Abstract
Complete and partial sequences of mariner-like elements (MLEs) have been reported for hundreds of species of animals, but only two have been identified in plants. On the basis of these two plant MLEs and several related sequences identified by database searches, plant-specific degenerate primers were derived and used to amplify a conserved region of MLE transposase genes from a variety of plant genomes. Positive products were obtained for 6 dicots and 31 monocots of 54 plant species tested. Phylogenetic analysis of 68 distinct MLE transposase sequences from 25 grass species is consistent with vertical transmission and rapid diversification of multiple lineages of transposases. Surprisingly, the evolution of MLEs in grasses was accompanied by repeated and independent acquisition of introns in a localized region of the transposase gene.
Movement of class 2 (DNA) transposable elements from one chromosomal site to another is catalyzed by an element-encoded transposase. Class 2 transposons found in various species are grouped into superfamilies according to similarities and/or specific signatures in the transposase. Tc1/mariner is one of the most diverse and widespread superfamilies in eukaryotes (for review, see refs. 1 and 2). Tc1/mariner transposons have been studied extensively in animals and have been crafted into valuable tools for gene manipulation and genetic analysis in invertebrates, vertebrates, and bacteria (2–6).
The Tc1/mariner superfamily of transposases shares a common amino acid motif called the “DDE/D” signature. This motif is part of the catalytic domain of the transposase and consists of two aspartic acid residues and a glutamic acid residue (or a third D) with characteristic spacing (2, 7, 8). A domain of ≈150 amino acids surrounding the acidic triad is relatively well-conserved and thus has served to establish the evolutionary relationships of Tc1/mariner elements (1, 2, 7, 9). On the basis of these studies, three distinct monophyletic groups have been distinguished: Tc1-like, mariner-like, and pogo-like (1, 2, 7, 9).
Complete and partial sequences of mariner-like elements (MLEs) have been reported for hundreds of species of animals, but only two have been identified in plants (9–11). Soymar1 was isolated from soybean (10), and a related 5.2-kb element was recently identified in a rice bacterial artificial chromosome clone (11). Both elements possess a long ORF with similarity to animal mariner transposases (10, 11). It was previously noted that these plant MLEs also share structural features (similarity in terminal inverted-repeat sequences and flanking TA target site duplications) with a large family of miniature inverted-repeat transposable elements called Stowaway (12, 13). Unlike MLEs, Stowaway elements have no coding capacity yet reside in the genomes of a wide variety of flowering plants (12). If Stowaway elements were mobilized by a transposase encoded in trans by MLEs, it follows that MLEs should also be widespread in plant genomes.
In this study, we combined database searches and PCR with newly designed degenerate primers to demonstrate that MLEs are present in a wide range of flowering plants. Phylogenetic analyses indicate that multiple divergent lineages of MLE transposases can coexist within a single plant species. Results also suggest that the evolution of MLEs in grasses was accompanied by repeated and independent acquisition of introns in a localized region of the transposase gene.
Materials and Methods
Plant Material and Extraction of Genomic DNAs.
Species examined along with their taxonomic classification and origin are published as Table 1 in the supporting information on the PNAS web site (www.pnas.org). Genomic DNA was extracted by using the hexadecyltrimethyl-ammonium bromide (CTAB) method (14) or was provided by groups listed in Table 1.
PCR Amplification of Plant MLE Transposase Sequences.
Primers were derived from regions encoding the amino acid motifs IDEKWF (MLE5A; 5′-ATHGATGARAARTGGTTC-3′) and IQQDNA (MLE3A; 5′-GCATTRTCYTGYTGDAT-3′) conserved in Soymar1, Osmar1, Osmar2, and in most plant MLE transposases mined in databases (see Table 2 in the supporting information, www.pnas.org). PCR amplifications were performed with 10–100 ng of genomic DNA in 25-μl reactions. Cycling conditions were 94°C for 5 min, followed by 30 cycles of 94°C for 45 s, 47°C for 1 min, 72°C for 1 min, and ending with 72°C for 10 min. Ten microliters of the PCR reaction was visualized on a 1% agarose gel. Products were cloned by using 2–4 μl of a 25-μl PCR reaction in a TOPO-TA cloning procedure (Invitrogen). Sequencing was carried out by the Molecular Genetics Instrumentation Facility of the University of Georgia.
