Abstract
Apc is a gene associated with familial adenomatous polyposis coli (FAP) and its inactivation is a critical step in colorectal tumor formation. The protein product, adenomatous polyposis coli (APC), acts to down-regulate intracellular levels of β-catenin, a key signal transducer in the Wnt signaling. Conditional targeting of Apc in the neural crest of mice caused massive apoptosis of cephalic and cardiac neural crest cells at about 11.5 days post coitum, resulting in craniofacial and cardiac anomalies at birth. Notably, the apoptotic cells localized in the regions where β-catenin had accumulated. In contrast to its role in colorectal epithelial cells, inactivation of APC leads to dysregulation of β-catenin/Wnt signaling with resultant apoptosis in certain tissues including neural crest cells.
The Apc gene encoding a tumor suppressor protein, adenomatous polyposis coli (APC), is known to be responsible for familial adenomatous polyposis coli (FAP), an autosomal-dominant inherited disorder characterized by multiple colorectal tumors (1, 2). Because Apc mutations are also frequently identified in sporadic colorectal tumors (3), the functional loss of APC is likely to be a key process in the colorectal tumorigenesis, especially during the early stages of tumor development. Indeed, conditional targeting of Apc in the colorectal epithelium initiated colorectal adenoma formation in mice (4).
The biological function of APC is not fully understood, however it is thought to be a multifunctional protein and plays an important role in the regulation of growth, differentiation, and/or migration of epithelial cells (5). Overexpression of APC can block cell cycle progression from G0/G1 to S phase in NIH 3T3 cells (6). APC concentrates near the ends of microtubules that extend into actively migrating regions of epithelial cell membrane (7), and recently it was suggested that APC might regulate the actin cytoskeleton through Asef, a novel APC-binding protein (8). Moreover, the nuclear export function of APC has been reported by Rosin-Arbesfeld et al. (9).
Accumulating evidence has indicated that APC acts as a negative regulator of the Wnt signaling cascade through down-regulation of β-catenin (10, 11). Functional loss of APC causes stabilization and accumulation of β-catenin (12), allowing it to form a complex with Tcf/Lef transcription factors to activate Tcf target genes (13–15). Mutant mice lacking certain Wnt functions exhibited abnormal phenotypes involving embryonic induction, cell polarity, and cell fate determination in various tissues during development (16–20). In particular, injection of mRNA for β-catenin or that encoding a mutant form of Tcf-3 into zebrafish embryos affected the Wnt signaling and resulted in abnormal development of lateral and medial neural crest cells (21).
To elucidate the function of APC in mammalian neural crest, in the present study we generated mutant mice harboring Apc disrupted specifically in the neural crest by using the Cre-loxP recombination system. We found that the mutant mice displayed severe craniofacial and cardiac defects due to massive apoptosis in the neural crest during development, highlighting the differential functions of APC in the neural crest and colorectal epithelial cells.
Materials and Methods
Genotyping.
Genotypes of mice or mouse embryos were determined by PCR of tail DNA or yolk sac DNA as described (4).
Skeleton Study.
Skin-removed and eviscerated embryos at 18.5 days post coitum (dpc) were fixed in 95% ethanol for 5 days and stained in Alcian Blue 8GX (Sigma) solution for 1 day. They were then washed in 95% ethanol, cleared in 1% KOH (potassium hydroxide), and counterstained in Alizarin Red (Sigma) in 2% KOH for 3 h, before being cleared in 2% KOH for 3 days, treated serially with 80:20, 60:40, 40:60, and 20:80 mixtures of 2% KOH and glycerol, and stored in the 20:80 mixture.
Histology and Immunohistochemistry.
