Abstract
Eusociality is a major evolutionary innovation involving alterations in life history, morphology, and behavior. Advanced eusocial insects, such as ants, termites, and corbiculate bees, cannot provide insights into the earliest stages of eusocial evolution because eusociality in these taxa evolved long ago (in the Cretaceous) and close solitary relatives are no longer extant. In contrast, primitively eusocial insects, such as halictid bees, provide insights into the early stages of eusocial evolution because eusociality has arisen recently and repeatedly. By mapping social behavior onto well-corroborated phylogenies, I show that eusociality has arisen only three times within halictid bees (contrary to earlier estimates of six or more origins). Reversals from eusocial to solitary behavior have occurred as many as 12 times, indicating that social reversals are common in the earliest stages of eusocial evolution. Important attributes of social complexity (e.g., colony size, queen/worker dimorphism) show no obvious association with phylogeny, and some reversals to solitary nesting are related to host-plant switches (from polylecty to oligolecty). These results provide a glimpse of social evolution in its earliest stages and provide insights into the early evolution of advanced eusocial organisms.
The evolutionary origin of eusociality, with overlapping generations, cooperative brood care, and reproductive division of labor (1–3), is considered to be one of the major transitions in evolution (4). Understanding the evolutionary origins of eusociality has been a focus of evolutionary and ethological research for over 40 years. The evolutionary origins of eusociality can best be analyzed in taxa with variable levels of sociality (those taxa including both solitary and eusocial species) and in taxa in which eusociality has had a relatively recent origin. Few lineages of eusocial insects are sufficiently variable in social behavior or sufficiently recent in origin to provide detailed insights into the early stages of eusocial evolution and the evolutionary transitions that occur early in the transformation from solitary living to eusociality. Although ants, termites, and corbiculate bees are model organisms for understanding the organization of eusocial insect colonies and the maintenance of eusociality (2, 3), these groups provide few insights into the evolutionary origins of eusociality (5). Ants, termites, and corbiculate bees are all of Cretaceous age (6–8), and closely related solitary taxa have long since gone extinct. Furthermore, advanced eusociality is fixed in these groups and flexibility in social behavior is limited.
Two groups of social Hymenoptera are ideal for investigating the evolutionary origins of eusociality: the wasp family Vespidae (9, 10) and the bee family Halictidae (11, 12). Halictid bees are a model group for understanding the evolutionary origins of eusociality for several reasons. First, halictid bees exhibit substantial diversity in social behavior among species. Species within the subfamily Halictinae (>2,500 species worldwide) exhibit solitary nesting (13), communal nesting (14), primitive eusociality (15), cleptoparasitism (Sphecodes), and social parasitism (16). Second, among the eusocial species there is substantial variation in the degree of sociality, with annual colonies of a queen and ≤5 workers to colonies of over 500 workers and perennial life cycles (17). Finally, some widespread species exhibit intraspecific variation in sociality associated with changes in altitude or latitude (18). These factors have led previous authors to conclude that eusociality is extraordinarily labile in halictid bees and that social diversity in halictid bees is attributable to numerous (≥6) independent origins of eusociality (11, 17, 19, 20).
However, previous hypotheses concerning the history of social evolution in halictid bees have not been based on explicit phylogenies at appropriate levels, and thus are highly speculative. Phylogenies for the halictid subfamilies, tribes, and genera, combined with phylogenies at the subgeneric and species levels for the predominantly eusocial groups, would allow us to accurately infer the number of origins of eusociality as well as identify reversals in social behavior (21). Phylogenies for the halictid bees also would allow us to infer the pattern of social evolution in its earliest stages and to investigate the evolutionary transitions that characterize rapidly diversifying eusocial lineages in the earliest stages of eusocial evolution.
To firmly establish the historical pattern of social evolution in the halictid bees, I reconstructed the higher-level phylogenetic relationships by using a large nucleotide data set of more than 1,600 aligned nucleotide sites spanning three exons and two introns of the nuclear, protein-encoding gene elongation factor-1α (22).
DNA extraction, PCR of mitochondrial and nuclear genes, and sequencing followed protocols detailed in refs. 22–25. Alignments were performed with the lasergene software package and improved by eye. Phylogenetic analyses based on parsimony and maximum likelihood were performed by using paup* version 4.0b8 (26), and characters were mapped onto trees by using macclade version 3.07 (27). Bootstrap values were calculated based on 100 replicates with 10 random sequence additions per replicate. paup Nexus files including DNA sequence alignments are available as supporting information on the PNAS web site, www.pnas.org. Specimen locality data, voucher codes, and Genbank accession numbers are available in Tables 2 and 3, which are published as supporting information on the PNAS web site.