Sequence and Phylogenetic Analyses.
Transposase sequences obtained by PCR or database searches were conceptually translated and aligned with clustalw (15) by using default parameters. When necessary, frameshifts were judiciously introduced to maintain aligned reading frames. Three different methods were used to generate phylogenetic trees: neighbor joining (NJ), maximum parsimony (MP), and maximum likelihood (ML). NJ and MP analyses were performed with paup* Version 4.0b8 (16) with default parameters and rooted with the ciliate TBE1 transposase (7). ML star decomposition was carried out by using protml from the molphy 2.3b3 package (17). Trees generated by the three methods were also reswapped with nearest neighbor interchange by using jtt transition matrix (17). Rearranged topologies were very similar to those initially generated by paup* with NJ and MP analyses, at least for the groupings discussed in this study. Introns were predicted by using netgene2 (http://www.cbs.dtu.dk; ref. 18) and fgenesh (http://genomic.sanger.ac.uk/gf/gf.html; ref. 19) programs. For all cases examined, introns were predicted with more than 85% confidence. Homologous introns in closely related sequences were inferred by visual inspection of multiple alignments.
Results and Discussion
Identification of New Plant MLEs by Database Searches.
Two MLEs have been previously reported in plants: Soymar1 from soybean (10) and an element recently identified in a rice bacterial artificial chromosome clone that we have named Osmar1 (11). Both contain long ORFs encoding putative transposases that were used as queries in tblastn searches of the GenBank database. These searches revealed five related coding sequences in rice and nine in Arabidopsis (Table 2 in supporting information, www.pnas.org). The matching regions included the presumed transposase catalytic domain, which was characterized by a “DD39D” signature (10).
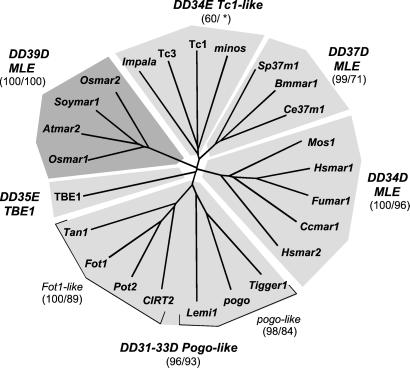
Presumed “DDE/D” regions (≈150 amino acids) of Arabidopsis, soybean, and rice MLE transposases were aligned with those of various representatives of the Tc1/mariner superfamily from a wide range of organisms (alignment available on request). Phylogenetic analyses based on this alignment show that the superfamily can be divided into six major groups with distinct “DDE/D” signatures (Fig. 1). Tc1-like, pogo-like, and animal mariner-like transposases group into three distinct monophyletic clades, as previously shown (1, 2, 9). An additional group is composed of three invertebrate MLEs sharing a “DD37D” motif, two of which were previously described as basal mariners (9). The ciliate TBE1 represents a more distantly related member of the superfamily with a “DD35E” signature (7). Finally, the plant MLE transposases, with their unique “DD39D” signature, group into a distinct monophyletic clade of the Tc1/mariner superfamily.
Figure 1.
Relationship of plant MLE transposases with other members of the Tc1/mariner superfamily. This unrooted NJ tree was generated from a multiple alignment of the “DDE” domain (≈150 amino acids) of various representatives of the Tc1/mariner superfamily of transposases (see Materials and Methods). Bootstrap values (1,000 replications) supporting these groupings are indicated in parentheses for NJ and MP analyses, respectively. The asterisk refers to a bootstrap value <50%. Accession numbers and corresponding species are given in Table 2 (www.pnas.org), except: A. thaliana Lemi1 (AC006161); fungi Fusarium oxysporum Impala (AF282722) and Fot1(X70186); Magnoporte grisea Pot2 (Z33638); Aspergillus awamori Tan1 (U58946); Candida albicans CIRT2(AF205929); cnidaria Fungia sp. Fumar1 (AB055188); C. elegans Tc1 (X01005) and Tc3 (S27787); D. hydei minos (X61695); D. melanogaster pogo (X59837); Homo sapiens Tigger1 (U49973).
PCR Amplification of MLE Transposase Sequences in Diverse Flowering Plants.