The embryos were trimmed, fixed in Bouin, embedded in paraffin, and sectioned at 5 μm thickness. The sections were stained in hematoxylin/eosin. Schwann cells in peripheral nerves were counted under the microscope on four or five sections from each of three independent wild-type or mutant mice. The number of dorsal root ganglia (DRG) neurons was counted on six or more sections of DRG at the thoracic level from each mouse. For immunohistochemistry, embryos at 11.5 dpc were fixed in 4% paraformaldehyde/PBS at 4°C overnight. Fixed embryos were washed in PBS, incubated serially in 12, 15, and 18% sucrose, embedded in OCT compound, and sectioned at 7 μm thickness. The sections were washed in PBS-TN (0.01% Triton-X/0.02% sodium azide in PBS), blocked with 10% normal horse serum in PBS at room temperature for 1 h and then incubated with primary antibody at 4°C overnight. The primary antibodies used were anti-β-catenin mouse monoclonal Ab (1:50, Transduction Laboratories, Lexington, KY). The sections were again washed in PBS-TN and β-catenin immunoreactivities were visualized by incubation with secondary antibodies (FITC-labeled anti-mouse IgG1, 1:50) at room temperature for 1.5 h. For paraffin-embedded samples, embryos were fixed in 10% neutral buffered formalin, embedded in paraffin, and sectioned at 5 μm thickness. The sections were deparaffinized and epitope retrieval was performed by autoclave in 10 mM citrate buffer pH 6.0. After treating in hydrogen peroxide, the sections were incubated with primary antibody, visualized by VECTASTAIN Elite ABC kit (POD; Vector Laboratories), and counterstained in hematoxylin. The primary antibodies used were anti-β-catenin rabbit polyclonal Ab (1:1200, H102, Santa Cruz Biotechnology).
X-gal Staining.
Mouse embryos were fixed in 4% paraformaldehyde/PBS on ice for 2–3 h. They were washed in PBS, and stained in X-gal solution (0.1 M phosphate buffer/2 mM MgCl2/0.01% sodium deoxycholate/0.02% Nonidet P-40/5 mM potassium ferricyanide/5 mM potassium ferrocyanide/20 mM Tris⋅HCl/1 mg/ml X-gal) at 37°C overnight. After staining, some embryos were refixed and frozen in OCT compound, and cryostat sections (10 μm) were taken for microscopic analysis.
Terminal Deoxynucleotidyltransferase-Mediated dUTP Nick End Labeling (TUNEL).
An In Situ Cell Death Detection Kit (Fluorescence or POD; Boehringer Mannheim) was used in accordance with the manufacturer's instructions. TUNEL-positive cells were counted in five sections from each of three independent control or mutant embryos for the experiments shown in Fig. 3. For the experiments to elucidate p53 involvement on apoptosis, TUNEL-positive cells were counted within the region where β-catenin had accumulated from three sections of each of two independent embryos. TUNEL-positive cells in DRG were counted in eight or more sections from each mouse.
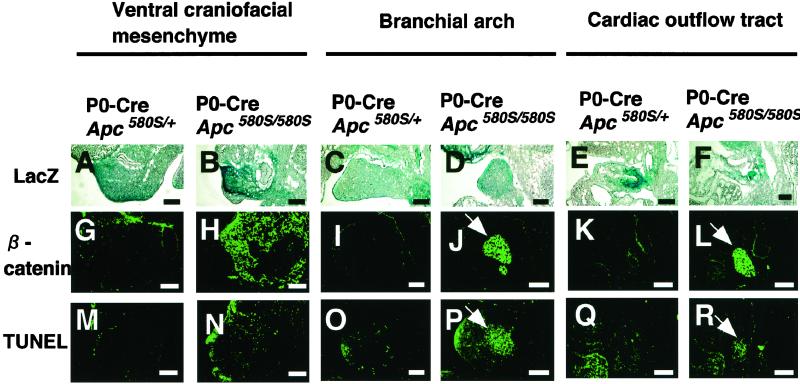
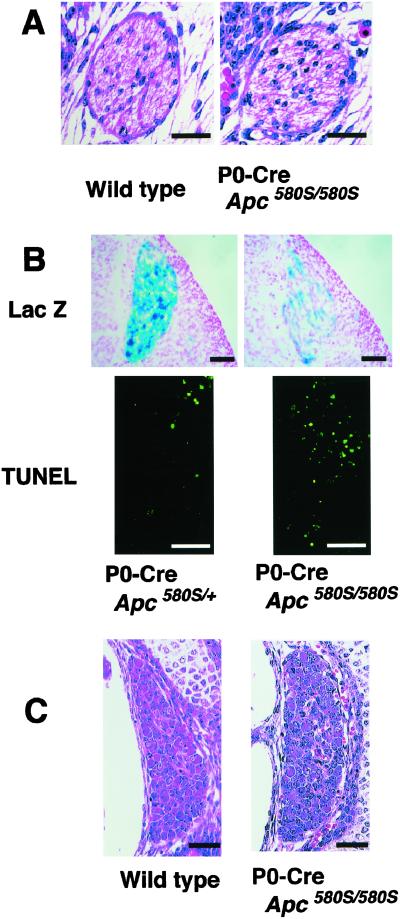
Figure 3.