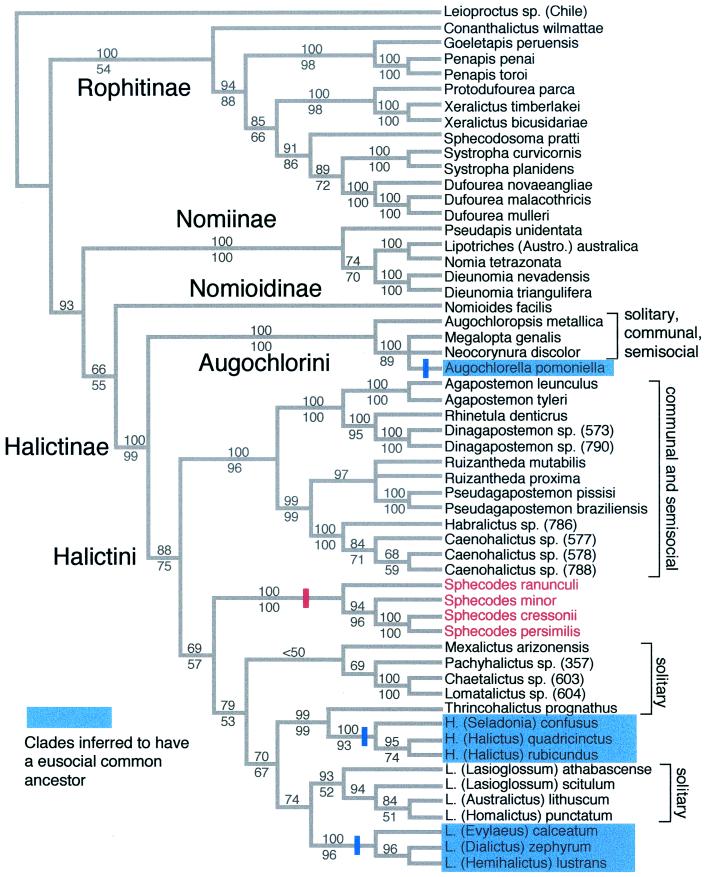
Taxa were sampled from all subfamilies, tribes, and most genera of halictid bees. Particular effort was made to provide extensive taxon sampling from the subfamily Halictinae, which includes all of the known genera of eusocial halictid bees. Analysis of this data set by equal weights parsimony yielded just six trees (Fig. 1). Virtually all nodes show bootstrap support >70%, whether introns are included (bootstrap values above the branches) or excluded (bootstrap values below the branches), and relationships among subfamilies are congruent with a recent morphological analysis (28). Analysis of the data set by maximum likelihood under various models yielded the same overall topology. All of the currently recognized subfamilies and tribes are recovered with high bootstrap support, and within the largest and behaviorally most important group (the tribe Halictini) generic and subgeneric relationships are strongly supported and congruent with morphological data.
Figure 1.
Phylogeny of the halictid subfamilies, tribes, and genera. Strict consensus of six trees based on equal weights parsimony analysis of the entire data set (spanning positions 145-1271 in the coding region of the Apis mellifera F2 copy). The data set includes three exons and two introns. Two regions within the introns were excluded because they could not be aligned unambiguously. Gaps coded as a fifth state or according to the methods described in ref. 23 yielded the same six trees. Bootstrap values above the nodes indicate bootstrap support based on the exons + introns data set. Bootstrap values below the nodes indicate support based on an analysis of exons only. For the exons + introns analysis the data set included 1,541 total aligned sites (619 parsimony-informative sites), the trees were 3,388 steps in length, and the consistency index (excluding uninformative sites) was 0.3831. For the exons-only analysis there were 1,127 aligned sites (327 parsimony-informative sites), the trees were 1,648 steps in length, and the consistency index (excluding uninformative sites) was 0.3195. The tree was rooted by using a colletid bee (Leioproctus sp.). Maximum-likelihood analyses using the K2P model with rate heterogeneity accounted for by site-specific rates yielded the same overall tree topology irrespective of whether exons or exons + introns were analyzed. Clades inferred to have a eusocial common ancestor (see text) are shown in blue. Cleptoparasitic taxa (Sphecodes) are shown in red. Three origins of eusociality (indicated by blue bars) are indicated on the cladogram.