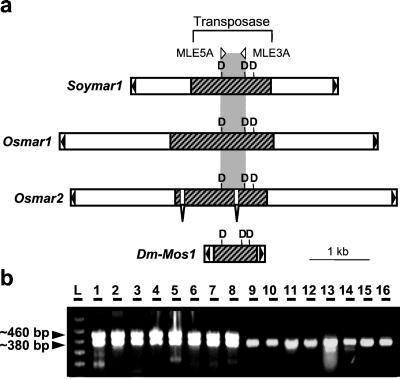
PCR primers used successfully to amplify transposase genes from a wide variety of animal MLEs failed to amplify products in plant genomes (P. Capy and H. M. Robertson, personal communication). Guided by the alignment of plant MLEs, a pair of plant-specific degenerate primers was designed to amplify a fragment of the transposase gene located between the first two Ds of the “DD39D” domain (Fig. 2a). PCR products of the expected size (≈380 bp) were obtained for 31 of 33 monocots and for 6 of 10 eudicots tested (Fig. 2b and Table 1 in supporting information, www.pnas.org). Monocot products were from the Poaceae and Iridaceae families, whereas the eudicot products represented the Solanaceae, Brassiceae, Fabaceae, and the more ancestral Caryophyllaceae. No products were obtained from the more basal angiosperms such as the Laurales, Magnoliales, Piperales, Nympheales, or from gymnosperms and ferns. For all grass species tested (except for three Bambuseae species), PCR amplification yielded a second product (≈470 bp) of approximately equal intensity (Fig. 2b, lanes 1–8).
Figure 2.
Detection of MLE transposase genes in plants by using PCR with degenerate primers. (a) Genetic organization of plant MLEs and design of PCR primers. Soymar1 and Osmar1 were identified in soybean and rice, respectively. Both possess a long uninterrupted ORF (hatched box) with similarity to animal mariner transposases (10, 11). Osmar2 was identified after database searches by using the Osmar1 putative transposase as a query (see Table 2, www.pnas.org). Two short introns are predicted to interrupt the transposase gene of Osmar2. The Mos1 mariner from Drosophila mauritiana is shown for comparison with plant MLEs. Black triangles represent terminal inverted repeats. The “DDD” triad is schematically depicted as well as the position of the degenerate primers (MLE5A, MLE3A) used to detect MLE transposases in other plant species. (b) PCR amplification products in diverse plant species. Shown are examples of amplification products from: 1, O. sativa (cultivated rice); 2, O. nivara (wild rice); 3, Pariana radiciflora (bamboo); 4, Cynodon dactylon (bermuda grass); 5, Setaria italica (foxtail millet); 6, Sorghum bicolor; 7, Zea diploperennis (teosinte); 8, Z. mays mays (maize); 9, Iris brevicaulis; 10, Iris verna; 11, N. longifolia; 12, A. thaliana; 13, Medicago sativa (alfalfa); 14, Glycine max (soybean); 15, Nicotiana tabacum (tobacco); 16, Dianthus sp. (carnation); L, 100-bp ladder. Two bands of approximately equal intensity were obtained for grass species (lanes 1–8), whereas amplification from Iridaceae and dicot species yielded a single band (lanes 9–16).
For each plant (or at least for one representative of each tribe), PCR products were cloned, and from 2 to 12 independent clones were randomly chosen to sequence. Comparison of the sequences with previously identified plant MLEs confirmed that most PCR products (≈95%) represented the same region of a mariner-like transposase gene. Larger products from the grasses contained insertions that were determined to be introns in the transposase sequence (see below).
Taken together, 37 of the 54 plant species sampled harbored mariner-like coding sequences in their genome (Table 1). This is a conservative estimate of the presence of MLE transposases in plant genomes, because failure to detect a PCR product from a particular species may be because of a single amino acid replacement in the motif recognized by the primers. Furthermore, in most species tested, all positive PCR clones turned out to be unique. For example, in barley, seven of the seven PCR products cloned and sequenced were from a different MLE transposase gene (see Fig. 3), indicating that there are multiple transposase genes in all genomes tested and that our sampling within any one organism is most likely incomplete.