Apoptosis and β-catenin accumulation in neural crest-derived tissues of mutant mice. Cre expression confirmed by LacZ staining (A–F), β-catenin immunoreactivity (G–L), and TUNEL stains (M–R) of the sections of ventral craniofacial mesenchyme (VCM), branchial arch (BA), and cardiac outflow tract (COT) from mutant and P0-Cre/Apc580S/+ embryos at 11.5 dpc are shown. Apoptotic cells are clustered in the branchial arch and cardiac outflow tract in one mutant (arrows in P and R). The number of apoptotic cells in VCM, BA, and COT of P0-Cre/Apc580S/+ mice was 32.4 ± 5.9, 6.2 ± 3.0, and 30.6 ± 19.1, respectively, and in mutants was 107.6 ± 15.8, >200, and 65.4 ± 21.4, respectively (mean ± SD, n = 3). β-catenin accumulated in TUNEL-positive cell-clustered regions (arrows in J and L). (Scale bars, 0.1 mm.)
In Situ Hybridization.
In situ hybridization was done as described (22).
Results and Discussion
To address the role of Apc in neural crest cell development, site-specific gene targeting was conducted using the Cre-loxP recombination system. For neural crest-specific ablation of Apc, Apc580S mice carrying a pair of loxP genes, inserted into introns flanking exon 14 of Apc (4), were mated with Protein 0 (P0)-Cre transgenic (Tg) mice that express Cre recombinase specifically in neural crest-derived cells (23). P0-Cre Tg mice harbors the Cre gene driven by the promoter of P0 glycoprotein, a cell adhesion molecule of the Ig superfamily. In these Tg lines, Cre is expressed in tissues derived from neural crest cells, such as spinal dorsal root ganglia, sympathetic nervous system, enteric nervous system, mesenchyme located within the outflow tract of the heart, and ventral craniofacial mesenchyme at stages later than 9.0 dpc (23). Cre expression in Apc580S mice results in the premature termination of the APC protein at codon 580 (4). P0-Cre/Apc580S/+ mice were born without any apparent abnormal phenotype. To generate Apc homozygous mutants with P0-Cre, P0-Cre/Apc580S/+ mice were crossed with Apc580S/+ mice carrying a CAG-CAT-Z transgene (24), in which loxP-flanked chloramphenicol acetyltransferase (CAT) gene is inserted between the ubiquitously active cytomegalovirus immediate early enhancer-chicken β-actin hybrid (CAG) promoter and a LacZ gene. Cre-mediated excision of CAT gene induces conditional activation of LacZ gene. This reporter system allows us to trace Cre expression and to identify cells or tissues where target gene is deleted in vivo.
