. Development (Cambridge, UK) 1995;121:1775–1785. doi: 10.1242/dev.121.6.1775. [DOI] [PubMed] [Google Scholar]
- 4.Kim-Ha J, Kerr K, Macdonald P M. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 5.Gebauer F, Corona D F, Preiss T, Becker P B, Hentze M W. EMBO J. 1999;18:6146–6154. doi: 10.1093/emboj/18.21.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richter J D. Microbiol Mol Biol Rev. 1999;63:446–456. doi: 10.1128/mmbr.63.2.446-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheets M D, Fox C A, Hunt T, Vande Woude G, Wickens M. Genes Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- 8.Sheets M D, Wu M, Wickens M. Nature (London) 1995;374:511–516. doi: 10.1038/374511a0. [DOI] [PubMed] [Google Scholar]
- 9.Paris J, Osborne H B, Couturier A, Le Guellec R, Philippe M. Gene. 1988;72:169–176. doi: 10.1016/0378-1119(88)90139-4. [DOI] [PubMed] [Google Scholar]
- 10.Paris J, Philippe M. Dev Biol. 1990;140:221–224. doi: 10.1016/0012-1606(90)90070-y. [DOI] [PubMed] [Google Scholar]
- 11.Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne H B. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salles F J, Lieberfarb M E, Wreden C, Gergen J P, Strickland S. Science. 1994;266:1996–1999. doi: 10.1126/science.7801127. [DOI] [PubMed] [Google Scholar]
- 13.Huarte J, Stutz A, O'Connell M L, Gubler P, Belin D, Darrow A L, Strickland S, Vassalli J D. Cell. 1992;69:1021–1030. doi: 10.1016/0092-8674(92)90620-r. [DOI] [PubMed] [Google Scholar]
- 14.Bouvet P, Omilli F, Arlot-Bonnemains Y, Legagneux V, Roghi C, Bassez T, Osborne H B. Mol Cell Biol. 1994;14:1893–1900. doi: 10.1128/mcb.14.3.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capri M, Santoni M J, Thomas-Delaage M, Ait-Ahmed O. Mech Dev. 1997;68:91–100. doi: 10.1016/s0925-4773(97)00130-5. [DOI] [PubMed] [Google Scholar]
- 16.Ait-Ahmed O, Bellon B, Capri M, Joblet C, Thomas-Delaage M. Mech Dev. 1992;37:69–80. doi: 10.1016/0925-4773(92)90016-d. [DOI] [PubMed] [Google Scholar]
- 17.Sleigh M J. Anal Biochem. 1986;156:251–256. doi: 10.1016/0003-2697(86)90180-6. [DOI] [PubMed] [Google Scholar]
- 18.Rio D C, Rubin G M. Mol Cell Biol. 1985;5:1833–1838. doi: 10.1128/mcb.5.8.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanes J R, Rubenstein J L, Nicolas J F. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 21.Maniatis T, Fritsh E, Sambrook J. Molecular Cloning, A Laboratory Manual. Plainveiw, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 22.Audic Y, Omilli F, Osborne H B. Mol Cell Biol. 1998;18:6879–6884. doi: 10.1128/mcb.18.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santoni M J, Ait-Ahmed O, Marilley M. Biochim Biophys Acta. 1998;1399:117–125. doi: 10.1016/s0167-4781(98)00093-1. [DOI] [PubMed] [Google Scholar]
- 24.Lie Y S, Macdonald P M. Development (Cambridge, UK) 1999;126:4989–4996. doi: 10.1242/dev.126.22.4989. [DOI] [PubMed] [Google Scholar]
- 25.Chagnovich D, Lehmann R. Proc Natl Acad Sci USA. 2001;98:11359–11364. doi: 10.1073/pnas.201284398. . (First Published September 18, 2001; 10.1073/pnas.201284398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webster P J, Liang L, Berg C A, Lasko P, Macdonald P M. Genes Dev. 1997;11:2510–2521. doi: 10.1101/gad.11.19.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson S R, Goodwin E B, Wickens M. Mol Cell Biol. 2000;20:2129–2137. doi: 10.1128/mcb.20.6.2129-2137.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verrotti A C, Thompson S R, Wreden C, Strickland S, Wickens M. Proc Natl Acad Sci USA. 1996;93:9027–9032. doi: 10.1073/pnas.93.17.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]