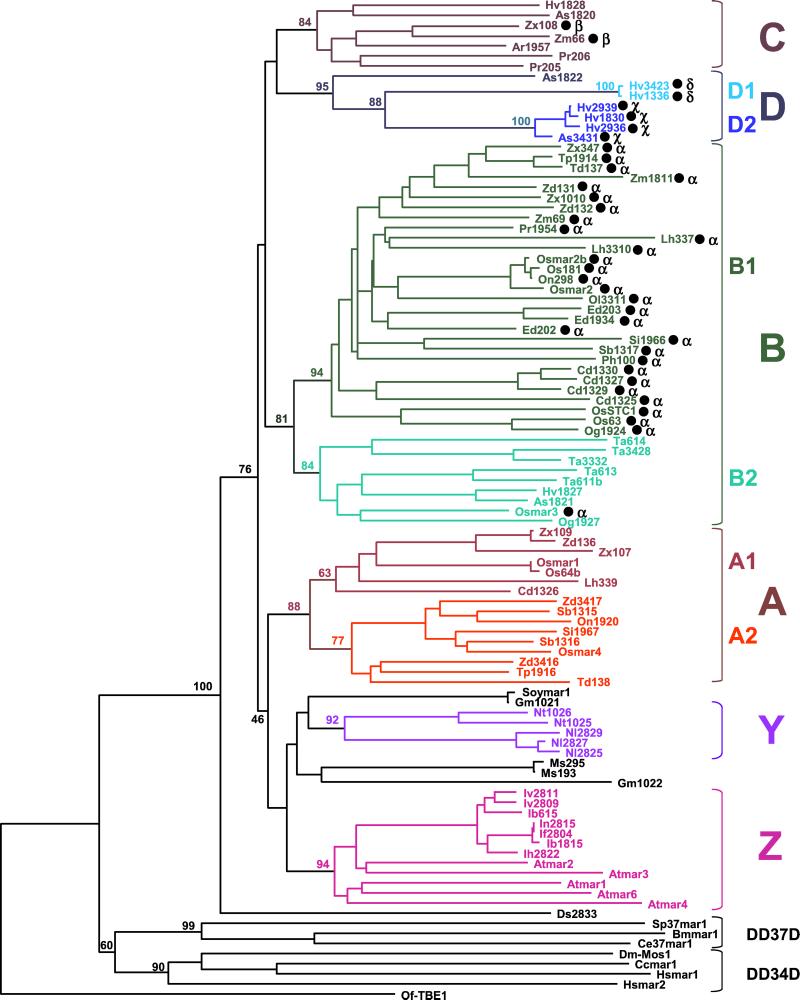
Figure 3.
Phylogenetic relationships of 91 plant MLE transposase fragments. This NJ tree was generated from a multiple alignment of conceptually translated transposase sequences, including 81 plant transposase fragments isolated by PCR (GenBank accession nos. AF432513–AF432593) and 10 obtained from database searches [Osmars, Soymars, and Atmars; see Table 2 (www.pnas.org) for accession nos.]. The corresponding regions of several animal MLE transposases from diverse subfamilies (9) and the distantly related Tc1/mariner-like transposase from the ciliate transposon TBE1 (7) were also included in this analysis [see Table 2 (www.pnas.org) for description]. Sequences were named according to the species initials followed by the number of the clone or the suffix -mar, following the nomenclature introduced by Robertson et al. (9) [see Tables 1 and 2 (www.pnas.org) for abbreviations]. A black dot after a name indicates that a predicted intron was removed from the nucleotide sequence. Introns occurred at four positions (α, β, χ, δ ; see text and Fig. 4b). Bootstrap values (1,000 replicates) are shown for the major nodes of the tree. Groupings defining lineages and sublineages of plant MLE transposases are emphasized by capital letters and different colors. Monocots, Poaceae: Ph, P. lappulaceus; Pr, P. radiciflora; Ar, Arundinaria simonii; Lh, Litachne humilis; Ed, Ehrarhta dura; Os, O. sativa, On, O. nivara; Og, O. grandiglumis; Ta, Triticum aestivum; Hv, Hordeum vulgare; As, Avena sativa; Cd, C. dactylon; Si, S. italica; Sb, S. bicolor; Td, Tripsacum dactyloides; Tp, Tripsacum pilosum; Zd, Z. diploperennis, Zx, Z. mays mexicana; Zm, Z. mays mays. Monocots Iridaceae: Nl, N. longifolia; Ib, I. brevicaulis; If, I. fulva; Ih, I. hybrida; In, I. nelsonii; Iv, I. verna. Dicots: Ds, Dianthus sp.; Gm, Glycine max; Ms, Medicago sativa; At, A. thaliana; Nt, N. tabacum. Insects Diptera Sp, Sarcophaga peregrina; Cc, Ceratitis capitata; insect Lepidoptera: Bm, Bombyx mori; nematode Ce, C. elegans; Hs, H. sapiens; ciliate: Of, Oxytricha fallax.