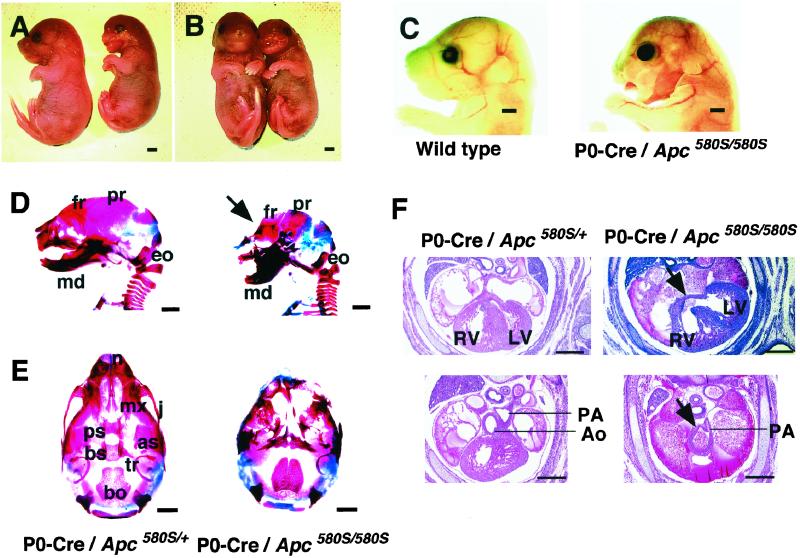
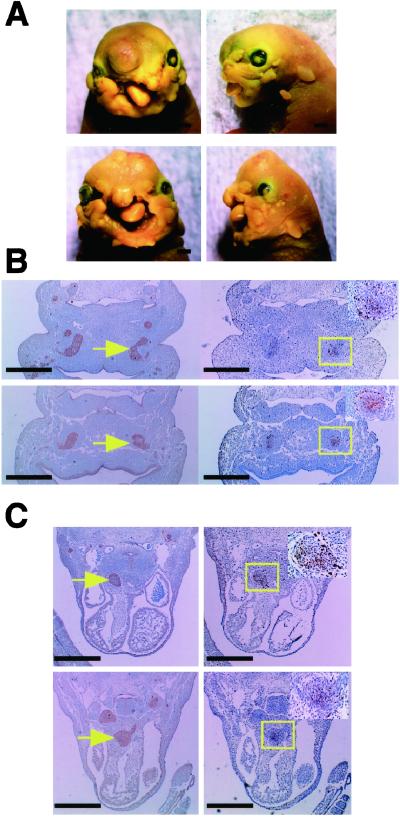
As shown in Table 1, no mutants (P0-Cre/Apc580S/580S) were found among 105 offspring at 4 weeks of age, irrespective of the presence of CAG-CAT-Z. When genotyping was carried out at postnatal day 0 (or 18.5 dpc), three mutants were identified among 51 offspring (5.9%). These mice exhibited severe craniofacial defects and died immediately after birth (Fig. 1 A–C). As shown in Fig. 1 A–C, the frontonasal region and both upper and lower jaws were severely malformed, and their tongues were prolapsed. Skeletal studies confirmed that the craniofacial defects were due to a disturbance in the achordal skeletogenesis (Fig. 1 D and E; refs. 25 and 26). Development of almost all of the bones derived from the cephalic neural crest was affected, to various degrees. The nasal, jugal, presphenoid, and frontal bones, in particular, were completely or partially missing. The remaining bones, including the maxillary, palatine, basisphenoid, alisphenoid, and mandibullar bones, and the tympanic ring, were significantly malformed but to a lesser degree (Fig. 1 D and E). Among the neural crest derivatives, only the parietal bone remained intact. The skeletal abnormalities were confirmed by histological analysis (data not shown). In contrast, skull bones of non-neural crest origin, such as the exoccipital and basioccipital bones, were not affected in the mutants (Fig. 1 D and E).
Table 1.
Genotype analysis of offspring of P0-Cre/Apc580S/+ male mice × CAG-CAT-Z/Apc580S/+ female mice
| Stage | Genotype
|
Total | |||||
|---|---|---|---|---|---|---|---|
| P0-Cre Apc+/+ | P0-Cre Apc580S/+ | P0-Cre Apc580S/580S | Apc+/+ | Apc580S/+ | Apc580S/580S | ||
| 11.5 dpc | 7 | 12 | 5 | 6 | 5 | 7 | 42 |
| 15.5 dpc | 6 | 9 | 3 | 7 | 6 | 5 | 36 |
| 18.5 dpc, post natal day 0 | 6 | 19 | 3 | 6 | 13 | 4 | 51 |
| 4 weeks | 13 | 28 | 0 | 15 | 33 | 16 | 105 |
Figure 1.