This tree, combined with published results for the Augochlorini (29, 30), the genus Halictus (23), and results for Lasioglossum presented below, indicates that eusociality has had just three independent origins (Fig. 1, indicated in blue) within the subfamily Halictinae: (i) in the Augochlorini [in the common ancestor of the monophyletic group including Augochlora and Augochlorella (29)], (ii) in the common ancestor of the genus Halictus (23), and (iii) in the common ancestor of the weak-veined Lasioglossum (including the subgenera Evylaeus, Dialictus, Hemihalictus, Paralictus, Sphecodogastra, and Sudila). The placement of Thrincohalictus [a monotypic and presumably solitary or communal genus of Middle Eastern bees (31)] as sister to Halictus, and the placement of the exclusively solitary and communal strong-veined Lasioglossum (referred to as the Lasioglossum series in ref. 31) as sister to the primarily eusocial weak-veined Lasioglossum (referred to as the Hemihalictus series in ref. 31), confirms that eusociality in Halictus and Lasioglossum has had independent origins.
Analyses of relationships within the three eusocial clades reveals that eusociality has arisen just once in each and that reversals to solitary nesting have occurred repeatedly.
Tribe Augochlorini.
Within the Augochlorini two genera are known to exhibit eusocial behavior: Augochlora and Augochlorella [comprising a total of 140 species, many of which are eusocial (31)]. Recent analyses of the generic relationships in the tribe determined that these two genera are sister groups [referred to as the “Augochlora group” (29)] and that one reversal to solitary nesting occurs within the genus Augochlora (29), implying a single origin of eusociality and one reversal.
Genus Halictus.
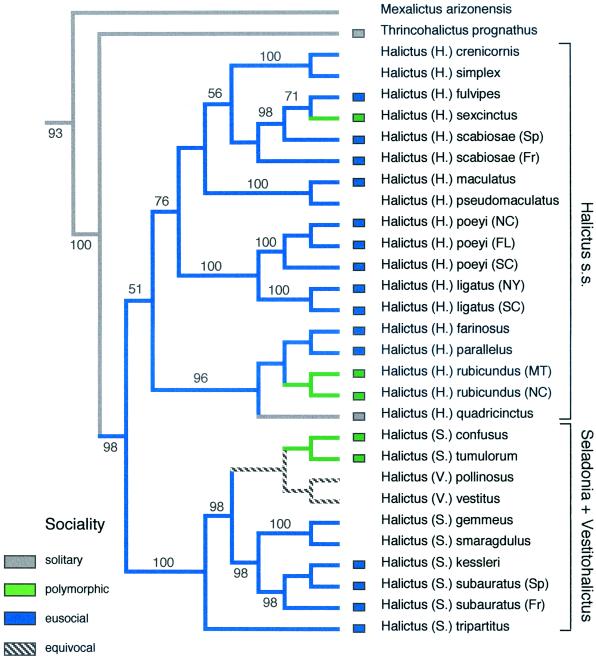
Phylogeny of the subgenera and species of Halictus [comprising 217 species, most of which are eusocial or socially polymorphic (31)] was recently analyzed and unambiguously supported the hypothesis that eusociality was present in the common ancestor of Halictus and later underwent multiple (4–6) reversals to solitary nesting within the genus (23) (Fig. 2), a result consistent with allozyme data (32).
Figure 2.
Phylogeny of the species and subgenera of Halictus based on elongation factor-1α. Details of the phylogenetic analysis are presented in ref. 23. Social behavior was mapped onto the tree by using macclade version 3.07 (27). Socially polymorphic taxa are those in which solitary as well as eusocial populations are known to occur within the same species. Equivocal branches are those in which ancestral character states could not be unambiguously inferred given the tree topology and available behavioral data.
Genus Lasioglossum.