Evolution and Diversity of Plant MLE Transposases.
One hundred and one distinct transposase fragments were conceptually translated. After removal of predicted introns, 51 of these contained insertions, deletions (1–33 bp), or substitutions that introduced stop codons or led to frameshifts. Therefore, a large proportion of plant MLEs probably contain inactive transposases, which is likely to be an underestimate because only about one-third of transposase genes were amplified (Fig. 2a). Thus, most of the plant MLE transposase genes may now be evolving as pseudogenes, accumulating mutations neutrally and thus rapidly within each species (8, 9).
Evolutionary relationships for 91 plant MLE transposases (10 sequences were too severely mutated to be included) were investigated by constructing multiple amino acid alignments and generating phylogenetic trees (Fig. 3). We conclude from these data that plant transposase sequences are monophyletic and extremely heterogeneous. Pairwise amino acid identities, even between sequences isolated from the same or a closely related species, can be as low as 35–45%. Consequently, sequences from two relatively distant species can be more similar to each other than those from two sibling species or from the same genome. For example, Osmar4 from cultivated rice (Oryza sativa) shares 87% amino acid identity with Sb1316 from sorghum but only 44% identity with Og1924, a sequence obtained from the wild rice Oryza grandiglumis. Furthermore, pairwise comparisons among Osmar1, Osmar2, Osmar3, Osmar4, and Os63, all from O. sativa, revealed only 46–61% identity. These results can be explained by postulating the existence of multiple ancestral lineages of transposases that diversified early in evolution [at least 50 million years (Myr) ago in the case of sorghum and rice (20, 21)] and were vertically inherited (1, 8, 9).
Six transposase lineages are shown (A–D, Y, and Z in Fig. 3) and are defined as the largest well-supported monophyletic group of sequences obtained from phylogenetic trees generated by three distinct methods (bootstrap values >60%; see Materials and Methods). Further support for these groupings comes from additional synapomorphic characteristics such as amino acid insertion/deletion and the presence of introns at the same location. Most of the sequences are from grass species, which were sampled most heavily because their evolutionary history and phylogenetic relationships have been the focus of several recent reports (for review, see refs. 20, 21). These studies provide a useful framework to analyze the molecular evolution of plant MLEs over a relatively long period.
Four major MLE transposase lineages can be defined in the grasses on the basis of criteria described above (A–D in Fig. 3; Fig. 4a). Lineage A can be further divided into two sublineages, each containing sequences from very distant grasses such as rice and maize [divergence time 50–70 Myr (20, 21)]. Lineage B also has a wide distribution as it includes sequences from 20 of 21 grasses sampled, representing all of the major subfamilies and including Pharus lappulaceus, one of the most ancestral species (20). Lineage C, with fewer sequences than lineages A and B, still includes sequences from three distant grass subfamilies (Bambusoideae, Poideae, and Panicoideae), also indicating an ancient origin. The wide distribution of distinct lineages in the grasses suggests that there first was extensive diversification of MLEs during early grass evolution and that this was followed by a period of stable maintenance for at least the last 50 Myr (Fig. 4a). Lineage D, on the other hand, contains sequences isolated from only the Poideae subfamily (oat and barley), suggesting either a more recent origin for this lineage [oat and barley diverged about 25 Myr ago (20, 21)] or its loss from the other grasses.
