Abstract
The cancers of the gastrointestinal (GI) tract have become a common diagnosis worldwide contributing to a large number of mortalities. Though potentially curable they are mostly fatal due to late diagnosis and lack of accurate diagnostic markers. microRNA, micromanagers of gene expression have been associated to have distinct roles as oncogenes or tumour suppressors in several cancers including GI cancers. These miRNAs are known to harbour single nucleotide polymorphisms (SNPs) that lead to loss or gain of its functions and have been found to be associated with altering susceptibility of several cancers. The current study aimed to investigate the role of miRSNPs in common gastrointestinal cancers. A case control study was designed which included 210 GI cancer cases and 230 cancer free controls. The miRSNPs were successfully genotyped using MassARRAY technique. Association analysis revealed that miR-196a; rs11614913, pre-mir-423; rs6505162, pre-mir-605; rs2043556, pre-mir-149; rs2292832 and pri-mir-30c; rs928508 polymorphisms significantly altered the risk of common GI cancers. Multifactor dimensionality reduction analysis demonstrated that miRSNPs alter GI cancer risk by interacting with exposures like diabetes mellitus, alcohol consumption, diet and socioeconomic status in the study subjects. In conclusion it was found that presence of miRNA polymorphism and certain lifestyle factors alters susceptibility to GI cancers significantly.
Keywords: Gastrointestinal cancers, MicroRNA, Single nucleotide polymorphisms, Multifactor dimensionality reduction, Lifestyle factors, MassARRAY
Graphical abstract
Highlights
-
•
miRNA polymorphisms significantly altered the risk of common GI cancers.
-
•
Diabetes mellitus, alcohol consumption, diet and socioeconomic status added to the risk of GI cancers along with the SNPs.
-
•
Multifactor dimensionality reduction analysis revealed intriguing interactions among miRSNPs and between lifestyle factors.
1. Introduction
The GI tract has an actively renewing epithelium with the most rapid turnover rate in the entire human body, making it more prone to neoplastic changes [1]. Malignancies of the GI tract such as tumors of the primary GI tract like colon, rectum, stomach, esophagus, anus and tumors of the secondary GI tract like gallbladder, pancreas, liver and bile duct amount for 23 % of the world's cancer deaths. After years of research on understanding these tumors, they still exhibit high mortality rates despite being highly curable. Factors such as late diagnosis, improper staging and resistance to treatments contribute to this irony. The 2022 GLOBOCAN official release on cancer statistics lists GI cancers, viz. colorectal cancer (CRC), gastric cancer (GC) and esophageal cancer (EC) as the top contributors to cancer deaths worldwide with CRC and GC occupying the top 5 and EC finding its place among top 10. When considering the incidence and mortality rates of common GI cancers such as CRC, GC and EC it was noted that India was the fourth most affected country.
microRNAs (miRNAs) are small but indispensable molecules of the cellular machinery that orchestrate the expression of almost one third of the human genome. They are noncoding RNA molecules transcribed by the nuclear genome and measure up to 22 nucleotides. They carry out their regulatory function by often binding to the 3′UTR of mRNA and destabilizing them leading to mRNA degradation or repression of translation. Ever since their discovery, miRNAs have been frequently associated with cancers, this was supported by the evidence that more than half of the known miRNAs are located in the fragile regions of genome associated with cancers [2]. miRNAs are said to play dual roles as oncogenes and tumor suppressors in cancer. As an oncogene, they regulate genes involved in tumorigenesis, proliferation, apoptosis, angiogenesis, invasion and migration and facilitate the progression of cancers. As tumor suppressors, miRNAs downregulate oncogenes or upregulate tumor suppressor genes. Several miRNA signatures have been observed in many cancers including breast, leukaemia, lung, thyroid and stomach.
Single nucleotide polymorphisms (SNPs) are single base variations abundantly present in the genome whose effect varies depending on their genomic location. Generally, polymorphisms do not cause a disease but may increase or decrease the susceptibility to a disease [3]. miRNAs are well appreciated for their stringent conservation across several organisms making it difficult for SNPs to occur. However, the past decade has witnessed several research studies reporting the presence of polymorphisms in miRNAs. These microRNA polymorphisms (miRSNPs) constitute a novel class of polymorphisms with functional effects due to their sequence dependent regulatory system. Based on the region where the polymorphisms exist, they can be classified into, miRNA gene polymorphisms, SNPs in miRNA binding regions in mRNAs (binding site polymorphisms) and polymorphisms in miRNA biogenesis pathways. Presence of SNPs in miRNAs lead to loss of function or gain in function of miRNAs leading to dysregulation of gene expression causing disease phenotypes [4].
Carlo Corce et al., were the forerunners who found a germline mutation in pri-mir-16-1 that was associated with familial chronic lymphocytic leukaemia and found to decrease expression of the miR-16-1 [5]. This was followed by a study on miR-146a polymorphism rs2910164 in papillary thyroid carcinoma (PTC) where the polymorphism increased cancer susceptibility by affecting miRNA expression [6] and till date a few miRNAs and their polymorphisms have been investigated in many cancers. Polymorphisms in miR-196a, miR-499a, miR-149, miR-27a, miR-423 and miR-let-7a2 have been found to be associated with the risk of common GI cancers but limited mostly to European and Chinese populations [7,8]. Thus, it is clear that microRNAs and their variants play an important role in altering the susceptibility to various cancers in different populations. They have immense potential to serve as population based genetic markers to predict risk, for early diagnosis and better management of disease outcome. In our previous studies we found that miR-146a rs2910164 polymorphism significantly increased the risk of CRC, whereas, miR-27a gene polymorphisms rs895819 and rs11671784 were not associated with GI cancers [9,10]. Therefore, we aimed to explore the role of various other microRNA polymorphisms in gastrointestinal cancers and analyse how they present themselves along with other environmental factors in subjects from the southern states of India.
2. Materials and methods
2.1. Study population and sample collection
The study was approved by the Institutional Ethics Committee of Sri Ramachandra Institute of Higher Education and Research (Deemed to be University) IEC-NI/13/APR/33/39 and was conducted as per ICMR ethical guidelines for biomedical research and the declaration of Helsinki.
The recruitment of subjects has been elaborately described in our earlier publication [10]. A total of 210 GI cancer patients and 230 cancer free controls were recruited for the study.
Around 3 mL of venous blood collected for routine clinical investigations was received from the Central laboratory of Sri Ramachandra Medical Centre and clinical profile of subjects was retrieved from the hospital medical records department. Basic characteristics of the subjects were obtained by interviewing them by means of a structured questionnaire. The identity and privacy of the participants were preserved throughout the study.
2.2. Selection of microRNA polymorphisms for the study
A total of 18 SNPs in 17 microRNA genes were identified from miRNA databases and literature. All the miRNAs were regulators of genes involved in cancer pathways. The polymorphisms selected were limited to the miRNA genes and flanking regions. The primary miRNA energy, SNP miRNA energy and △G was obtained from miRNA SNP database V2.0.
2.3. SNP genotyping
Genotyping was done by using MassARRAY kits purchased from Agena Bioscience Inc, San Diego, CA, USA. The MassARRAY system implies the Matrix Assisted Laser Desorption/Ionisation-Time of Flight methodology (MALDI-TOF) [11]. The major steps involved in it are multiplex PCR, shrimp alkaline phosphatase (SAP) reaction, Extension reaction and MALDITOF MS.
The SNP IDs selected for the study were submitted to the assay design software for primer and assay design. The extension and PCR primers sequences were retrieved from the software and procured commercially from Integrated DNA Technologies, Inc. USA.
The multiplex PCR was set up with 4 μL of multiplex PCR cocktail and 2 μL of the appropriate genomic DNA sample (5–10 ng/μL) in each well of a 384-well microtiter plate. The plate was then centrifuged at 10,000 rpm for 1 min and was run in the thermocycler with the following reaction conditions; initial denaturation at 95 °C for 15 min, annealing at 56 °C for 30 s, extension at 72 °C for 1 min. Steps 1–3 were repeated for 45 cycles following which final extension was carried out at 72 °C for 3 min. The SAP reaction was done by adding 2 μL of the SAP mixture to each sample in the 384-well microtiter plate. The plate was incubated in a thermocycler at 37 °C for 40 min, 55 °C for 5 min and stored at 4 °C.
The iPLEX Gold reaction mixture was prepared as per manufacturer's instructions and 2 μL of it was added to each sample in the 384-well microtiter plate. The plate was sealed with the sealant and cycled in a ABI thermo cycler with the following reaction conditions; 94 °C for 30 s, 94 °C for 5 s, 5 cycles at 52 °C for 5 s and 80 °C for 5 s, 72 °C for 3 min and stored at 4 °C after completion of 40 cycles.
The samples were conditioned with 6 mg of clean resin per well to remove unwanted ions that could interfere with the MALDI-TOF MS analysis. The samples were then dispensed on to the SpectroCHIP by the Nano dispenser. The SpectroCHIP spotted with the samples was placed in the chip slot of the mass spectrometer. The results were viewed as cluster plots using Typer Tool 4.0.
2.4. Statistical analysis
The concordance of genotype frequencies with Hardy – Weinberg Equilibrium (HWE) was calculated using the chi-square test. SNPs with P value more that 0.05 was considered to be concurrent with HWE. The association of the genotypes and alleles with GI cancer was done by calculating the odds ratio with 95 % CI through Binary logistic regression while P value less than 0.05 was considered significant. The crude odds ratio was adjusted for covariates using multinomial regression. The statistical tests were done using SPSS v16.0. The interaction between the SNPs and environmental parameters were tested using Multifactor dimensionality reduction (MDR) analysis by MDR software v3.2 [12].
3. Results
3.1. SNPs selected for the study
A total of 18 miRSNPs; miR-196a; rs11614913, miR-499a; rs3746444, miR-149; rs2292832, miR-25; rs41274221, miR-26a-1; rs7372209, miR-218-2; rs11134527, miR-124-1; rs531564, miR-92a-1; rs9589207, miR-608; rs4919510, miR-605; rs2043556, miR-34b/c; rs4938723, miR-603; rs11014002, miR-30c; rs928508, miR-423; rs6505162, miR-Let7a2; rs629367, miR-27a; rs895819, rs11671784 and miR-146a; rs2910164 were identified from miRNA databases and literature. . SNPs rs2910164, rs11671784 and rs895819 were analysed in our earlier study [9,10]. In the current study the results will be included in the MDR analysis to check for SNP-SNP interactions and Genotype-Phenotype interactions. The results of the 15 SNPs have been presented below.
3.2. Distribution of miRNA polymorphisms and compliance with hardy weinberg equilibrium
Features of the study population such as age, sex and basic characteristics have been represented already. The genotype and allele frequencies of all the 16 miRSNPs were calculated and tested for HWE using chi square test. All the polymorphisms which were concordant with HWE in the control group were included for the study. It was found that polymorphisms rs3746444, rs531564, rs4919510 and rs9589207 present in miR-499, miR-124-1, miR-608 and miR-92a-1 genes respectively were not in HWE in the control group and were excluded from further analysis. HWE concordance in the control group is the standard criteria for association analysis. The HWE deviation could be due to genotyping error or population stratification effect. All the other polymorphisms were in HWE. The miR-25 rs41274221 polymorphism was not observed in our study population (Table 1).
Table 1.