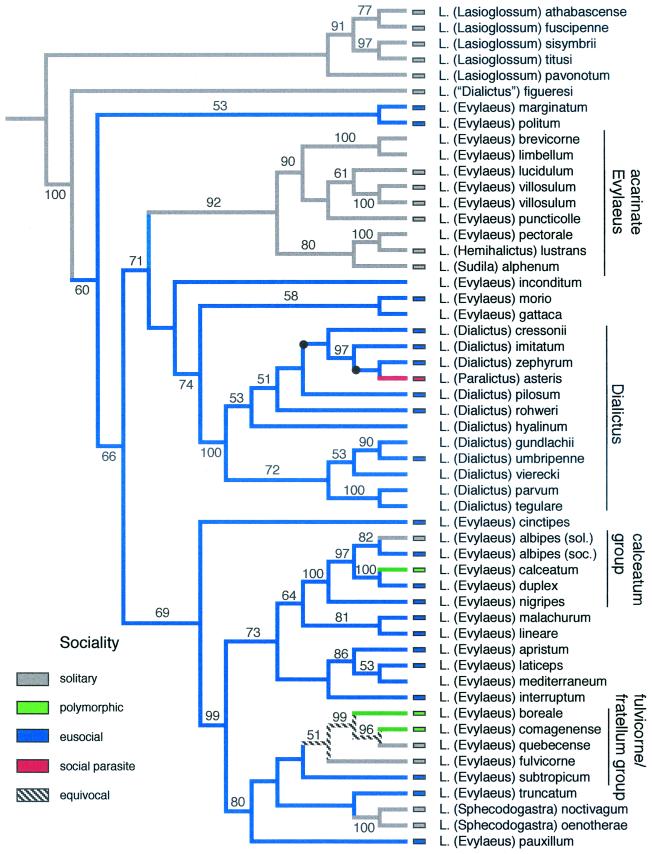
The phylogeny of Lasioglossum based on mitochondrial (24) as well as nuclear genes (25) recently has been analyzed, but results based on either gene alone were inconclusive. When additional species are added to the data set (especially “acarinate” Evylaeus) and relationships within the weak-veined subgenera of Lasioglossum [comprising 544 total species, most of which are eusocial (31)] are analyzed by using a combined nuclear and mitochondrial data set (Fig. 3), one obtains a clear hypothesis of relationships with virtually every node supported by >50% bootstrap support. By analyzing the distribution of eusociality on this cladogram it is evident that eusociality within Lasioglossum has had a single origin with multiple reversals to solitary nesting: (i) in the “acarinate Evylaeus” (including the subgenera Hemihalictus and Sudila), (ii) twice in the calceatum group of Evylaeus, (iii) in the fulvicorne/fratellum group of Evylaeus, and (iv) in the subgenus Sphecodogastra. Social parasitism arose once in the subgenus Paralictus from within the lineage of its hosts, Dialictus (16, 24).
Figure 3.
Phylogeny of the eusocial clade of Lasioglossum. One of six trees based on equal weights parsimony analysis of the combined cytochrome oxidase I (COI) (24) and elongation factor-1α (25) data set. (Nodes that collapse in the strict consensus are marked with a black dot and do not alter the character mapping.) Third position sites in COI were excluded (downweighting yielded the same overall tree topologies) because this data partition showed a highly skewed base composition (91% A/T), significant base compositional heterogeneity among taxa (P ≤ 0.001), and an 8-fold higher rate of substitution than any of the other six data partitions. The total data set included 2,734 aligned nucleotide sites (2,321 with COI nucleotide 3 excluded; 394 parsimony informative sites). Trees were 1,765 steps in length and the consistency index was 0.3669. The tree was rooted by using five species of Lasioglossum s.s. The tree recovers several of the recognized subgenera of Lasioglossum (including Sphecodogastra and Dialictus) as well as many of the species groups of Evylaeus. Social behavior and parasitism (obtained from published studies listed in ref. 37) were mapped onto the tree by using macclade version 3.07 (27). Eusociality arose once in the common ancestor of the ingroup species excluding L. (“Dialictus”) figueresi. Reversals to either solitary nesting or social polymorphism (eusocial and solitary populations of the same species) occurred as many as five times. Social parasitism (in the subgenus Paralictus) is inferred to have arisen once. Methods are outlined in the text.
Among the most striking patterns that emerge from analysis of social evolution in Lasioglossum is the lack of correspondence between the cladogram and the correlates of social “complexity” (33, 34). Colony size, for example, is enormous in L. (E.). marginatum, a relatively basal branch of the tree. Likewise, the magnitude of queen/worker dimorphism also shows no clear phylogenetic pattern. The Evylaeus calceatum group shows weak queen/worker dimorphism and yet is sister to the Evylaeus malachurum group [L. (Evylaeus) malachurum + L. (Evylaeus) lineare], which shows strong queen/worker dimorphism (34). The pattern of social evolution is one of increased and decreased social complexity independent of the phylogeny. These results are consistent with Michener's argument that eusociality need not arise through a gradual accumulation of more and more complex social behaviors (35), but may arise (and disappear) without invoking species intermediate in social behavior.