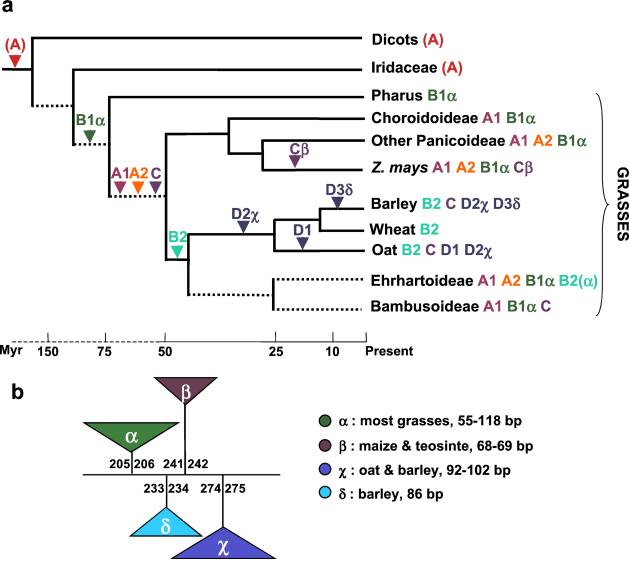
Figure 4.
Evolution of grass MLE transposases and accompanying intron gains. (a) Diversification of plant MLEs and possible intron gains during grass evolution. Relationships among grass taxa and estimated divergence times are from refs. 20 and 21. Uncertain divergence times are represented by dashed lines. MLE lineages detected in each taxa are shown by their respective capital letters. Arrowheads indicate the presence of a particular lineage before the divergence of the descending species. It was assumed that sequences from dicots and Iridaceae are related to lineage A of grasses, although this relationship was not strongly supported by bootstrap analysis (see Fig. 2). (b) Positions of introns in different grass MLE transposases. Introns are depicted as triangles. Numbers above or below the triangles refer to the position of coding nucleotides interrupted by the intron. The distribution and range of lengths for each type of intron are shown (see also Fig. 2).
In contrast, no clear relationships emerged from the sequences derived from dicot species. To illustrate the problem, note that sequences from Arabidopsis were more closely related to those isolated from the Louisiana iris complex than to any other dicot sequences (62–70% amino acid identity). Similarly, two sequences from tobacco clustered with products obtained from Neomarica longifolia, a close relative of Iris (62–68% identity). Both groupings have strong bootstrap support in all phylogenies; therefore, they can be considered as two separate lineages (Y and Z in Fig. 3). Given the close taxonomic relationship between the Iris and Neomarcia genus in the Iridaceae family, one would expect that their MLE transposases would group together rather than with sequences obtained from two different dicots. Such phylogenetic discrepancies might be because of the horizontal transfer of MLEs between these species or between their ancestors. Horizontal transfer across wide taxonomic gaps has been well documented for animal MLEs and is believed to be important for the maintenance and propagation of DNA transposons (1, 8, 9). Resolving the dicot lineages and detecting horizontal transfer of plant MLEs will require a larger sampling of species as well as obtaining additional sequences per species.
Acquisition of Introns in Grass MLE Transposase Genes.
Few Tc1/mariner transposase genes are known to contain introns. Examples include Tc1-like transposons from Drosophila hydei and Caenorhabditis elegans and pogo-like elements from Drosophila melanogaster and Arabidopsis thaliana (22–24). Despite the vast number of species examined, no animal MLE transposases are known to contain introns (9). It was, for this reason, surprising that more than half of the MLE transposase fragments isolated from grasses were predicted to contain an intron. As expected for introns, there is little conservation in length and sequences among species, except between closely related species (e.g., 93% similarity between O. sativa Os63 and O. grandiglumis Og1924).
There is also variation in intron insertion sites within the amplified transposase fragment with four sites of insertion that define four different introns (Fig. 4b). The most common insertion site (intron α) is in sequences obtained from species representing five different subfamilies of grasses (Figs. 3 and 4), suggesting that this intron was present in a transposase gene in a common ancestor of the grasses ≈55–70 Myr ago (Fig. 4a). In our phylogenetic analyses, sequences containing intron α fell into lineage B of grass MLE transposases. However, intron α is lacking in lineage B sequences isolated from oat, wheat, barley (Poideae subfamily), and from the rice species O. grandiglumis (Fig. 3). These sequences form a well-supported monophyletic group within lineage B (called sublineage B2), along with a sequence that does display intron α (Osmar3, Fig. 3). This suggests that intron α may have been lost sporadically during MLE transposase evolution.