Frequency distribution of the genotypes and HWE analysis of the selected miRNA polymorphisms.
| microRNA |
SNP | Base Change | Control Genotypes N (%) |
MAF |
HWE |
Case Genotypes N (%) |
MAF |
HWE |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wt | Hz | Mu | Χ2 (p value) | Wt | Hz | Mu | Χ2 (p value) | ||||||
| miR-92a-1 | rs9589207 | G/A | 199(99.5) | 0(0) | 1(0.5) | 0.01 | 200(0.00) | 198(99) | 0(0) | 2(1) | 0.01 | 200(0.000) | |
| miR-608 | rs4919510 | C/G | 93(46.5) | 77(38.5) | 29(15.0) | 0.34 | 4.22(0.040) | 93(46.7) | 87(43.7) | 19(9.5) | 0.31 | 0.04(0.835) | |
| miR-605 | rs2043556 | T/C | 118(59) | 65(32.5) | 17(8.5) | 0.25 | 3.25(0.071) | 126(63) | 67(33.5) | 7(3.5) | 0.20 | 0.28(0.6) | |
| miR-34b/c | rs4938723 | C/T | 123(61.5) | 45(22.5) | 5(2.5) | 0.22 | 0.53(0.467) | 152(76) | 45(22.5) | 3(1.5) | 0.26 | 0.026(0.873) | |
| miR-603 | rs11014002 | C/T | 157 (159.3) | 43(38.4) | 0 (2.3) | 0.11 | 2.902(0.089) | 155(77.5) | 37(18.5) | 8(4.0) | 0.46 | 7.625(0.006) | |
| miR-30c | rs928508 | A/G | 69(34.5) | 92(46.0) | 39(19.5) | 0.43 | 0.692(0.401) | 54(27.0) | 106(53.0) | 40(20.0) | 0.47 | 0.851(0.356) | |
| miR-423 | rs6505162 | A/C | 63(31.7) | 89(44.7) | 47(23.6) | 0.46 | 1.98(0.1596) | 72(36.2) | 82(41.2) | 45(22.6) | 0.43 | 5.121(0.024) | |
| miR-Let7a2 | rs629367 | A/C | 129(64.5) | 62(31) | 9(4.5) | 0.20 | 0.195(0.659) | 121(60.5) | 65(32.5) | 14(7) | 0.23 | 1.597(0.206) | |
| miR-27a | rs11671784 | G/A | 183(96.3) | 7(3.7) | 0(0) | 0.02 | 0.07 (0.796) | 172(94) | 10(5.5) | 1(0.5) | 0.19 | 3.51 (0.061) | |
| miR-196a | rs11614913 | C/T | 97(48.5) | 79(39.5) | 24(12) | 0.32 | 1.569(0.06) | 115(57.5) | 73(36.5) | 12(6.0) | 0.24 | 0.0008(0.927) | |
| miR-499 | rs3746444 | A/G | 91(45.5) | 10(5) | 99(49.5) | 0.52 | 161.94(0.00) | 104(52) | 5(2.5) | 91(45.5) | 0.47 | 180.42(0.000) | |
| miR-149 | rs2292832 | T/C | 62(31) | 90(45) | 48(24) | 0.47 | 1.83(0.177) | 64(32) | 90(45) | 46(23) | 0.46 | 1.72(0.190) | |
| miR-25 | rs41274221 | C/T | 200(100) | 0(0) | 0(00) | 0.10 | 220(0.000) | 202(100) | 0(0) | 0(0) | 0.00 | – | |
| miR-26a-1 | rs7372209 | C/T | 150(75) | 45(22.5) | 5(2.5) | 0.14 | 0.53(0.467) | 152(76) | 45(22.5) | 3(1.5) | 0.13 | 0.03(0.873) | |
| miR-218-2 | rs11134527 | A/G | 60(30) | 95(47.5) | 45(22.5) | 0.46 | 0.398(0.528) | 60(30.0) | 92(46) | 48(24) | 0.47 | 1.176(0.278) | |
| miR-124-1 | rs531564 | C/G | 188(94) | 10(5) | 2(1) | 0.04 | 13.5(0.000) | 179(89.5) | 19(9.5) | 2(1.0) | 0.06 | 3.05(0.081) | |
Wt, wild type, Hz, Heterozygous, Mu, Mutant, MAF, Minor allele frequency, HWE, Hardy Weingberg Equilibrium. bold p values in controls represent SNPs not in HWE.
3.3. Association of miRNA polymorphisms with common GI cancers
3.3.1. Association of miR-196a polymorphism rs11614913 with GI cancers
The dominant model showed an increase in GI cancer risk in model a (ORa = 1.52, pa = 0.051). Further the analyses revealed that ‘TT’, ‘T’ genotype and alleles significantly increased the risk of GI cancers (CC vs TT; OR = 2.35, p value = 0.024, ORa = 2.58, Pa = 0.020, ORb = 2.31, Pb = 0.035 & C Vs T; OR = 1.44, p value = 0.021, ORa = 1.52, Pa = 0.013, ORb = 1.4, pb = 0.041) while the recessive model showed that the ‘CC’ genotype had a protective effect (Recessive; OR = 0.47, p value = 0.041, ORa = 0.44, pa = 0.038, ORb = 0.48, pb = 0.050) against GI cancer. In the subgroup'others’ we found that the ‘TT’ genotypes significantly increased the risk of these cancers. However, due to the small numbers further warranting of these results is needed for the others group (Table 2). The allele frequencies obtained from the study were compared with frequencies from worldwide populations obtained from 1000 genomes Phase 3 project and it showed that allele frequency recorded in the study population was found similar to the South Asian populations (Supplementary Fig. 1).
Table 2.
Association analysis of miR-196a rs11614913 SNP with GI cancers.
| Cancer Site | Model | OR | 95 % CI | p Value | ORa | 95 % CI | pa Value | ORb | 95 % CI | pb Value |
|---|---|---|---|---|---|---|---|---|---|---|
| GIC | Dominant | 1.42 | 096–2.11 | 0.079 | 1.52 | 1–2.31 | 0.051 | 1.36 | 0.90–2.05 | 0.142 |
| Recessive | 0.47 | 0.23–0.97 | 0.041 | 0.44 | 0.21–0.96 | 0.038 | 0.48 | 0.23–1.00 | 0.050 | |
| CC Vs CT | 1.27 | 0.84–1.93 | 0.259 | 1.35 | 0.87–2.10 | 0.180 | 1.21 | 0.78–1.87 | 0.391 | |
| CC Vs TT | 2.35 | 1.12–4.95 | 0.024 | 2.58 | 1.16–5.75 | 0.020 | 2.31 | 1.06–5.04 | 0.035 | |
| C Vs T | 1.44 | 1.06–1.97 | 0.021 | 1.52 | 1.09–2.11 | 0.013 | 1.4 | 1.01–1.93 | 0.041 | |
| GC | Dominant | 1.21 | 0.71–2.05 | 0.479 | 1.3 | 0.72–2.34 | 0.381 | 1.18 | 0.69–2.02 | 0.557 |
| Recessive | 0.3 | 0.09–1.02 | 0.053 | 0.34 | 0.08–1.31 | 0.117 | 0.3 | 0.1–1.04 | 0.058 | |
| CC Vs CT | 1.01 | 0.59–1.75 | 0.966 | 1.15 | 0.62–2.13 | 0.653 | 0.99 | 0.57–1.74 | 0.977 | |
| CC Vs TT | 3.38 | 0.96–11.8 | 0.057 | 3.1 | 0.78–12.2 | 0.107 | 3.26 | 0.91–11.7 | 0.069 | |
| C Vs T | 1.37 | 0.90–2.09 | 0.140 | 1.38 | 0.87–2.21 | 0.172 | 1.35 | 0.88–2.06 | 0.175 | |
| CRC | Dominant | 1.51 | 1–2.56 | 0.122 | 1.64 | 0.95–2.83 | 0.075 | 1.56 | 0.91–2.68 | 0.110 |
| Recessive | 0.7 | 0.29–1.7 | 0.436 | 0.59 | 0.24–1.50 | 0.274 | 0.7 | 0.28–1.74 | 0.445 | |
| CC Vs CT | 1.47 | 0.84–2.59 | 0.179 | 1.54 | 0.86–2.75 | 0.144 | 1.53 | 0.85–2.75 | 0.157 | |
| CC Vs TT | 1.67 | 0.67–4.13 | 0.275 | 2.12 | 0.79–5.64 | 0.134 | 1.78 | 0.69–4.59 | 0.233 | |
| C Vs T | 1.4 | 0.92–2.11 | 0.116 | 1.52 | 0.99–2.34 | 0.056 | 1.43 | 0.93–2.19 | 0.107 | |
| EC | Dominant | 1.16 | 0.49–2.75 | 0.739 | 1.26 | 0.52–3.04 | 0.604 | 1.22 | 0.49–3.03 | 0.664 |
| Recessive | 0.69 | 0.15–3.17 | 0.642 | 0.66 | 0.14–3.08 | 0.597 | 0.7 | 0.15–3.30 | 0.655 | |
| CC Vs CT | 1.09 | 0.44–2.71 | 0.860 | 1.21 | 0.48–3.06 | 0.694 | 1.15 | 0.44–3.03 | 0.772 | |
| CC Vs TT | 1.49 | 0.31–7.08 | 0.62 | 1.46 | 0.3–7.16 | 0.639 | 1.58 | 0.30–8.21 | 0.587 | |
| C Vs T | 1.18 | 0.60–2.32 | 0.629 | 1.26 | 0.63–2.49 | 0.516 | 1.22 | 0.60–2.49 | 0.580 | |
| Others | Dominant | 2.97 | 1.03–8.57 | 0.044 | 2.87 | 0.98–8.36 | 0.054 | 2.73 | 0.92–8.13 | 0.071 |
| CC Vs CT | 2.28 | 0.79–6.61 | 0.129 | 2.29 | 0.78–6.7 | 0.131 | 2.07 | 0.69–6.22 | 0.194 | |
| C Vs T | 3.07 | 1.17–8.05 | 0.023 | 2.88 | 1.09–7.61 | 0.033 | 2.88 | 1.08–7.7 | 0.035 |
CI; Class Interval, OR; Crude Odds Ratio, ORa, Pa; adjusted for age and sex, ORb, Pb; adjusted for Diabetes, diet and region, p value - significant values are bold. GIC; Gastrointestinal cancers, GC; Gastric cancer, CRC; Colorectal cancer, EC; Esophageal cancer.
3.3.2. Association of pre-mir-149 polymorphism rs2292832 with GI cancers
Association analysis showed that the ‘CC’, ‘C’ genotype and alleles of this polymorphism significantly increased the risk of GI cancers on the whole in both unadjusted and adjusted models (TT Vs CC; crude OR = 1.80, p = 0.032 & T Vs C; crude OR = 1.38, p = 0.024). The recessive model indicated that the wild type genotype had a positive effect on the carriers in the adjusted models (ORb = 0.63, pb = 0.05). Further, the subgroup analysis done to check the effect of the polymorphism on the individual GI cancer groups showed that the polymorphism influenced the risk of CRC. The ‘CC’ genotype and ‘C’ allele increased the risk of CRC significantly in the adjusted (model b) and unadjusted models (TT Vs CC; ORb = 2.45, pb = 0.026 & T Vs C; crude OR = 1.44, p = 0.052, ORb = 1.54, pb = 0.027). Moreover, the heterozygous genotype ‘TC’ was found to increase the risk of esophageal cancer when adjusted for DM, region and diet, but the results were only marginally significant (TT Vs TC; ORb = 2.92, pb = 0.068) (Table 3). The allele frequencies of the control groups were compared to the 1000 genomes Phase 3 worldwide populations and it was found to be same as the Gujarati Indian population (Supplementary Fig. 2).
Table 3.