Craniofacial and cardiac defects in mutant mice (P0-Cre/Apc580S/580S). (A and B) Lateral (A) and frontal (B) views of a wild-type littermate (Left) and a mutant (Right) at birth. (C) Lateral views of faces of a wild-type littermate (Left) and a mutant (Right). (D) Lateral views of skeletal preparations of a P0-Cre/Apc580S/+ mouse (Left) and a mutant (Right) at 18.5 dpc. Part of the frontal bone is abnormal in mutants (arrow). fr, frontal; pr, parietal; eo, exoccipital; md, mandibullar bone. (E) Ventral views of skeletal preparations of a P0-Cre/Apc580S/+ mouse (Left) and a mutant (Right) at 18.5 dpc. n, nasal; mx, maxillary; j, jugal; ps, presphenoid; bs, basisphenoid; as, alisphenoid; tr, tympanic ring; bo, basioccipital bone. (F) Coronal sections of the trunk of P0-Cre/Apc580S/+ mice and mutants at 15.5 dpc. A ventricular septal defect (VSD) is shown in the heart of a mutant (arrow in Upper Right). The aorta and pulmonary artery are still adjoined and the pulmonary artery comes directly from the truncus arteriosus (arrow in Lower Right) in another mutant. RV, right ventricle; LV, left ventricle; Ao, aorta; PA, pulmonary artery. [Scale bars, 1 mm (A–E) and 0.5 mm (F).]
We next carried out a morphological examination of the hearts of the mutants at 15.5 dpc, because neural crest cells are known to populate the cardiac outflow tract and proximal great vessels (27, 28). As expected, Cre expression was detectable in mesenchyme located within the corresponding regions of P0-Cre mice (23). Five of the seven mutants exhibited ventricular septal defects (VSD) and one also had a persistent truncus arteriosus (PTA) (Fig. 1F). As with the cephalic defects, the cardiac anomalies were observed exclusively in the mutants, indicating that these phenotypes were due to the homozygous disruption of Apc in cephalic and cardiac neural crest cells.
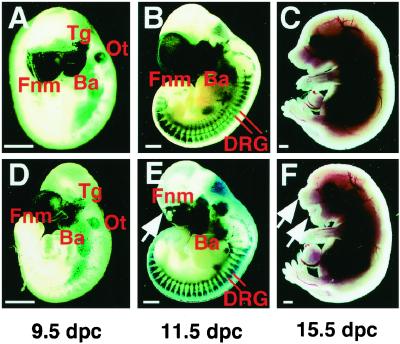
To analyze the process of neural crest development in the mutants, embryos at 9.5 and 11.5 dpc were stained with X-gal to trace Cre expression (Fig. 2 A, B, D, and E). At 9.5 dpc, Cre expression was detectable in the frontonasal mass, trigeminal ganglia, and branchial arches. The otocyst, a non-neural crest derivative, also was stained with X-gal, suggesting that there was leaky Cre expression in this cell type. However, there was no significant difference in the staining pattern between the mutants and their heterozygous littermates carrying P0-Cre at this stage (Fig. 2 A and D). The mutants became distinguishable at 11.5 dpc by the presence of defects in the frontonasal mass (indicated by arrow in Fig. 2E). Their heterozygous littermates carrying P0-Cre revealed intense X-gal staining in the corresponding region (Fig. 2B). Despite intense staining with X-gal, the development of the branchial arch at 11.5 dpc appeared to be normal, but at 15.5 dpc all mutants exhibited extensive defects in the craniofacial region derived from the first branchial arch including the lower jaw (indicated by arrow in Fig. 2F).
Figure 2.
Embryonal development of mutant and P0-Cre/Apc580S/+mice. Appearance of P0-Cre/Apc580S/+ embryos (A–C) and mutants (D–F) at various developmental stages indicated below the panels. Embryos at 9.5 and 11.5 dpc were stained with X-gal. In 11.5 dpc P0-Cre/Apc580S/+ embryos, frontonasal mass, trigeminal ganglia, branchial arches, and DRG were positive for LacZ. Fnm, frontonasal mass; Tg, trigeminal ganglia; Ba, branchial arch; Ot, otocyst; DRG, dorsal root ganglia. (Scale bars, 0.5 mm.)