The other three types of introns were detected only in one or two of the 25 grass species examined. Intron β was predicted in two sequences isolated from two subspecies of Zea mays that clustered in lineage C (Figs. 3 and 4). Intron χ was predicted in sequences from oat and barley (sublineage D2), and intron δ was found only in barley sequences (sublineage D1) (Figs. 3 and 4).
That each type of intron is restricted to a particular lineage (or sublineage) of transposase strongly suggests they represent four independant introns (i.e., nonhomologous). Alternatively, the four extant introns may have derived from a single ancestral intron that moved to their present positions, possibly by a mechanism akin to “intron sliding” (25). This latter scenario is considered unlikely, as studies have shown that intron sliding is a rare event, and sliding by only one base has received statistical support (25–28).
Two contrasting hypotheses provide explanations for the introns seen in the MLE transposases of extant grasses. The first hypothesis posits that the introns are relics of an ancestral intron-rich MLE transposase gene that was subject to massive intron loss during its diversification. Alternatively, transposase diversification in the course of grass evolution was accompanied by independent gains of introns. The first hypothesis, which is consistent with the “intron-early” theory (29), is considered less likely for the following reasons. First, sequences with more than one intron were not detected (i.e., no intermediate state of intron loss). Second, small exons are rare in plant genes (30). An ancestral intron-rich transposase gene would have had a region of 69 bp broken up into exons as small as 28, 8, and 33 bp (Fig. 4b). Third, each intron is found in a different lineage of transposase (or a sublineage) and, in three cases (β, χ, δ), their phylogenetic distribution is restricted to one or two closely related species (Fig. 4b). Such a patchy but nonrandom distribution is more parsimoniously explained by the independent addition of introns to exon sequences (see Fig. 4a).
There is now compelling evidence for the gain of spliceosomal introns during eukaryotic evolution (26, 31). Introns can be viewed as selfish elements that spread neutrally in genomes, but it is also likely that the addition of introns can be positively selected if intron gain confers an advantage to the host. The prevalence of alternative and differential splicing in eukaryotes may provide support for this hypothesis (31). More relevant to our results is the example of differential splicing of the P element transposase gene in D. melanogaster tissues. Retention of the third intron in somatic cells prevents the production of an active transposase and reduces the detrimental effects of P transposition (32). Whether intron acquisition in grass MLEs could have been selected to regulate transposase activity is an intriguing hypothesis that is now under investigation.
Supplementary Material
Acknowledgments
We are grateful to colleagues who kindly provided plant tissues or genomic DNAs. We thank D. Holligan and T. Stevenson for technical assistance, N. Jiang for help in mining rice MLEs, and B. Gaut, E. Kellogg, E. Kentner, and C. Lawrence for comments and suggestions. This work was supported by grants from the National Science Foundation Plant Genome Initiative and the National Institutes of Health to S.R.W. C.F. was supported in part by the University of Georgia Research Foundation.
Abbreviations
- MLE
mariner-like elements
- Myr
million years
- NJ
neighbor joining
- MP
maximum parsimony
Footnotes
References
- 1.Capy P, Bazin C, Higuet D, Langin T. Dynamics and Evolution of Transposable Elements. Austin, TX: Springer; 1998. [Google Scholar]
- 2.Plasterk R H A, Izsvák Z, Ivics Z. Trends Genet. 1999;15:326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 3.Catteruccia F, Nolan T, Loukeris T G, Blass C, Savakis C, Kafatos F C, Crisanti A. Nature (London) 2000;405:959–962. doi: 10.1038/35016096. [DOI] [PubMed] [Google Scholar]