Association analysis of Pre-mir-149 rs2292832 SNP with GI cancers.
| Cancer Site | Model | OR | 95 % CI | p Value | ORa | 95 % CI | pa Value | ORb | 95 % CI | pb Value |
|---|---|---|---|---|---|---|---|---|---|---|
| GIC | Dominant | 1.5 | 0.97–2.33 | 0.070 | 1.4 | 0.89–2.25 | 0.145 | 1.68 | 1.06–2.63 | 0.029 |
| Recessive | 0.67 | 0.43–1.05 | 0.077 | 0.64 | 0.40–1.02 | 0.063 | 0.63 | 0.39–1.0 | 0.05 | |
| TT Vs TC | 1.35 | 0.84–2.17 | 0.218 | 1.25 | 0.76–2.05 | 0.383 | 1.44 | 0.87–2.38 | 0.153 | |
| TT Vs CC | 1.8 | 1.05–3.07 | 0.032 | 1.77 | 1.01–3.11 | 0.046 | 2.1 | 1.18–3.75 | 0.012 | |
| T Vs C | 1.38 | 1.04–1.82 | 0.024 | 1.37 | 1.02–1.83 | 0.036 | 1.47 | 1.1–1.97 | 0.009 | |
| GC | Dominant | 1.43 | 0.80–2.56 | 0.224 | 1.37 | 0.71–2.63 | 0.353 | 1.62 | 0.89–2.95 | 0.117 |
| Recessive | 0.73 | 0.40–1.33 | 0.301 | 0.74 | 0.38–1.43 | 0.369 | 0.73 | 0.4–1.34 | 0.307 | |
| TT Vs TC | 1.32 | 0.71–2.48 | 0.383 | 1.27 | 0.63–2.56 | 0.499 | 1.5 | 0.78–2.89 | 0.226 | |
| TT Vs CC | 1.63 | 0.80–3.32 | 0.177 | 1.61 | 0.74–3.49 | 0.23 | 1.82 | 0.87–3.82 | 0.111 | |
| T Vs C | 1.3 | 0.90–1.90 | 0.155 | 1.28 | 0.84–1.93 | 0.249 | 1.37 | 0.94–2.01 | 0.104 | |
| CRC | Dominant | 1.53 | 0.86–2.69 | 0.147 | 1.36 | 0.75–2.45 | 0.307 | 1.68 | 0.92–3.05 | 0.091 |
| Recessive | 0.6 | 0.33–1.11 | 0.104 | 0.59 | 0.32–1.12 | 0.109 | 0.55 | 0.29–1.05 | 0.068 | |
| TT Vs TC | 1.32 | 0.72–2.43 | 0.377 | 1.18 | 0.63–2.22 | 0.601 | 1.37 | 0.72–2.6 | 0.341 | |
| TT Vs CC | 1.98 | 0.96–4.05 | 0.063 | 1.79 | 0.85–3.78 | 0.124 | 2.45 | 1.11–5.37 | 0.026 | |
| T Vs C | 1.44 | 1–2.09 | 0.052 | 1.38 | 0.95–2.01 | 0.096 | 1.54 | 1.05–2.26 | 0.027 | |
| EC | Dominant | 2.04 | 0.83–5 | 0.121 | 1.84 | 0.73–4.6 | 0.195 | 2.32 | 0.87–6.16 | 0.092 |
| Recessive | 0.97 | 0.38–2.49 | 0.956 | 0.98 | 0.38–2.52 | 0.965 | 1.01 | 0.38–2.66 | 0.985 | |
| TT Vs TC | 2.41 | 0.85–6.88 | 0.1 | 2.33 | 0.80–6.73 | 0.119 | 2.92 | 0.92–9.22 | 0.068 | |
| TT Vs CC | 1.66 | 0.58–4.78 | 0.347 | 1.46 | 0.49–4.36 | 0.498 | 1.81 | 0.59–5.57 | 0.303 | |
| T Vs C | 1.37 | 0.74–2.53 | 0.314 | 1.31 | 0.70–2.43 | 0.4 | 1.39 | 0.73–2.63 | 0.318 | |
| Others | Dominant | 1.13 | 0.39–3.30 | 0.822 | 1.13 | 0.38–3.35 | 0.828 | 1.68 | 0.54–5.26 | 0.370 |
| Recessive | 0.42 | 0.12–1.49 | 0.177 | 0.42 | 0.12–1.51 | 0.185 | 0.27 | 0.06–1.21 | 0.087 | |
| TT Vs TC | 0.85 | 0.28–2.6 | 0.778 | 0.87 | 0.28–2.73 | 0.811 | 1.11 | 0.35–3.54 | 0.865 | |
| TT Vs CC | 2.15 | 0.49–9.46 | 0.31 | 2.26 | 0.51–10.1 | 0.284 | 4.47 | 0.77–25.9 | 0.095 | |
| T Vs C | 1.42 | 0.73–2.78 | 0.303 | 1.42 | 0.72–2.79 | 0.315 | 1.83 | 0.90–3.73 | 0.095 |
CI; Class Interval, OR; Crude Odds Ratio, ORa, Pa; adjusted for age and sex, ORb, Pb; adjusted for Diabetes, diet and region, p values-significant values are bold. GIC; Gastrointestinal cancers, GC; Gastric cancer, CRC; Colorectal cancer, EC; Esophageal cancer.
3.3.3. Association of pre-mir-423 polymorphism rs6505162 with GI cancers
The rs6505162 polymorphism had no association with GI cancers as a whole in any of the genetic models. However, the analysis with subgroup cancers showed a strong association between CRC and the polymorphism. The ORs indicated that the ‘AA’ genotype of the SNP increased the risk of CRC in the adjusted model (ORb = 2.39; pb = 0.037). Similarly, the allelic model also suggested that the ‘A’ allele significantly influenced the risk of CRC (Crude OR = 1.44, p = 0.045, ORb = 1.6; pb = 0.013) (Table 4). The allele frequencies of the control groups were compared to the 1000 genomes Phase 3 worldwide populations and found similar to the Indian Telugu in UK, Punjab from Lahore and Srilankan Tamil in UK populations (Supplementary Fig. 3).
Table 4.
Association analysis of Pre-mir-423 rs6505162 SNP with GI cancers.
| Cancer Site | Model | OR | 95 % CI | p Value | ORa | 95 % CI | Pa Value | ORb | 95 % CI | pb Value |
|---|---|---|---|---|---|---|---|---|---|---|
| GIC | Dominant | 1.21 | 0.80–1.84 | 0.359 | 1.12 | 0.72–1.73 | 0.618 | 1.29 | 0.84–1.99 | 0.243 |
| Recessive | 0.93 | 0.58–1.49 | 0.762 | 0.98 | 0.60–1.61 | 0.944 | 0.82 | 0.50–1.34 | 0.434 | |
| CC Vs AC | 1.22 | 0.78–1.92 | 0.385 | 1.14 | 0.71–1.82 | 0.599 | 1.26 | 0.79–2.0 | 0.342 | |
| CC Vs AA | 1.20 | 0.71–2.05 | 0.498 | 1.09 | 0.62–1.92 | 0.765 | 1.45 | 0.82–2.56 | 0.207 | |
| C Vs A | 1.12 | 0.86–1.12 | 0.401 | 1.05 | 0.8–1.39 | 1.054 | 1.22 | 0.92–1.59 | 0.166 | |
| GC | Dominant | 1.24 | 0.71–2.15 | 0.446 | 1.17 | 0.64–2.17 | 0.609 | 1.33 | 0.75–2.35 | 0.323 |
| Recessive | 0.87 | 0.46–1.65 | 0.665 | 1.05 | 0.51–2.15 | 0.897 | 0.79 | 0.41–1.53 | 0.482 | |
| CC Vs AC | 1.22 | 0.67–2.22 | 0.522 | 1.23 | 0.63–2.41 | 0.545 | 1.28 | 0.69–2.36 | 0.434 | |
| CC Vs AA | 1.29 | 0.63–2.64 | 0.495 | 1.13 | 0.51–2.51 | 0.758 | 1.64 | 0.76–3.58 | 0.211 | |
| C Vs A | 1.12 | 0.86–1.46 | 0.401 | 1.05 | 0.79–1.39 | 0.712 | 1.22 | 0.92–1.59 | 0.166 | |
| CRC | Dominant | 1.34 | 0.78–2.31 | 0.282 | 1.28 | 0.74–2.24 | 0.379 | 1.53 | 0.87–2.68 | 0.139 |
| Recessive | 0.57 | 0.29–1.15 | 0.118 | 0.57 | 0.28–1.17 | 0.126 | 0.51 | 0.24–1.05 | 0.69 | |
| CC Vs AC | 1.17 | 0.66–2.07 | 0.603 | 1.12 | 0.62–2.03 | 0.698 | 1.26 | 0.71–2.35 | 0.410 | |
| CC Vs AA | 1.90 | 0.88–4.08 | 0.101 | 1.81 | 0.82–4.01 | 0.145 | 2.39 | 1.05–5.4 | 0.037 | |
| C Vs A | 1.44 | 1.01–2.05 | 0.045 | 1.41 | 0.98–2.03 | 0.067 | 1.596 | 1.1–2.3 | 0.013 | |
| EC | Dominant | 1.37 | 0.56–3.32 | 0.491 | 1.3 | 0.53–3.20 | 0.566 | 1.39 | 0.53–3.6 | 0.502 |
| Recessive | 2.09 | 0.85–5.14 | 0.107 | 2.22 | 0.89–5.53 | 0.087 | 2.06 | 0.8–5.3 | 0.134 | |
| CC Vs AC | 2.50 | 0.80–7.82 | 0.115 | 2.5 | 0.79–7.93 | 0.120 | 2.44 | 0.74–8.1 | 0.145 | |
| CC Vs AA | 0.73 | 0.27–1.99 | 0.544 | 0.64 | 0.23–1.79 | 0.398 | 0.74 | 2.5–2.195 | 0.592 | |
| C Vs A | 0.76 | 0.43–1.34 | 0.347 | 0.72 | 0.41–1.28 | 0.267 | 0.76 | 0.41–1.38 | 0.362 | |
| Others | Dominant | 0.57 | 0.18–1.78 | 0.330 | 0.52 | 0.16–1.65 | 0.267 | 0.68 | 0.21–2.23 | 0.521 |
| Recessive | 1.90 | 0.71–5.1 | 0.203 | 2.1 | 0.76–5.74 | 0.148 | 1.62 | 0.55–4.75 | 0.383 | |
| CC Vs AC | 0.70 | 0.20–2.41 | 0.566 | 0.65 | 0.19–2.29 | 0.502 | 0.88 | 0.24–3.19 | 0.841 | |
| CC Vs AA | 0.42 | 0.12–1.52 | 0.185 | 0.38 | 0.10–1.39 | 0.145 | 0.48 | 0.12–2.03 | 0.320 | |
| C Vs A | 0.59 | 0.31–1.11 | 0.099 | 0.54 | 0.28–1.03 | 0.062 | 0.67 | 0.34–1.30 | 0.234 |
CI; Class Interval, OR; Crude Odds Ratio, ORa, Pa; adjusted for age and sex, ORb, Pb; adjusted for Diabetes, diet and region, p values-significant values are bold. GIC; Gastrointestinal cancers, GC; Gastric cancer, CRC; Colorectal cancer, EC; Esophageal cancer.
3.3.4. Association of pri-mir-30c polymorphism rs928508 with GI cancers
The association analysis showed that AA & AG genotypes reduced GI cancer risk (Dominant; ORb = 0.64, pb = 0.048 & GG Vs AG; ORb = 0.62, pb = 0.045). In the gastric cancer subgroup the dominant model showed a similar risk reducing effect (OR = 0.49, p = 0.024; ORa = 0.51, pa = 0.052; ORb = 0.47, pb = 0.019) The heterozygote ‘AG’ genotype also reduced the risk of gastric cancer in our study population in the co dominant model (GG Vs AG; OR = 0.47, p = 0.022). The results were consistent even after adjusting for covariates (GG Vs AG; ORa = 0.51, pa = 0.052, ORb = 0.47, pb = 0.019). There was no association of the polymorphism with other GI cancers (Table 5). In the esophageal cancer subgroup the recessive model showed an increased risk (ORb = 2.7, pb = 0.045) and therefore due to small size of the EC subgroup this results need to be validated further for consideration. The allele frequencies derived from the current study were similar to those of the South Asians (Supplementary Fig. 4).