The craniofacial and cardiac defects in the mutants could be due to impaired proliferation, retarded migration, and/or augmented apoptosis of neural crest cells. We evaluated the proliferation potential and apoptosis in cells of the ventral craniofacial mesenchyme, first branchial arch, and cardiac outflow tract in mutant embryos at 11.5 dpc. Levels of 5-bromo-2′-deoxyuridine (BrdUrd) incorporation in these tissues were indistinguishable between the mutants and their heterozygous littermates (data not shown), indicating that the growth potential of the cells was unlikely to be affected. In contrast, TUNEL staining revealed a significant increase in the number of apoptotic cells in the tissues of the mutants (Fig. 3 M–R). The first branchial arch and cardiac outflow tract in particular had clusters of TUNEL-positive cells (Fig. 3 P and R). In the ventral craniofacial mesenchyme, extensive apoptosis was detectable within the tissues derived from the cephalic neural crest, but not in non-neural crest derivatives such as skin and olfactory epithelium (Fig. 3N). LacZ staining confirmed Cre expression in the regions where apoptotic cells were present (Fig. 3 A–F). These findings strongly suggest a close link between functional loss of APC and apoptosis of neural crest cells, leading to the craniofacial and cardiac defects.
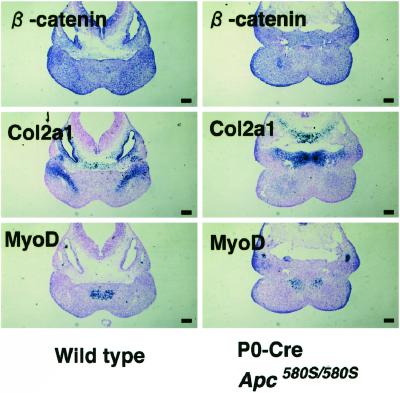
APC is known to control the Wnt signaling through down-regulation of β-catenin (5). To further elucidate mechanisms of apoptosis induced by Apc inactivation, we examined the expression of β-catenin in cephalic and cardiac neural crest cells of embryos at 11.5 dpc by immunohistochemistry (Fig. 3 G–L). Accumulation of β-catenin was evident in the ventral craniofacial mesenchyme, first branchial arch, and cardiac outflow tract of mutant embryos (Fig. 3 H, J, and L). In branchial arch and cardiac outflow tract, β-catenin was accumulated exactly in the region where TUNEL-positive cells were clustered (Fig. 3 J, L, P, and R). On the other hand, apoptotic cells were rather scattered in ventral craniofacial mesenchyme, but localized exclusively within the region where β-catenin were also accumulated (Fig. 3 H and N). No β-catenin accumulation was found in any tissues of the heterozygous littermates carrying P0-Cre (Fig. 3 G, I, and K). Because the expression of β-catenin mRNA was revealed to be ubiquitous throughout the branchial arch by in situ hybridization (Fig. 4), the loss of APC may result in the β-catenin accumulation at posttranslational levels in certain cells of the branchial arch. In a wild-type mouse embryo, neural crest-derived cells committed to bone and cartilage tissues, identifiable by the presence of collagen type 2a1 (Col 2a1) mRNA (29), were linearly distributed in the central area of the bilateral branchial arches (Fig. 4). In the mutants, however, Col 2a1-positive cells at the corresponding area of the branchial arch were missing and replaced by apoptotic cells. In contrast, the number and distribution of MyoD-positive cells (30), non-neural crest derivatives, remained unaffected on the medial region of the first branchial arch (Fig. 4). These data suggested that neural crest derived cells committed to bone and cartilage tissues might suffer apoptosis caused by the loss of Apc function.
Figure 4.
Collagen type 2a1-positive cells are lost and enter into apoptosis in the first branchial arch in the mutant mice. In situ hybridization for β-catenin, collagen type 2a1 (Col2a1), and MyoD on sections of the first branchial arch of wild-type (Left) and mutant mice (Right). (Scale bars, 0.1 mm.)
Neural crest cells are multipotent stem cells and give rise to a variety of other cell-types, including peripheral nerves, Schwann cells, and melanocytes (31). However, no defects other than the craniofacial and cardiac anomalies were observed in the mutants. As shown in Fig. 5A, Schwann cells also appeared to be histologically normal. In the DRG of mutant embryos at 11.5 dpc, despite strong expression of Cre, cells aligned normally and their gross appearance was indistinguishable from that of the heterozygous embryos (Fig. 2 B and E). Examination of DRG with TUNEL and X-gal staining showed that the number of apoptotic cells was increased and the number of Lac Z-positive cells was reduced in the mutants at 11.5 dpc compared with the heterozygous embryos (Fig. 5B). The number of DRG cells in the mutants, however, had reached a level equivalent to that of wild-type mice at 15.5–18.5 dpc (Fig. 5C), and so it is likely that APC functions to suppress apoptosis in DRG neurons during development, but its functional loss is not sufficient to result in neuronal anomalies in the mutants.