Interestingly, of the reversals to solitary nesting, at least two (Hemihalictus and Sphecodogastra) are correlated with switches from polylectic (generalist) to oligolectic (specialist) pollen foraging [Hemihalictus is oligolectic on Aplopappus (Compositae) (13) and Sphecodogastra is oligolectic on Oenothera and related genera (Onagraceae) (36)]. Narrow host plant preferences are generally considered to preclude bivoltine life cycles and hence eusociality.
These phylogenies indicate that eusociality has had just three independent origins within halictine bees, and that diversity in sociality is primarily attributable to reversals to solitary nesting within predominantly eusocial clades (Table 1). By resolving the tribal, generic, and subgeneric relationships within halictine bees, these results provide the best estimate of the number of independent origins of eusociality within halictid bees. Furthermore, these phylogenies provide important insights into behavioral and ecological correlates of eusociality (10). Traits that appear to promote eusociality in halictine bees include adult diapause (restricted within Halictidae to the subfamily Halictinae), and a primarily northern Hemisphere, temperate distribution (which characterizes the two largest eusocial genera, Halictus and Lasioglossum). Communal and semisocial nesting [which are primarily restricted to the agapostemonine genera (14) and the Australian subgenera of Lasioglossum (25)] appear unrelated to the evolutionary origins of eusociality, thus favoring the subsocial rather than parasocial “route” to eusociality (as suggested by refs. 12 and 34). Finally, these results provide insights into the evolutionary transitions that may have occurred early in the evolution of ancient, advanced eusocial lineages, such as ants, termites, and corbiculate bees. The initial stages of social evolution may be characterized by few origins of eusociality and multiple reversals back to solitary nesting.
Table 1.
Origins and losses of eusociality in halictid bees
Supplementary Material
Acknowledgments
I am grateful to the following collaborators for providing specimens for this study: J. Ascher, M. Ayasse, C. Eardley, M. Engel, P. Kukuk, P. Lincoln, Y. Maeta, R. Miyanaga, J. Neff, B. Norden, A. Pauly, L. Packer, J. G. Rozen, Jr., R. Snelling, W. Wcislo, and D. Yanega. L. Packer, C. Plateaux-Quénu, K. Walker, and A. Ebmer provided crucial taxa, identifications, and additional help. S. Ji helped collect much of the DNA sequence data. Helpful comments on earlier drafts of this paper were provided by S. Brady, M. Caillaud, C. D. Michener, T. Seeley, S. Sipes, and two anonymous reviewers. This project was supported by National Science Foundation Grants DEB-9508647 and DEB-9815236.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database. All accession nos. can be found in the supporting information, which is published on the PNAS web site, www.pnas.org.