Table 5.
Association analysis of Pri-mir-30c rs928508 SNP with GI cancers.
| Cancer Site | Model | OR | 95 % CI | p Value | ORa | 95 % CI | pa Value | ORb | 95 % CI | pb Value |
|---|---|---|---|---|---|---|---|---|---|---|
| GIC | Dominant | 0.69 | 0.45–1.06 | 0.90 | 0.68 | 0.44–1.06 | 0.095 | 0.64 | 0.41–0.99 | 0.048 |
| Recessive | 1.04 | 0.63–1.71 | 0.880 | 1 | 0.59–1.68 | 0.999 | 1.04 | 0.62–1.74 | 0.881 | |
| GG Vs AG | 0.67 | 0.43–1.05 | 0.082 | 0.65 | 0.41–1.06 | 0.082 | 0.62 | 0.38–0.98 | 0.045 | |
| GG Vs AA | 0.75 | 0.43–1.33 | 0.327 | 0.76 | 0.41–1.39 | 0.375 | 0.71 | 0.39–1.28 | 0.255 | |
| G Vs A | 0.84 | 0.64–1.11 | 0.229 | 0.85 | 0.63–1.14 | 0.272 | 0.82 | 0.61–1.08 | 0.166 | |
| GC | Dominant | 0.49 | 0.26–0.91 | 0.024 | 0.51 | 0.25–1.01 | 0.052 | 0.47 | 0.25–0.88 | 0.019 |
| Recessive | 1.12 | 0.58–2.15 | 0.738 | 1.02 | 0.49–2.10 | 0.955 | 1.15 | 0.59–2.24 | 0.683 | |
| GG Vs AG | 0.47 | 0.24–0.85 | 0.022 | 0.48 | 0.23–0.99 | 0.046 | 0.45 | 0.23–0.88 | 0.019 | |
| GG Vs AA | 0.54 | 0.25–1.21 | 0.133 | 0.58 | 0.24–1.43 | 0.237 | 0.5 | 0.22–1.14 | 0.100 | |
| G Vs A | 0.84 | 0.64–1.11 | 0.229 | 0.85 | 0.63–1.14 | 0.272 | 0.82 | 0.61–1.09 | 0.166 | |
| CRC | Dominant | 0.80 | 0.46–1.39 | 0.424 | 0.77 | 0.43–1.37 | 0.372 | 0.76 | 0.43–1.36 | 0.358 |
| Recessive | 0.83 | 0.41–1.65 | 0.591 | 0.82 | 0.40–1.67 | 0.586 | 0.79 | 0.39–1.61 | 0.515 | |
| GG Vs AG | 0.73 | 0.41–1.32 | 0.302 | 0.71 | 0.38–1.29 | 0.259 | 0.7 | 0.38–1.28 | 0.247 | |
| GG Vs AA | 1.00 | 0.46–2.19 | 0.996 | 0.99 | 0.43–2.25 | 0.974 | 1 | 0.45–2.23 | 0.998 | |
| G Vs A | 0.96 | 0.66–1.38 | 0.808 | 0.94 | 0.64–1.38 | 0.761 | 0.95 | 0.65–1.39 | 0.795 | |
| EC | Dominant | 1.19 | 0.49–2.90 | 0.695 | 1.21 | 0.49–2.95 | 0.680 | 0.92 | 0.36–2.35 | 0.856 |
| Recessive | 2.27 | 0.90–5.75 | 0.083 | 2.24 | 0.87–5.75 | 0.093 | 2.73 | 1.02–7.26 | 0.045 | |
| GG Vs AG | 1.97 | 0.67–5.8 | 0.217 | 1.99 | 0.67–5.91 | 0.216 | 1.53 | 0.49–4.84 | 0.467 | |
| GG Vs AA | 0.61 | 0.22–1.71 | 0.349 | 0.65 | 0.23–1.88 | 0.429 | 0.48 | 0.16–1.42 | 0.183 | |
| G Vs A | 0.79 | 0.43–1.46 | 0.45 | 0.799 | 0.43–1.48 | 0.476 | 0.65 | 0.34–1.24 | 0.191 | |
| Others | Dominant | 0.66 | 0.23–1.92 | 0.448 | 0.74 | 0.25–2.16 | 0.582 | 0.59 | 0.20–1.80 | 0.356 |
| Recessive | 0.50 | 0.11–2.26 | 0.370 | 0.45 | 0.09–2.05 | 0.302 | 0.54 | 0.12–2.54 | 0.439 | |
| GG Vs AG | 0.55 | 0.18–1.63 | 0.278 | 0.59 | 0.19–1.78 | 0.353 | 0.49 | 0.15–1.52 | 0.215 | |
| GG Vs AA | 1.36 | 0.25–7.33 | 0.723 | 1.48 | 0.27–8.21 | 0.655 | 0.77 | 0.12–5.35 | 0.786 | |
| G Vs A | 1.00 | 0.51–1.95 | 0.990 | 1.1 | 0.55–2.14 | 0.818 | 0.93 | 0.45–1.89 | 0.830 |
CI; Class Interval, OR; Crude Odds Ratio, ORa, Pa; adjusted for age and sex, ORb, Pb; adjusted for Diabetes, diet and region, p values-significant values are bold. GIC; Gastrointestinal cancers, GC; Gastric cancer, CRC; Colorectal cancer, EC; Esophageal cancer.
3.3.5. Association of pre-mir-605 polymorphism rs2043556 with GI cancers
The ‘CC’ genotype of rs2043556 SNP was found to increase the risk of GI cancers drastically (OR = 2.57, p = 0.043) whereas the ‘TT’ genotype was found to play a protective role (OR = 0.39, p = 0.042). Similar outcomes were observed in the adjusted ORs (Recessive; ORa = 0.29, pa = 0.011, ORb = 0.35, pb = 0.027 & TT Vs CC; ORa = 3.25, pa = 0.016, ORb = 2.93, pb = 0.028). There was no association of the polymorphisms with the sub group cancers (Table 6). The allele frequencies of the control groups were compared to the 1000 genomes Phase 3 worldwide populations and found similar to South Asian populations (Supplementary Fig. 5).
Table 6.
Association analysis of Pre-mir-605 rs2043556 SNP with GI cancers.
| Cancer Site | Model | OR | 95 % CI | p Value | ORa | 95 % CI | Pa Value | ORb | 95 % CI | Pb Value |
|---|---|---|---|---|---|---|---|---|---|---|
| GIC | Dominant | 1.17 | 0.79–1.76 | 0.435 | 1.17 | 0.76–1.78 | 0.481 | 1.17 | 0.77–1.78 | 0.451 |
| Recessive | 0.39 | 0.16–0.97 | 0.042 | 0.29 | 0.11–0.76 | 0.011 | 0.35 | 0.14–0.89 | 0.027 | |
| TT Vs TC | 1.03 | 0.67–1.57 | 0.899 | 0.97 | 0.62–1.52 | 0.885 | 1.01 | 0.65–1.57 | 0.956 | |
| TT Vs CC | 2.57 | 1.03–6.43 | 0.043 | 3.25 | 1.25–8.48 | 0.016 | 2.93 | 1.12–7.66 | 0.028 | |
| T Vs C | 1.29 | 0.92–1.8 | 0.138 | 1.34 | 0.94–1.91 | 0.102 | 1.31 | 0.93–1.88 | 0.124 | |
| GC | Dominant | 1.09 | 0.64–1.86 | 0.757 | 0.94 | 0.52–1.71 | 0.840 | 1.2 | 0.69–2.10 | 0.517 |
| Recessive | 0.44 | 0.12–1.53 | 0.196 | 0.27 | 0.07–1.05 | 0.058 | 0.37 | 0.10–1.32 | 0.124 | |
| TT Vs TC | 0.96 | 0.55–1.68 | 0.884 | 0.72 | 0.38–1.37 | 0.318 | 1.04 | 0.58–1.86 | 0.892 | |
| TT Vs CC | 2.26 | 0.63–8.06 | 0.210 | 3.4 | 0.85–13.67 | 0.084 | 2.69 | 0.73–9.93 | 0.136 | |
| T Vs C | 1.29 | 0.92–1.8 | 0.138 | 1.34 | 0.94–1.91 | 0.102 | 1.31 | 0.93–1.85 | 0.124 | |
| CRC | Dominant | 1.22 | 0.72–2.09 | 0.463 | 1.2 | 0.69–2.09 | 0.510 | 1.27 | 0.73–2.20 | 0.403 |
| Recessive | 0.57 | 0.19–1.74 | 0.321 | 0.47 | 0.15–1.48 | 0.195 | 0.49 | 0.15–1.56 | 0.224 | |
| TT Vs TC | 1.12 | 0.64–1.98 | 0.686 | 1.06 | 0.59–1.897 | 0.855 | 1.15 | 0.64–2.06 | 0.650 | |
| TT Vs CC | 1.84 | 0.59–5.73 | 0.295 | 2.14 | 0.67–6.88 | 0.200 | 2.05 | 0.62–6.74 | 0.237 | |
| T Vs C | 1.27 | 0.81–1.98 | 0.299 | 1.3 | 0.82–2.06 | 0.260 | 1.33 | 0.84–2.10 | 0.222 | |
| EC | Dominant | 1.08 | 0.45–2.62 | 0.863 | 1.05 | 0.43–2.57 | 0.911 | 1.15 | 0.46–2.87 | 0.774 |
| TT Vs TC | 0.86 | 0.35–2.09 | 0.734 | 0.79 | 0.32–1.96 | 0.614 | 0.87 | 0.34–2.20 | 0.770 | |
| T Vs C | 1.35 | 0.63–2.90 | 0.438 | 1.38 | 0.64–2.97 | 0.417 | 1.48 | 0.67–3.24 | 0.330 | |
| Others | Dominant | 1.51 | 0.55–4.12 | 0.426 | 1.37 | 0.495–3.80 | 0.544 | 1.03 | 0.37–2.91 | 0.951 |
| TT Vs TC | 1.19 | 0.43–3.29 | 0.732 | 1.03 | 0.37–2.91 | 0.951 | 1.12 | 0.39–3.22 | 0.839 | |
| T Vs C | 1.75 | 0.71–4.32 | 0.222 | 1.68 | 0.65–4.17 | 0.261 | 1.92 | 0.76–4.84 | 0.170 |
CI; Class Interval, OR; Crude Odds Ratio, ORa, Pa; adjusted for age and sex, ORb, Pb; adjusted for Diabetes, diet and region, p values-significant values are bold. GIC; Gastrointestinal cancers, GC; Gastric cancer, CRC; Colorectal cancer, EC; Esophageal cancer.