Figure 5.
Apoptosis and morphological appearance of Schwann cells and DRG in mutant mice. (A) Peripheral nerves in the legs of wild-type (Left) and mutant mice (Right) at 15.5 dpc. The number of Schwann cells in peripheral nerves at 18.5 dpc is 34.0 ± 11.5 in wild-type mice and 37.5 ± 1.9 in the mutants (mean ± SD, n = 3). (B) LacZ expression (Cre expression) and TUNEL in 11.5 dpc DRG. The number of LacZ-positive cells per DRG is 176.5 ± 23.9 in heterozygotes, and 50.1 ± 27.7 in the mutants (mean ± SD, n = 3). The number of apoptotic cells per DRG is 8.2 ± 5.6 in heterozygotes, and 19.9 ± 9.9 in mutants (mean ± SD, n = 3). (C) Whole views of DRG from 15.5 dpc wild-type (Left) and mutant mice (Right). The number of DRG neurons at 18.5 dpc is 116.7 ± 10.8 in wild-type and 105.8 ± 14.7 in the mutant mice (mean ± SD, n = 3). [Scale bars, 25 μm (A) and 50 μm (B and C).]
Damalas et al. (32) reported that overexpression of β-catenin resulted in the accumulation of a transcriptionally active p53 tumor suppressor protein, through interference with its proteolytic degradation. p53 is known to become activated in response to a variety of cellular stressors including DNA damage and hypoxia, and induces growth arrest and/or apoptosis (33). It is therefore conceivable that inactivation of Apc and the subsequent accumulation of β-catenin may qualify as a stress signal, triggering p53 activation, and leading to apoptosis in the cephalic and cardiac neural crest cells. To elucidate the involvement of p53 in the apoptosis observed in neural crest derived tissues, we first examined the expression of p53 protein. However, no p53 immunostaining was detected in branchial arch and cardiac outflow tract of mutants (data not shown). We next generated conditional mutants on p53 knock-out background (P0-Cre/Apc580S/580S/p53−/−) by crossing them with p53 mutants (34). These mice displayed similar phenotypes to mutants carrying wild-type p53 allele (Fig. 6 A–C) and the numbers of TUNEL-positive cells in neural crest-derived tissues of these mice were comparable to those of p53 heterozygous mutants (P0-Cre/Apc580S/580S/p53+/−). These data strongly indicate that the apoptosis of neural crest-derived cells induced by the loss of Apc was not mediated by p53.
Figure 6.
Effects of p53 deficiency on phenotypes of conditional mutants. (A) Frontal (Left) and lateral (Right) views of a P0-Cre/Apc580S/580S/p53+/+ (Upper) and a P0-Cre/Apc580S/580S/p53−/− (Lower) at birth. (B and C) β-catenin accumulation (Left) and TUNEL (Right) in branchial arch (B) and cardiac outflow tract (C) of mutant mice on p53 heterozygous (Upper) or homozygous (Lower) background. Yellow arrows indicate accumulation of β-catenin. (Insets) Magnified views of apoptotic lesions (yellow box). The number of TUNEL-positive cells within the region where β-catenin was accumulated in branchial arch is 48.4 ± 1.9 in P0-Cre/Apc580S/580S/p53+/− and 50.7 ± 4.7 in P0-Cre/Apc580S/580S/p53−/− (mean ± SD, n = 2, P < 0.05). In cardiac outflow tract, the number is 96.7 ± 1.9 in P0-Cre/Apc580S/580S/p53+/− and 76.2 ± 7.3 in P0-Cre/Apc580S/580S/p53−/− (mean ± SD, n = 2, P < 0.05). [Scale bars, 1 mm (A) and 0.25 mm (B and C).]
It is well known that accumulated β-catenin forms Tcf/Lef transcription factor complexes, resulting in the activation of Tcf/Lef target genes (13–15). Interestingly, Ahmed et al. (35) found neuronal apoptosis during retinal development in Drosophila, devoid of the Drosophila homolog of APC (D-APC) and either reduction of Armadillo (Arm) activity by replacing one wild-type copy of the Arm gene with null allele or dTCF mutation rescued neuronal apoptosis in the homozygous D-APC mutant (35). Because Arm and dTCF are Drosophila homologues of mammalian β-catenin and Tcf/Lef, respectively, it would thus be conceivable that unidentified Wnt-target genes directly activated by β-catenin/Tcf complexes are involved in the apoptosis in the Apc mutant mice.