References
- 1.Batra S W T. Indian J Entomol. 1966;28:375–393. [Google Scholar]
- 2.Michener C D. Annu Rev Entomol. 1969;14:299–342. [Google Scholar]
- 3.Wilson E O. The Insect Societies. Cambridge, MA: Belknap; 1971. [Google Scholar]
- 4.Maynard Smith J, Szathmáry E. The Major Transitions in Evolution. Oxford: Oxford Univ. Press; 1997. pp. 255–278. [Google Scholar]
- 5.Bourke A F G, Franks N R. Social Evolution in the Ants. Princeton: Princeton Univ. Press; 1995. pp. 72–73. [Google Scholar]
- 6.Hölldobler B, Wilson E O. The Ants. Cambridge, MA: Harvard Univ. Press; 1990. [Google Scholar]
- 7.Emerson A E. Psyche. 1968;74:276–289. [Google Scholar]
- 8.Michener C D, Grimaldi D A. Proc Natl Acad Sci USA. 1988;85:6424–6426. doi: 10.1073/pnas.85.17.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross K G, Mathews R W, editors. The Social Behavior of Wasps. Ithaca, NY: Comstock; 1991. [Google Scholar]
- 10.Hunt J H. Evolution. 1999;53:225–237. doi: 10.1111/j.1558-5646.1999.tb05348.x. [DOI] [PubMed] [Google Scholar]
- 11.Seger J. In: Behavioural Ecology: An Evolutionary Approach. 3rd Ed. Krebs J R, Davies N B, editors. London: Blackwell Scientific; 1991. pp. 338–373. [Google Scholar]
- 12.Crespi B J. In: Phylogenies and the Comparative Method in Animal Behavior. Martins E P, editor. New York: Oxford Univ. Press; 1996. pp. 253–287. [Google Scholar]
- 13.Daly H V. J Kansas Entomol Soc. 1961;34:134–140. [Google Scholar]
- 14.Abrams J, Eickwort G C. Insectes Soc. 1981;28:105–116. [Google Scholar]
- 15.Michener C D. In: Social Insects: An Evolutionary Approach to Castes and Reproduction. Engels W, editor. New York: Springer; 1990. pp. 77–121. [DOI] [PubMed] [Google Scholar]
- 16.Wcislo W T. Ethology. 1997;103:1–11. [Google Scholar]
- 17.Michener C D. The Social Behavior of the Bees. Cambridge, MA: Belknap; 1974. [Google Scholar]
- 18.Sakagami S F, Munakata M. J Fac Sci Hokkaido Univ Ser 6 (Zool) 1972;18:411–439. [Google Scholar]
- 19.Eickwort G C. Fla Entomol. 1984;69:742–754. [Google Scholar]
- 20.Packer L. In: Queen Number and Sociality in Insects. Keller L, editor. Oxford: Oxford Univ. Press; 1993. pp. 214–233. [Google Scholar]
- 21.Wcislo W T, Danforth B N. Trends Ecol Evol. 1997;12:468–474. doi: 10.1016/s0169-5347(97)01198-1. [DOI] [PubMed] [Google Scholar]
- 22.Danforth B N, Ji S. Mol Biol Evol. 1998;15:225–235. doi: 10.1093/oxfordjournals.molbev.a025920. [DOI] [PubMed] [Google Scholar]
- 23.Danforth B N, Sauquet H, Packer L. Mol Phylo Evol. 1999;13:605–618. doi: 10.1006/mpev.1999.0670. [DOI] [PubMed] [Google Scholar]
- 24.Danforth B N. Syst Entomol. 1999;24:377–393. [Google Scholar]
- 25.Danforth B N, Ji S. Syst Biol. 2001;50:268–283. [PubMed] [Google Scholar]
- 26.Swofford D L. PAUP* Phylogenetic Analysis Using Parsimony (*and other Methods) Sunderland, MA: Sinauer; 1999. [Google Scholar]
- 27.Maddison W P, Maddison D R. MacClade. Sunderland, MA: Sinauer; 1992. , version 3.07. [Google Scholar]
- 28.Pesenko Y A. J Kansas Entomol Soc. 1999;72:104–123. [Google Scholar]
- 29.Danforth B N, Eickwort G C. In: The Evolution of Social Behavior in Insects and Arachnids. Crespi B J, Choe J C, editors. Cambridge, MA: Cambridge Univ. Press; 1997. pp. 270–292. [Google Scholar]
- 30.Engel M S. Bull Am Mus Natural History. 2000;250:1–89. [Google Scholar]
- 31.Michener C D. The Bees of the World. Baltimore: Johns Hopkins Univ. Press; 2000. [Google Scholar]
- 32.Richards M H. Insectes Soc. 1994;41:315–325. [Google Scholar]
- 33.Breed M D. Evolution. 1976;30:234–240. doi: 10.1111/j.1558-5646.1976.tb00906.x. [DOI] [PubMed] [Google Scholar]
- 34.Packer L, Knerer G. Behav Ecol Sociobiol. 1985;17:143–150. [Google Scholar]
- 35.Michener C D. In: Experimental Behavioral Ecology and Sociobiology. Hölldobler B, Lindauer M, editors. Sunderland, MA: Sinauer; 1985. pp. 293–306. [Google Scholar]
- 36.Kerfoot W B. Anim Behav. 1967;15:479–486. doi: 10.1016/0003-3472(67)90047-4. [DOI] [PubMed] [Google Scholar]
- 37.Yanega D. In: The Evolution of Social Behavior in Insects and Arachnids. Crespi B J, Choe J C, editors. Cambridge: Cambridge Univ. Press; 1997. pp. 293–315. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.