3.4. Lack of association of miR polymorphisms with GI cancers
The association analysis of polymorphisms miR-26a-1; rs7372209, miR-218-2 rs11134527, miR-34b/c rs4938723, miR-603 rs11014002 and mir-Let7a2 rs629367 revealed that they were not associated with altering the susceptibility of GI cancers or its cancer sub groups (Supplementary Table 1 - Supplementary Table 5)
3.5. Multifactor dimensionality reduction analysis to identify interaction between miRSNPs
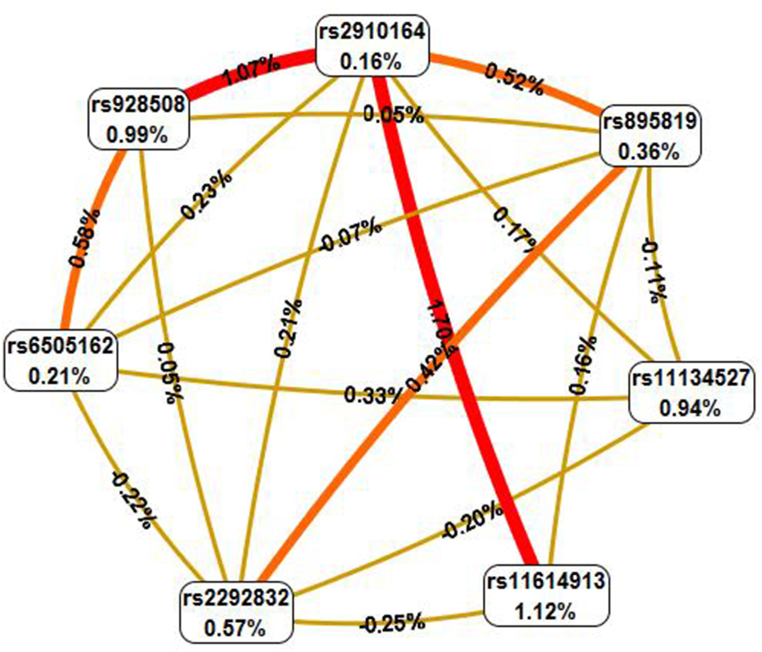
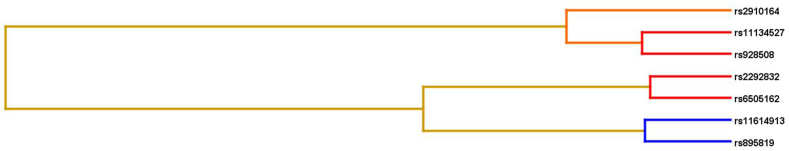
MDR analysis was conducted to identify models that elevate the risk of gastrointestinal cancers. This analysis produced the best-fit models, with total risk quantified as odds ratios (ORs) and p-values. Entropy-based interaction graphs were generated from the MDR analysis for each dataset. The entropy values in the cells corresponding to individual SNPs represent their primary independent effects. The entropy values marked on the lines connecting two SNPs indicate the nature of their interaction: blue lines denote a high degree of redundancy, green lines indicate a reduced level of redundancy, and gold lines signify independence or additivity. Additionally, interaction dendrograms were created for each MDR analysis dataset. These dendrogram interaction graphs were constructed using hierarchical cluster analysis to reveal the presence, strength, and nature of epistatic effects. If the line connecting two factors is further to the right, it indicates a stronger interaction effect, represented by a black line in figures. A red line between factors signifies a high degree of synergy, while an orange line indicates a lesser degree of synergy.
MDR analysis was done to detect SNP-SNP interactions that might possibly increase the risk of GI cancers and subgroup cancers. In the GI cancer analysis, it was observed that polymorphisms rs11134527, rs2292832, rs6505162, rs928508, rs895819 increased the odds of GI cancer for a person having these polymorphisms together. Table 7 clearly illustrates the significant models and the consistent rise in odds ratio (OR) values for each model as SNPs are added sequentially to the analysis, highlighting the effect of SNP-SNP interactions. The entropy and dendrogram figures also showed the possible interactions between the SNPs to increase the risk of GI cancers (Fig. 1A, Fig. 1BA & B).
Table 7.
Interaction analysis of miRSNPs in GI cancers.
| Factors | Bal. Acc. CV training | Bal. Acc. CV testing | CV consistency | OR | CI | p Value |
|---|---|---|---|---|---|---|
| rs11614913 | 0.5537 | 0.4681 | 6/10 | 1.48 | 0.99–2.36 | 0.0586 |
| rs11614913, rs2910164 | 0.5918 | 0.4842 | 5/10 | 2.3 | 1.44–63.67 | 0.0004 |
| rs11614913, rs2292832, rs2910164 | 0.6415 | 0.4719 | 3/10 | 3 | 1.9–4.40 | <0.0001 |
| rs2292832, rs629367, rs6505162, rs928508 | 0.7036 | 0.5811 | 7/10 | 5.39 | 3.45–8.40 | <0.0001 |
| rs11134527, rs2292832, rs6505162, rs928508, rs895819 | 0.7865 | 0.4744 | 4/10 | 12.66 | 7.63–21.02 | <0.0001 |
CV; Cross validation, Bal; Balanced, Acc; Accuracy, OR; Odds Ratio, CI; Class interval. Significant p values are bold.
Fig. 1A.
Entropy diagram depicting interactions between miRSNPs in GI cancers. Values inside SNP cells indicate the main independent effects. Values marked connecting lines between two SNPs represent the entropy of interaction. Blue lines indicate a high degree of redundancy, green lines a reduced degree of redundancy and gold lines represent independence or additivity.
Fig. 1B.
Dendrogram depicting interactions between miRSNPs in GI cancers. Strongly interacting SNPs are placed closer and weekly interacting SNPs are placed far apart.
MDR modelling was done for GC, CRC and EC subgroups. Table 8, Table 9, Table 10 represent the SNP-SNP interactions observed in GC, CRC and EC respectively. The results were further elaborated in the entropy and dendrograms. The rs11134527, rs2292832, rs6505162, rs928508, rs895819 polymorphisms showed significant interaction towards increasing the risk for GC (Fig. 2A, Fig. 2BA & B). SNPs rs11134527, rs11614913, rs2292832, rs6505162, rs895819 showed significant
Table 8.
Interaction analysis of miRSNPs in Gastric cancers.
| Model | Bal. Acc. CV training | Bal. Acc. CV testing | CV consistency | OR | CI | p Value |
|---|---|---|---|---|---|---|
| rs11134527 | 0.5741 | 0.4348 | 3/10 | 1.65 | 0.95–2.84 | 0.0724 |
| rs11614913, rs2910164 | 0.6136 | 0.4518 | 3/10 | 2.74 | 1.52–4.93 | 0.0006 |
| rs11134527, rs928508, rs2910164 | 0.6759 | 0.494 | 7/10 | 4.00 | 2.25–7.11 | <0.0001 |
| rs11134527, rs6505162, rs928508, rs2910164 | 0.7607 | 0.4711 | 3/10 | 10.61 | 5.24–21.48 | <0.0001 |
| rs11134527, rs2292832, rs6505162, rs928508, rs895819 | 0.8558 | 0.4276 | 4/10 | 41.93 | 15.98–110.0 | <0.0001 |
CV; Cross validation, Bal; Balanced, Acc; Accuracy, OR; Odds Ratio, CI; Class interval. Significant p values are bold.
Table 9.
Interaction analysis of miRSNPs in Colorectal cancers.
| Model | Bal. Acc. CV training | Bal. Acc. CV testing | CV consistency | OR | CI | p Value |
|---|---|---|---|---|---|---|
| rs2292832, rs6505162 | 0.6228 | 0.471 | 4/10 | 2.77 | 1.46–5.28 | 0.0015 |
| rs11134527, rs2292832, rs928508 | 0.7046 | 0.6209 | 10/10 | 5.65 | 2.99–10.68 | <0.0001 |
| rs11134527, rs2292832, rs928508, rs2910164 | 0.7758 | 0.497 | 3/10 | 12.32 | 5.56–27.32 | <0.0001 |
| rs11134527, rs11614913, rs2292832, rs6505162, rs895819 | 0.8699 | 0.4908 | 8/10 | 64.46 | 19.33–215 | <0.0001 |
CV; Cross validation, Bal; Balanced, Acc; Accuracy, OR; Odds Ratio, CI; Class interval. Significant p values are bold.
Table 10.
Interaction analysis of miRSNPs in Esophageal cancers.
| Model | Bal. Acc. CV training | Bal. Acc. CV testing | CV consistency | OR | CI | P Value |
|---|---|---|---|---|---|---|
| rs2043556 | 0.6028 | 0.4089 | 4/10 | 2.39 | 0.85–6.75 | 0.0906 |
| rs4919510, rs928508 | 0.686 | 0.4571 | 4/10 | 5.41 | 1.77–16.57 | 0.0012 |
| rs2043556, rs2292832, rs4919510 | 0.7824 | 0.4147 | 2/10 | 15.45 | 3.51–68.00 | <0.0001 |
| rs11134527, rs6505162, rs928508, rs895819 | 0.8866 | 0.43 | 6/10 | ∞ | – | <0.0001 |
| rs11134527, rs2292832, rs6505162, rs928508, rs895819 | 0.9572 | 0.4729 | 8/10 | ∞ | – | <0.0001 |
CV; Cross validation, Bal; Balanced, Acc; Accuracy, OR; Odds Ratio, CI; Class interval. Significant p values are bold.
Fig. 2A.
Entropy diagram depicting interactions between miRSNPs in Gastric cancers. Values inside SNP cells indicate the main independent effects. Values marked connecting lines between two SNPs represent the entropy of interaction. Blue lines indicate a high degree of redundancy, green lines a reduced degree of redundancy and gold lines represent independence or additivity.
Fig. 2B.
Dendrogram depicting interactions between miRSNPs in Gastric cancers. Strongly interacting SNPs are placed closer and weekly interacting SNPs are placed far apart.
interaction towards increasing the risk for CRC (Fig. 3A, Fig. 3BA & B). MiRSNPs rs11134527, rs2292832, rs6505162, rs928508, rs895819 showed significant interaction towards increasing the risk for EC (Fig. 4A, Fig. 4BA & B). The analysis revealed risk instilling roles of several SNPs that were not observed in the earlier association analysis. This shows that polymorphisms act differently based on the presence of other SNPs and influence each other's role towards the development of the disease.
Fig. 3A.
Entropy diagram depicting interactions between miRSNPs in Colorectal cancers. Values inside SNP cells indicate the main independent effects. Values marked connecting lines between two SNPs represent the entropy of interaction. Blue lines indicate a high degree of redundancy, green lines a reduced degree of redundancy and gold lines represent independence or additivity.
Fig. 3B.
Dendrogram depicting interactions between miRSNPs in Colorectal cancers. Strongly interacting SNPs are placed closer and weekly interacting SNPs are placed far apart.
Fig. 4A.
Entropy diagram depicting interactions between miRSNPs in Esophageal cancers.
Fig. 4B.
Dendrogram depicting interactions between miRSNPs in Esophageal cancers. Strongly interacting SNPs are placed closer and weekly interacting SNPs are placed far apart.
3.6. Multifactor dimensionality reduction analysis to identify interaction between SNP and environmental factors
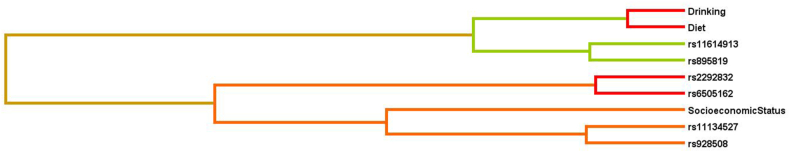
MDR analysis was performed to examine the environmental factors in the study that had the propensity to alter susceptibility to GI cancers. The analysis was done for GI cancers as a whole and for the individual subgroup GI cancers.
In GI cancers, socioeconomic status (SES) along with polymorphisms rs2292832, rs629367, rs6505162, rs928508 increased the risk of GI cancers. There was a steady increase in the OR values indicating the increase in risk of GI cancer with the increase in number of risk factors (Table 11). The entropy diagram and dendrogram showed diabetes mellitus (DM) and SES to be involved in GI cancer risk (Fig. 5A, Fig. 5BA and B).
Table 11.