- 4.Judson N, Mekalanos J J. Nat Biotechnol. 2000;18:740–745. doi: 10.1038/77305. [DOI] [PubMed] [Google Scholar]
- 5.Bessereau J L, Wright A, Williams D C, Schuske K, Davis M W, Jorgensen E M. Nature (London) 2001;413:70–74. doi: 10.1038/35092567. [DOI] [PubMed] [Google Scholar]
- 6.Zagoraiou L, Drabek D, Alexaki S, Guy J A, Klinakis A G, Langeveld A, Skavdis G, Mamalaki C, Grosveld F, Savakis C. Proc Natl Acad Sci USA. 2001;98:11474–11478. doi: 10.1073/pnas.201392398. . (First Published September 18, 2001; 10.1073/pnas.201392398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doak T G, Doerder F P, Jahn C L, Herrick G. Proc Natl Acad Sci USA. 1994;91:942–946. doi: 10.1073/pnas.91.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartl D L, Lohe A R, Lozovskaya E R. Annu Rev Genet. 1997;31:337–358. doi: 10.1146/annurev.genet.31.1.337. [DOI] [PubMed] [Google Scholar]
- 9.Robertson H M, Soto-Adames F N, Walden K O, Avancini R M, Lampe D J. In: Horizontal Gene Transfer. Syvanen M, Kido C I, editors. London: Chapman & Hall; 1998. pp. 268–284. [Google Scholar]
- 10.Jarvik T, Lark K G. Genetics. 1998;149:1569–1574. doi: 10.1093/genetics/149.3.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarchini R, Biddle P, Wineland R, Tingey S, Rafalski A. Plant Cell. 2000;12:381–391. doi: 10.1105/tpc.12.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bureau T E, Wessler S R. Plant Cell. 1994;6:907–916. doi: 10.1105/tpc.6.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turcotte K, Srinivasan S, Bureau T. Plant J. 2001;25:169–179. doi: 10.1046/j.1365-313x.2001.00945.x. [DOI] [PubMed] [Google Scholar]
- 14.Doyle J J, Doyle J L. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 15.Thompson J D, Desmond D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swofford D L. paup*: Phylogenetic Analysis Using Parsimony and Other Methods. Sunderland, MA: Sinauer; 1999. [Google Scholar]
- 17.Adachi J, Hasegawa M. molphy: Programs for Molecular Phylogenetics Based on Maximum Likelihood 2.3. Tokyo: The Institute of Statistical Mathematics; 1996. [Google Scholar]
- 18.Hebsgaard S M, Korning P G, Tolstrup N, Engelbrecht J, Rouze P, Brunak S. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salamov A A, Solovyev V V. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kellogg E A. Plant Physiol. 2001;125:1198–1205. doi: 10.1104/pp.125.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaut, B. S. (2001) New Phytol., in press.
- 22.Franz G, Loukeris T G, Dialektaki G, Thompson C R, Savakis C. Proc Natl Acad Sci USA. 1994;91:4746–4750. doi: 10.1073/pnas.91.11.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plasterk R H. Curr Top Microbiol Immunol. 1996;204:125–143. doi: 10.1007/978-3-642-79795-8_6. [DOI] [PubMed] [Google Scholar]
- 24.Feschotte C, Mouchès C. Mol Biol Evol. 2000a;17:730–737. doi: 10.1093/oxfordjournals.molbev.a026351. [DOI] [PubMed] [Google Scholar]
- 25.Stoltzfus A, Logsdon J M, Jr, Palmer J D, Doolittle W F. Proc Natl Acad Sci USA. 1997;94:10739–10744. doi: 10.1073/pnas.94.20.10739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Logsdon J M., Jr Curr Opin Genet Dev. 1998;8:637–648. doi: 10.1016/s0959-437x(98)80031-2. [DOI] [PubMed] [Google Scholar]
- 27.Rogozin I B, Lyons-Weiler J, Koonin E V. Trends Genet. 2000;16:430–432. doi: 10.1016/s0168-9525(00)02096-5. [DOI] [PubMed] [Google Scholar]
- 28.Sakharkar M K, Tan T W, de Souza S J. Bioinformatics. 2001;17:671–675. doi: 10.1093/bioinformatics/17.8.671. [DOI] [PubMed] [Google Scholar]
- 29.Gilbert W, de Souza S J, Long M. Proc Natl Acad Sci USA. 1997;94:7698–7703. doi: 10.1073/pnas.94.15.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filipowicz W, Gniadkowski M, Klahre U, Liu H X. In: Pre-mRNA Processing. Lamond A I, editor. Austin, TX: Landes; 1995. pp. 65–77. [Google Scholar]
- 31.Mattick J S, Gagen M J. Mol Biol Evol. 2001;18:1611–1630. doi: 10.1093/oxfordjournals.molbev.a003951. [DOI] [PubMed] [Google Scholar]
- 32.Laski F A, Rio D C, Rubin G M. Cell. 1986;44:7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.