Interaction analysis of miRSNPs and Environmental factors in GI Cancers.
| Model | Bal. Acc. CV training | Bal. Acc. CV testing | CV consistency | OR | CI | p Value |
|---|---|---|---|---|---|---|
| rs2292832, SES | 0.6035 | 0.4881 | 4/10 | 2.25 | 1.48–3.40 | 0.0001 |
| rs2043556, rs2292832, SES | 0.6446 | 0.503 | 4/10 | 3.33 | 2.15–5.14 | <0.0001 |
| rs2292832, rs6505162, rs928508 | 0.7038 | 0.5953 | 7/10 | 5.39 | 3.45–8.40 | <0.0001 |
| rs2292832, rs629367, rs6505162, rs928508, SES | 0.7813 | 0.5544 | 6/10 | 13.49 | 8.05–22.59 | <0.0001 |
SES; Socioeconomic Status, CV; Cross validation, Bal; Balanced, Acc; Accuracy, OR; Odds Ratio, CI; Class interval. Significant p values are bold.
Fig. 5A.
Entropy diagram depicting interactions between miRSNPs and Environmental factors in GI cancers.
Fig. 5B.
Dendrogram depicting interactions between miRSNPs and environmental factors in GI cancers.
Similarly, rs11134527, rs928508, rs2910164 and socioeconomic status of an individual determined the amount of risk towards GC (Table 12). The entropy diagram also revealed that sex interacted with rs629367 to increase GC risk. The entropy diagram and dendrogram provided a better view on the networking among the different SNPs and environmental factors (Fig. 6A, Fig. 6BA and B). Table 13 represents the models and respective ORs for CRC cancers. In CRC, polymorphisms rs11134527, rs11614913, rs2292832, rs6505162, rs895819 and factors like alcohol drinking, diet and SES were modelled in the entropy diagram and dendrogram (Fig. 7A, Fig. 7BA and B). The esophageal cancers showed no remarkable interactions between SNPs and environmental factors.
Table 12.
Interaction analysis of miRSNPs and Environmental factors in GC.
| Model | Bal. Acc. CV training | Bal. Acc. CV testing | CV consistency | OR | CI | p Value |
|---|---|---|---|---|---|---|
| rs6505162, SES | 0.6283 | 0.4827 | 4/10 | 2.78 | 1.58–4.89 | 0.0003 |
| rs629367, rs6505162, SES | 0.6895 | 0.4975 | 4/10 | 4.69 | 2.58–8.51 | <0.0001 |
| rs11134527, rs928508, rs2910164, SES | 0.7664 | 0.5233 | 4/10 | 12.52 | 5.88–26.67 | <0.0001 |
Socioeconomic Status, CV; Cross validation, Bal; Balanced, Acc; Accuracy, OR; Odds Ratio, CI; Class interval. Significant p values are bold.
Fig. 6A.
Entropy diagram depicting interactions between miRSNPs and Environmental factors in gastric cancers.
Fig. 6B.
Dendrogram depicting interactions between miRSNPs and environmental factors in gastric cancers.
Table 13.
Interaction analysis of miRSNPs and Environmental factors in CRC.
| Model | Bal. Acc. CV training | Bal. Acc. CV testing | CV consistency | OR | CI | p Value |
|---|---|---|---|---|---|---|
| Drinking, Diet | 0.6292 | 0.4692 | 3/10 | 3.57 | 1.72–7.43 | 0.0004 |
| rs11134527, rs2292832, rs928508 | 0.7053 | 0.5489 | 7/10 | 5.65 | 2.98–10.68 | <0.0001 |
| rs11134527, rs2292832, rs928508, SES | 0.8034 | 0.5931 | 7/10 | 18.91 | 8.14–43.9 | <0.0001 |
SES; Socioeconomic Status, CV; Cross validation, Bal; Balanced, Acc; Accuracy, OR; Odds Ratio, CI; Class interval. Significant p values are bold.
Fig. 7A.
Entropy diagram depicting interactions between miRSNPs and Environmental factors in Colorectal cancers.
Fig. 7B.
Dendrogram depicting interactions between miRSNPs and environmental factors in Colorectal cancers.
4. Discussion
The initial screening of the polymorphisms showed that miR-499; rs3746444, miR-124-1; rs531564, miR-608; rs4919510 and miR-92a-1; rs9589207 polymorphisms deviated from HWE and therefore were excluded from further investigations. Deviation from HWE mostly indicates errors in genotyping though theoretically it might be an indicator of inbreeding and population stratification [13,14]. Association analysis yielded quite a few interesting results that are discussed below.
miR-196a is widely recognized for its oncogenic role, as it affects carcinogenesis and tumor progression by regulating processes such as proliferation, invasion, metastasis, and apoptosis. It is mostly correlated with the prognosis of several cancers making it a potential diagnostic marker. miR-196a has been identified to have strong oncogenic roles in most of the cancers of the GI tract such as EC [15], GC [16] and CRC [17]. rs11614913 present in miR-196a is one of the earliest miRSNPs to be studied for its functional effects and role in altering susceptibility of malignant phenotypes. In the current study, we observed that the homozygous mutant genotype (TT) and allele (T) influenced the risk of GI cancers as a group which is an indicator of its probable role in the GI subgroups also. In addition, the recessive model showed that the ‘CC’ and ‘CT’ genotypes had a protective effect on its carriers. Likewise, the odds of acquiring gastric cancer were higher in the ‘TT’ carriers and lower in the ‘CC’/‘CT’ carriers and these results were almost statistically significant. Similarly, in the subgroup others it was seen that the ‘T allele increased cancer risk. The current study is a forerunner in bringing out the association of the SNP with GC in an Indian subpopulation.
Our findings align with several other studies that highlight the role of rs11614913 in influencing gastric cancer (GC) susceptibility, although a few studies dispute this role, noting that the effect alleles differ by population. The 'CC' genotype was associated with a higher risk of GC among Chinese individuals with a history of alcohol abuse, H. pylori infection, and familial cancer history [18]. This contrasts with a large study from the same population, which found that the genotype was linked to a lower risk of GC and better survival, supporting our results [19]. In addition, it is important to mention a study conducted in 2014 stated that the polymorphism had no role in being susceptible to GC in the Chinese [20]. Such seemingly farcical results from the same population leave us with an inconclusive role of the SNP in the Chinese populations. Further, the ‘CC’ genotype elevated the risk of GC in Greeks [21], Chinese [22], Korean females [23] and in aged Japanese [24] while, the polymorphism lacked any significance in the Romanian population [25]. A meta-analysis performed on the existing studies showed that the ‘CC’ genotype reduced the risk of GC [26] supporting our findings while 2 other meta-analysis suggested that the polymorphism had no effect on GC risk [27,28].
miR-423 has been recognized as a contributor to various normal lipid metabolism and cancer pathways. Expression studies have shown abnormal expression patterns of miR-423 in cancers such as OSCC and HNSCC. Similar to other miRNAs, miR-423 exhibits dual roles in different cancer tissues. It was found to be upregulated in the early stages of CRC and ovarian cancer, highlighting its potential as a diagnostic marker.The rs6505162 polymorphism present in this miRNA has been widely studied in ECs and has exhibited race dependant roles [[29], [30]]. In our study, the polymorphism had significant association with CRC where the ‘AA’ genotype actively increased the odds of acquiring CRC in its carriers in the adjusted models. Further, it was also noticed that the ‘A’ allele influenced the risk of CRC in all the models showing the prominent role of the SNP in CRC. A similar scenario was observed in the Chinese population where the ‘AA’ genotype was associated with the metastasis of CRC [31]. In addition, the study by Xing et al., showed the influence of the SNP on survival rate and recurrence free survival in CRC patients with higher BMI and TNM stages [32]. Though, there were studies where the SNP was associated with EC [33,34] it was not evident in our study. It is also important to note at this juncture that a single study on EC from a north Indian population also did not find any association of the SNP with EC risk [35]. Our study was the first to report the association of miR-423 SNP with risk of CRC and also to report the MAF from a South Indian population.
miR-30c, a member of the miR-30 family, is essential for lipid metabolism, adipogenesis, cell proliferation, and differentiation. It has a dual role in cancer, acting as both a tumor suppressor and an oncogene. miR-30c is actively involved in cancers affecting the breast, colon, bladder, lung, ovary, prostate, liver, endometrium, and Myc-induced B lymphoma. It is known to regulate the expression of the KRAS gene and inhibit the KRAS-MAPK pathway, which helps to impede cell proliferation, invasion, and metastasis [[36], [37]]. Several studies have found this polymorphism to be associated with risk, survival and chemoresistance of lung cancers [[38], [39], [40], [41]]. However, there is only a single report on the association of the polymorphism with GC. This Chinese study demonstrated that the ‘AA’ genotype instilled an increased risk of GC and also influenced upregulation of miR-30c expression [42]. In our study, the GIC group showed that the ‘AG’ genotype decreased the disease risk significantly in all the models. Moreover, the sub group cancer analysis disclosed new findings which were never reported before. The dominant and ‘AA vs AG’ models showed a significantly reduced risk of GC in all the models hinting a protective role of the ‘AG’ genotype in GC. Another interesting revelation was that the recessive model indicated a significant risk of EC in the adjusted model which was also the first report on the SNPs association with EC. At this point it is noteworthy to state that the current study was first to screen for this polymorphism in the Indian continent and observe its MAF and effects in GI cancers.
miR-605 is well acclaimed for its tumor suppressing role in multiple cancers by preventing expression of genes involved, in cell proliferation, invasion and metastasis. miR-605 primarily functions as a tumor suppressor, playing a key role in regulating various signaling pathways. It has been found to be downregulated in melanoma tissues, which helps to inhibit cancer progression. Additionally, miR-605 prevents epithelial-mesenchymal transition (EMT) and metastasis by interfering with TRAF6 in the NF-kB pathway [43]. The rs2043556 housed within this miRNA has been associated with risk of GI cancers and BC [[44], [45],46]. Similarly, the ‘AG’ and ‘GG’ genotypes increased the risk of GC in people of Brazil [47] indicative of a potential role of the SNP in GI cancers. In the current study, the ‘CC’ genotype remarkably increased the odds of the carrier to acquire GI cancers and on the other hand the recessive model (CC vs TT + TC) conferred a protective role against GI cancer on its carriers. Likewise, the ORs achieved in the GI cancer sub groups indicated an increase in disease risk of GC and CRC but the results were statistically non-significant. This is the 1st attempt to bring out the significance of miR-605 polymorphism in cancers from India. To the best of our knowledge, this study is the forerunner in screening for rs2043556 and reporting its MAF from an Indian sub population.
miRNA polymorphisms in miR-27a; rs895819 & rs11671784 [10] miR-26a-1; rs7372209, miR-218-2; rs11134527, miR-34b/c; rs4938723, miR-603; rs11014002 and Let-7a2; rs629367 were not associated with the risk of GI cancers or the subgroups GC, CRC, EC and ‘others. These results may seem to do away with the importance of these polymorphisms in predicting GI cancer risk but it is important to note that all these polymorphisms were well addressed for their role in GI cancers in several studies which is evident from the review of literature. The lack of association could perhaps reflect on underlying reasons like ethnic differences and dependence on other risk factors. The polymorphism rs41274221 in miR-25a had only the ‘CC’ genotype in the study population which was concordant with the allele frequencies depicted in the Ensemble database [48] but discordant with the study by Zhou et al., where the ‘A’ allele of the polymorphism reduced the risk of GC by breaking the oncogenic character of miR-25 [49].
In order to address an important objective of the study which was ‘to elucidate the interaction between miRSNPs and environmental factors’ we opted for Multifactor dimensionality reduction (MDR) analysis which was the first tool to detect non additive gene-gene interactions and genotype-phenotype interactions in polygenic human diseases [50]. It is basically a non-parametric method which uses a highly creative constructive induction algorithm which reduces the dimensions of the SNP data into a single feature making it easier to identify interactions that point out to two possible outcomes; high risk and low risk [51]. Moreover, MDR analysis is also a powerful tool in detecting valuable interaction even in the presence of defective data with genotyping error, missing data, phenocopy and genetic heterogeneity [[52], [53]]. There are several evidences which have shown the efficiency of MDR in detecting significant genotype phenotype interactions which were not observed in the conventional regression analysis [[53], [54], [55]].
The SNP-SNP interaction analysis conducted in this study examined the polymorphisms miR-218-2; rs11134527, miR-149; rs2292832, miR-423; rs6505162, miR-30c; rs928508 and miR-27A; rs895819 within a single model, which significantly elevated the risk of gastrointestinal cancers.This analysis brought to light, the polymorphisms which did not appear in the association analysis done previously. miR-218-2; rs11134527 and miR-27a; rs895819 were not associated with GI risk individually but in the presence of the above SNPs they worked towards increasing the individual's susceptibility to GI cancers. The ORs also showed a steady increase in risk with addition of each SNP to the model indicating how a person's risk towards GI cancer varies based on the presence of different miRSNPs. The MDR analysis done in the GC and EC group found that miR-218-2; rs11134527, miR-149; rs2292832, miR-423; rs6505162, miR-30c; rs928508 and miR-27a; rs895819 interacted together significantly to increase the risk of GC and EC. The MDR analysis done in the CRC group presented miR-SNPs miR-218-2; rs11134527, miR-196a; rs11614913, miR-149; rs2292832, miR-423; rs6505162, and miR-27a; rs895819, to have an increased risk of CRCs. It is important to highlight that the SNP miR-30c (rs928508) was excluded from the colorectal cancer (CRC) model, while miR-196a (rs11614913) was carefully included, illustrating the differences in the models based on the specific disease groups. Consequently, the MDR analysis produced novel SNP-SNP models that could potentially be used to quantitatively predict an individual's susceptibility in the future.
MDR analysis was also employed to detect interactions between miRSNPs and environmental factors recorded in the study. In GI cancer group socioeconomic status (SES) interacted with miR-149; rs2292832, Let-7a2; rs629367, miR-423; rs6505162, and miR-30c; rs928508 to increase the risk of GI cancer. It is important to note that Let-7a2; rs629367 was added only in this model and not in the SNP-SNP model showing that SES is an important influencer and determinant of cancer risk. Further the entropy diagram and dendrograms also exhibited mild interactions between diabetes mellitus and the polymorphisms. SES was also modelled along with miR-218-2; rs11134527, miR-30c; rs928508, miR-146a; rs2910164 and with Let-7a2; rs629367, miR-423; rs6505162 separately in the GC subgroup. Thus, there were two different models increasing the GC risk. Moreover, the entropy diagram and dendrogram showed that Let-7a2; rs629367 interacted with sex in GC. Likewise, miR-218-2; rs11134527, miR-149; rs2292832, miR-30c; rs928508 and SES were put together as a significant model affecting CRC risk. The entropy diagram and dendrogram revealed a strong interaction between miR-149; rs2292832 and diet while miR-34b/c; rs4938723 weakly interacted with alcohol drinking to result in elevated CRC risk. The impact of SES on cancer burden has been well established earlier. SES is an important factor that reflects on an individual's exposure to environmental pollutants, availability of medical facilities, cancer awareness, nutrition and various other lifestyle practices. Our study also showed that SES of a person magnified the effect of SNPs and increased an individual's cancer risk [[56], [57], [58], [59], [60]]. The MDR analysis yielded such pronounced results which were not demonstrated earlier making the study one of its kind. The importance of mathematical modelling in genomic datasets is well highlighted by the above results that can be employed for construction of cancer prediction kits for the future generations where genome sequencing will be a common method in medicine, made accessible to the common man.
5. Conclusion
The study demonstrated that miRSNPs alter GI cancer risk and it is enhanced by the presence of risk factors like socioeconomic status, diabetes mellitus, diet and alcohol consumption in the Indian subpopulation. However, the role of these polymorphisms requires a comprehensive study across various populations to arrive at the exact potential of these SNPs as SNP biomarkers for GI cancer risk prediction. Determining how SNPs influence the health of an individual and later transforming this knowledge into the development of new treatment approaches will revolutionize the management of polygenic diseases like cancers. Moreover, these results are sure to attract attention towards implying these polymorphisms for future research.
CRediT authorship contribution statement
Charles Emmanuel Jebaraj Walter: Writing – review & editing, Supervision, Formal analysis, Conceptualization. Zioni Sangeetha Shankaran: Writing – review & editing, Writing – original draft, Validation, Methodology, Formal analysis, Conceptualization. Sai Sushmitha Kontham: Formal analysis. Kotteeswaran Ramachandran: Formal analysis. Nandini Prakash: Formal analysis. Thanka Johnson: Validation, Conceptualization. Sri Nisha JR: Validation.
Ethics statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board. The study was approved by the Institutional Ethics Committee of Sri Ramachandra Institute of Higher Education and Research (Deemed to be University), Chennai, IEC-NI/13/APR/33/39. Written Informed consent was obtained after explaining the study to the participant before recruitment.
Data availability statement
Not Applicable.
Funding
This research received no external funding.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Charles Emmanuel Jebaraj W reports administrative support, article publishing charges, and equipment, drugs, or supplies were provided by Sri Ramachandra Institute of Higher Education and Research (Deemed to be University). If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the management of Sri Ramachandra Institute of Higher Education and Research for providing us the infrastructure to conduct the study. We thank Dr. B.W.C. Sathiysekaran, Dean Research, Sree Balaji Medical College & Hospital for his valuable inputs in statistical analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e41519.
Contributor Information
Charles Emmanuel Jebaraj Walter, Email: cejwalter@sriramachandra.edu.in.
Sri Nisha JR, Email: jsrinishapd@gmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Greenwood-Van Meerveld B., Johnson A.C., Grundy D. Gastrointestinal physiology and function. Handb. Exp. Pharmacol. 2017;239 doi: 10.1007/164_2016_118. [DOI] [PubMed] [Google Scholar]
- 2.Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., et al. Human MicroRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shastry B.S. SNPs. Impact on gene function and phenotype. Methods Mol. Biol. 2009;578:3–22. doi: 10.1007/978-1-60327-411-1_1. [DOI] [PubMed] [Google Scholar]
- 4.Nicoloso M.S., Sun H., Spizzo R., Kim H., Wickramasinghe P., Shimizu M., Wojcik S.E., Ferdin J., Kunej T., Xiao L., et al. Single-nucleotide polymorphisms inside MicroRNA target sites influence tumor susceptibility. Cancer Res. 2010;70:2789–2798. doi: 10.1158/0008-5472.CAN-09-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin G.A., Ferracin M., Cimmino A., Di Leva G., Shimizu M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M., et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 6.Jazdzewski K., Murray E.L., Franssila K., Jarzab B., Schoenberg D.R., de la Chapelle A. Common SNP in pre-MiR-146a decreases mature MiR expression and predisposes to papillary thyroid carcinoma. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Y., Yu C.-Y., Wang J.-L., Guan J., Chen H.-Y., Fang J.-Y. MicroRNA sequence polymorphisms and the risk of different types of cancer. Sci. Rep. 2014;4:3648. doi: 10.1038/srep03648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han B.W., Li Z.H., Liu S.F., Han H.B., Dong S.J., Zou H.J., Sun R.F., Jia J. A comprehensive review of MicroRNA-related polymorphisms in gastric cancer. Genet. Mol. Res. 2016;15 doi: 10.4238/gmr.15028289. [DOI] [PubMed] [Google Scholar]
- 9.Shankaran Z.S., Walter Charles Emmanuel Jebaraj, Ramanathan A., Chinambedu Dandapani M., Selvaraj S., Kontham S.S. MicroRNA-146a gene polymorphism alters human colorectal cancer susceptibility and influences the expression of its target genes in toll-like receptor (TLR) pathway. Meta Gene. 2020;24 doi: 10.1016/j.mgene.2020.100654. [DOI] [Google Scholar]
- 10.Shankaran Z.S., Walter C.E.J., Prakash N., Ramachandiran K., C G.P.D., Johnson T. Investigating the role of MicroRNA-27a gene polymorphisms and its interactive effect with risk factors in gastrointestinal cancers. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e03565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellis J.A., Ong B. The MassARRAY® system for targeted SNP genotyping. Methods Mol. Biol. 2017;1492:77–94. doi: 10.1007/978-1-4939-6442-0_5. [DOI] [PubMed] [Google Scholar]
- 12.Hahn L.W., Ritchie M.D., Moore J.H. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19(3):376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- 13.Hosking L., Lumsden S., Lewis K., Yeo A., McCarthy L., Bansal A., et al. Detection of genotyping errors by Hardy-Weinberg equilibrium testing. Eur. J. Hum. Genet. 2004;12(5):395–399. doi: 10.1038/sj.ejhg.5201164. [DOI] [PubMed] [Google Scholar]
- 14.Salanti G., Amountza G., Ntzani E.E., Ioannidis J.P.A. Hardy-Weinberg equilibrium in genetic association studies: an empirical evaluation of reporting, deviations, and power. Eur. J. Hum. Genet. 2005;13(7):840–848. doi: 10.1038/sj.ejhg.5201410. [DOI] [PubMed] [Google Scholar]
- 15.Hu C., Peng J., Lv L., Wang X., Zhou Y., Huo J., Liu D. MiR-196a regulates the proliferation, invasion and migration of esophageal squamous carcinoma cells by targeting ANXA1. Oncol. Lett. 2019;17:5201–5209. doi: 10.3892/ol.2019.10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai K.W., Liao Y.L., Wu C.W., Hu L.Y., Li S.C., Chan W.C., Ho M.R., Lai C.H., Kao H.W., Fang W.L., et al. Aberrant expression of MiR-196a in gastric cancers and correlation with recurrence. Genes Chromosom. Cancer. 2012;51:394–401. doi: 10.1002/gcc.21924. [DOI] [PubMed] [Google Scholar]
- 17.Schimanski C.C., Frerichs K., Rahman F., Berger M., Lang H., Galle P.R., Moehler M., Gockel I. High MiR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J. Gastroenterol. 2009;15:2089–2096. doi: 10.3748/wjg.15.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M., Li R.J., Bai H., Xiao P., Liu G.J., Guo Y.W., Mei J.Z. Association between the pre-MiR-196a2 Rs11614913 polymorphism and gastric cancer susceptibility in a Chinese population. Genet. Mol. Res. 2016;15 doi: 10.4238/gmr.15027516. [DOI] [PubMed] [Google Scholar]
- 19.Wang S., Tao G., Wu D., Zhu H., Gao Y., Tan Y., Wang M., Gong W., Zhou Y., Zhou J., et al. A functional polymorphism in MiR196a2 is associated with risk and prognosis of gastric cancer. Mol. Carcinog. 2013;52:87–95. doi: 10.1002/mc.22017. [DOI] [PubMed] [Google Scholar]
- 20.Pu J.-Y., Dong W., Zhang L., Liang W.-B., Yang Y., Lv M.-L. No association between single nucleotide polymorphisms in pre-MiRNAs and the risk of gastric cancer in Chinese population. Iran. J. Basic Med. Sci. 2014;17:128–133. PMID: 24711897. [PMC free article] [PubMed] [Google Scholar]
- 21.Dikeakos P., Theodoropoulos G., Rizos S., Tzanakis N., Zografos G., Gazouli M. Association of the MiR-146aC>G, MiR-149t>C, and MiR-196a2T>C polymorphisms with gastric cancer risk and survival in the Greek population. Mol. Biol. Rep. 2014;41:1075–1080. doi: 10.1007/s11033-013-2953-0. [DOI] [PubMed] [Google Scholar]
- 22.Peng S., Kuang Z., Sheng C., Zhang Y., Xu H., Cheng Q. Association of MicroRNA-196a-2 gene polymorphism with gastric cancer risk in a Chinese population. Dig. Dis. Sci. 2010;55:2288–2293. doi: 10.1007/s10620-009-1007-x. [DOI] [PubMed] [Google Scholar]
- 23.Ahn D.H., Rah H., Choi Y.K., Jeon Y.J., Min K.T., Kwack K., Hong S.P., Hwang S.G., Kim N.K. Association of the MiR-146aC>G, MiR-149t>C, MiR-196a2T>C, and MiR-499a>G polymorphisms with gastric cancer risk and survival in the Korean population. Mol. Carcinog. 2013;52:39–51. doi: 10.1002/mc.21962. [DOI] [PubMed] [Google Scholar]
- 24.Parlayan C., Ikeda S., Sato N., Sawabe M., Muramatsu M., Arai T. Association analysis of single nucleotide polymorphisms in MiR-146a and MiR-196a2 on the prevalence of cancer in elderly Japanese: a case-control study. Asian Pacific J. Cancer Prev. 2014;15:2101–2107. doi: 10.7314/APJCP.2014.15.5.2101. [DOI] [PubMed] [Google Scholar]
- 25.Rogoveanu I., Burada F., Cucu M.G., Vere C.C., Ioana M., Cîmpeanu R.A. Association of MicroRNA polymorphisms with the risk of gastric cancer in a Romanian population. J. Gastrointestin. Liver Dis. 2017;26:231–238. doi: 10.15403/jgld.2014.1121.263.rog. [DOI] [PubMed] [Google Scholar]
- 26.Ni Q., Ji A., Yin J., Wang X., Liu X. Effects of two common polymorphisms Rs2910164 in MiR-146a and Rs11614913 in MiR-196a2 on gastric cancer susceptibility. Gastroenterol. Res. Pract. 2015;2015 doi: 10.1155/2015/764163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei Y., Li L., Gao J. The association between two common polymorphisms (MiR-146a Rs2910164 and MiR-196a2 Rs11614913) and susceptibility to gastric cancer: a meta-analysis. Cancer Biomarkers. 2015;15:235–248. doi: 10.3233/CBM-150470. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L., Gao J., Zhou D., Bao F. Lack of association of two common polymorphisms rs2910164 and rs11614913 with susceptibility to gastric cancer: a meta-analysis. Turkish J. Gastroenterol. 2015;26:378–385. doi: 10.5152/tjg.2015.6603. [DOI] [PubMed] [Google Scholar]
- 29.Chen R., Zheng Y., Zhuo L., Wang S. The association between MiR-423 Rs6505162 polymorphism and cancer susceptibility: a systematic review and meta-analysis. Oncotarget. 2017;8:40204–40213. doi: 10.18632/oncotarget.16319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin J., Wang X., Zheng L., Shi Y., Wang L., Shao A., Tang W., Ding G., Liu C., Liu R., et al. Hsa-MiR-34b/c rs4938723 T>C and hsa-MiR-423 rs6505162 C>A polymorphisms are associated with the risk of esophageal cancer in a Chinese population. PLoS One. 2013;8 doi: 10.1371/journal.pone.0080570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nariman-Saleh-Fam Z., Bastami M., Somi M.H., Behjati F., Mansoori Y., Daraei A., Saadatian Z., Nariman-Saleh-Fam L., Mahmoodzadeh H., Makhdoumi Y., et al. MiRNA-related polymorphisms in MiR-423 (Rs6505162) and PEX6 (Rs1129186) and risk of esophageal squamous cell carcinoma in an Iranian cohort. Genet. Test. Mol. Biomarkers. 2017;21:382–390. doi: 10.1089/gtmb.2016.0346. [DOI] [PubMed] [Google Scholar]
- 32.Xing J., Wan S., Zhou F., Qu F., Li B., Myers R.E., Fu X., Palazzo J.P., He X., Chen Z., et al. Genetic polymorphisms in pre-MicroRNA genes as prognostic markers of colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 2012;21:217–227. doi: 10.1158/1055-9965.EPI-11-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin J., Wang X., Zheng L., Shi Y., Wang L., Shao A., et al. Hsa-miR-34b/c rs4938723 T>C and hsa-miR-423 rs6505162 C>A polymorphisms are associated with the risk of esophageal cancer in a Chinese population PLoS one. 2013;8(11) doi: 10.1371/journal.pone.0080570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y., Vogelsang M., Schäfer G., Matejcic M., Parker M.I. MicroRNA polymorphisms and environmental smoke exposure as risk factors for oesophageal squamous cell carcinoma. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0078520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Umar M., Upadhyay R., Prakash G., Kumar S., Ghoshal U.C., Mittal B. Evaluation of common genetic variants in pre-MicroRNA in susceptibility and prognosis of esophageal cancer. Mol. Carcinog. 2013;52:10–18. doi: 10.1002/mc.21931. [DOI] [PubMed] [Google Scholar]
- 36.Liu X., Li M., Peng Y., Hu X., Xu J., Zhu S., Yu Z., Han S. MiR-30c regulates proliferation, apoptosis and differentiation via the shh signaling pathway in P19 cells. Exp. Mol. Med. 2016;48 doi: 10.1038/emm.2016.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J.P., Liu Y., Hu Z Bin, Shen H.B. Single nucleotide polymorphism in flanking region of miR-30c influences the maturing process of miR-30c in lung carcinoma. Chin. J. Oncol. 2012;34(9):664–668. doi: 10.3760/cma.j.issn.0253-3766.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Hu Z., Shu Y., Chen Y., Chen J., Dong J., Liu Y., Pan S., Xu L., Xu J., Wang Y., et al. Genetic polymorphisms in the precursor MicroRNA flanking region and non-small cell lung cancer survival. Am. J. Respir. Crit. Care Med. 2011;183:641–648. doi: 10.1164/rccm.201005-0717OC. [DOI] [PubMed] [Google Scholar]
- 39.Yin Z., Cui Z., Ren Y., Xia L., Wang Q., Zhang Y., He Q., Zhou B. Association between polymorphisms in pre-MiRNA genes and risk of lung cancer in a Chinese non-smoking female population. Lung Cancer. 2016;94:15–21. doi: 10.1016/j.lungcan.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 40.Xia L., Yin Z., Li X., Ren Y., Zhang H., Zhao Y., Zhou B. Genetic polymorphisms in pre-MiRNAs predict the survival of non-small-cell lung cancer in Chinese population: a cohort study and a meta-analysis. Oncotarget. 2017;8:77963–77974. doi: 10.18632/oncotarget.20276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang C., Li X.P., Chen Y.X., Wu N.Y., Yin J.Y., Zhang W., Zhou H.H., Liu Z.Q. Functional MiRNA variants affect lung cancer susceptibility and platinum-based chemotherapy response. J. Thorac. Dis. 2018;10:3329–3340. doi: 10.21037/jtd.2018.05.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mu Y.P., Su X.L. Polymorphism in pre-MiR-30c contributes to gastric cancer risk in a Chinese population. Med. Oncol. 2012;29:1723–1732. doi: 10.1007/s12032-011-0115-6. [DOI] [PubMed] [Google Scholar]
- 43.Chen L., Cao Y., Rong D., Wang Y., Cao Y. MicroRNA-605 functions as a tumor suppressor by targeting INPP4B in melanoma. Oncol. Rep. 2017;38:1276–1286. doi: 10.3892/or.2017.5740. [DOI] [PubMed] [Google Scholar]
- 44.Kazemi A., Vallian S. Significant association of MiR-605 Rs2043556 with susceptibility to breast cancer. MicroRNA. 2019;9:133–141. doi: 10.2174/2211536608666190926155149. [DOI] [PubMed] [Google Scholar]
- 45.Ryan B.M., Robles A.I., Harris C.C. Genetic variation in microRNA networks: the implications for cancer research. Nat. Rev. Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alidoust M., Hamzehzadeh L., Rivandi M., Pasdar A. Polymorphisms in non-coding RNAs and risk of colorectal cancer: a systematic review and meta-analysis. Crit. Rev. Oncol. Hematol. 2018;132:100–110. doi: 10.1016/j.critrevonc.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 47.Poltronieri-Oliveira A.B., Madeira F.F., Nunes D.B.S.M., Rodrigues G.H., Lopes B.C., Manoel-Caetano F.S., Biselli J.M., Silva A.E. Polymorphisms of MiR-196a2 (Rs11614913) and MiR-605 (Rs2043556) confer susceptibility to gastric cancer. Gene Reports. 2017;7:154–163. doi: 10.1016/j.genrep.2017.04.006. [DOI] [Google Scholar]
- 48.Rs41274221 (SNP) - population genetics - Homo_sapiens - ensembl genome browser 101. https://asia.ensembl.org/Homo_sapiens/Variation/Population?db=core;r=7:100093077-100094077 Available online:
- 49.Zhou J., Zhou J., Wang W., Li W., Wu L., Li G., Shi J., Zhou S. The polymorphism in MiR-25 attenuated the oncogenic function in gastric cancer. Tumor Biol. 2016;37:5515–5520. doi: 10.1007/s13277-015-4376-0. [DOI] [PubMed] [Google Scholar]
- 50.Ritchie M.D., Hahn L.W., Roodi N., Bailey L.R., Dupont W.D., Parl F.F., Moore J.H. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am. J. Hum. Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dunlap J.C., Moore, Detecting J.H. Characterizing, and interpreting nonlinear gene–gene interactions using multifactor dimensionality reduction. Advances in Genetics, Academic Press. 2010;72:101–116. doi: 10.1016/B978-0-12-380862-2.00005-9. [DOI] [PubMed] [Google Scholar]
- 52.Ritchie M.D., Hahn L.W., Moore J.H. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet. Epidemiol. 2003;24:150–157. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- 53.Ritchie M.D., Hahn L.W., Roodi N., Bailey L.R., Dupont W.D., Parl F.F., et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am. J. Hum. Genet. 2001;69(1):138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kehm R.D., Spector L.G., Poynter J.N., Vock D.M., Osypuk T.L. Socioeconomic status and childhood cancer incidence: a population-based multilevel analysis. Am. J. Epidemiol. 2018;187(5):982–991. doi: 10.1093/aje/kwx322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagel G., Linseisen J., Boshuizen H.C., Pera G., Del Giudice G., Westert G.P., et al. Socioeconomic position and the risk of gastric and oesophageal cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST) Int. J. Epidemiol. 2007;36(1):66–76. doi: 10.1093/ije/dyl275. [DOI] [PubMed] [Google Scholar]
- 56.Mw Z., Mj J., Yx Y., Sc Z., B L., X J., Yf P., Ql L., Xy M., K C. Associations of lifestyle-related factors, hsa-MiR-149 and hsa-MiR-605 gene polymorphisms with gastrointestinal cancer risk. Mol. Carcinog. 2012;51:21–31. doi: 10.1002/mc.20863. [DOI] [PubMed] [Google Scholar]
- 57.Kehm R.D., Spector L.G., Poynter J.N., Vock D.M., Osypuk T.L. Socioeconomic status and childhood cancer incidence: a population-based multilevel analysis. Am. J. Epidemiol. 2018;187:982–991. doi: 10.1093/aje/kwx322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nagel G., Linseisen J., Boshuizen H.C., Pera G., Del Giudice G., Westert G.P., Bueno-de-Mesquita H.B., Allen N.E., Key T.J., Numans M.E., et al. Socioeconomic position and the risk of gastric and oesophageal cancer in the European prospective investigation into cancer and nutrition (EPIC-EURGAST) Int. J. Epidemiol. 2007;36:66–76. doi: 10.1093/ije/dyl275. [DOI] [PubMed] [Google Scholar]
- 59.Sharma R. An examination of colorectal cancer burden by socioeconomic status: evidence from GLOBOCAN 2018. EPMA J. 2020;11:95–117. doi: 10.1007/s13167-019-00185-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coughlin S.S. Social determinants of breast cancer risk, stage, and survival. Breast Cancer Res. Treat. 2019;177:537–548. doi: 10.1007/s10549-019-05340-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not Applicable.