Recent reports demonstrated that loss of APC caused chromosomal missegregation through β-catenin/Wnt-independent mechanisms (36, 37) and it is also possible that chromosomal missegregation may cause apoptosis (38, 39). Therefore, we cannot exclude an possibility that apoptosis described here might be independent from dysregulation of β-catenin/Wnt signaling. However, in the present study, apoptotic cells were observed exclusively in the regions where intracellular β-catenin had accumulated. It is thus most likely that β-catenin accumulation plays a crucial role in this apoptosis.
In this study, we demonstrated the evidence that APC inactivation leads to dysregulation of β-catenin/Wnt signaling with resultant apoptosis in certain tissues including neural crest. This contrasts with its role as a tumor suppressor in colorectal epithelial cells, and tissue-specific functions of APC may explain the colorectal epithelium-specific tumor spectrum in familial adenomatous polyposis coli (FAP) patients.
Acknowledgments
We thank N. Osumi for helpful discussion and J. Kuno and H. Yamanaka for technical assistance.
Abbreviations
- APC
adenomatous polyposis coli
- dpc
days post coitum
- DRG
dorsal root ganglia
- TUNEL
terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
- P0
Protein 0
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A, Koyama K, Utsunomiya J, Baba S, Hedge P. Science. 1991;253:665–669. doi: 10.1126/science.1651563. [DOI] [PubMed] [Google Scholar]
- 2.Joslyn G, Carlson M, Thliveris A, Albertsen H, Gelbert L, Samowitz W, Groden J, Stevens J, Spirio L, Robertson M, et al. Cell. 1991;66:601–613. doi: 10.1016/0092-8674(81)90022-2. [DOI] [PubMed] [Google Scholar]
- 3.Miyoshi Y, Nagase H, Ando H, Horii A, Ichii S, Nakatsuru S, Aoki T, Miki Y, Mori T, Nakamura Y. Hum Mol Genet. 1992;1:229–233. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- 4.Shibata H, Toyama K, Shioya H, Ito M, Hirota M, Hasegawa S, Matsumoto H, Takano H, Akiyama T, Toyoshima K, et al. Science. 1997;278:120–123. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- 5.Polakis P. Biochim Biophys Acta. 1997;1332:127–147. doi: 10.1016/s0304-419x(97)00008-5. [DOI] [PubMed] [Google Scholar]
- 6.Baeg G H, Matsumine A, Kuroda T, Bhattacharjee R N, Miyashiro I, Toyoshima K, Akiyama T. EMBO J. 1995;14:5618–5625. doi: 10.1002/j.1460-2075.1995.tb00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nathke I S, Adams C L, Polakis P, Sellin J H, Nelson W J. J Cell Biol. 1996;134:165–179. doi: 10.1083/jcb.134.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawasaki Y, Senda T, Ishidate T, Koyama R, Morishita T, Iwayama Y, Higuchi O, Akiyama T. Science. 2000;289:1194–1197. doi: 10.1126/science.289.5482.1194. [DOI] [PubMed] [Google Scholar]
- 9.Rosin-Arbesfeld R, Townsley F, Bienz M. Nature (London) 2000;406:1009–1012. doi: 10.1038/35023016. [DOI] [PubMed] [Google Scholar]
- 10.Cadigan K M, Nusse R. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 11.Clevers H, van de Wetering M. Trends Genet. 1997;13:485–489. doi: 10.1016/s0168-9525(97)01305-x. [DOI] [PubMed] [Google Scholar]
- 12.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 14.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich P, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 15.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 16.Takada S, Stark K L, Shea M J, Vassileva G, McMahon J P, McMahon A P. Genes Dev. 1994;8:174–189. doi: 10.1101/gad.8.2.174. [DOI] [PubMed] [Google Scholar]
- 17.Sterk K, Vainio S, Vassileva G, McMahon A P. Nature (London) 1994;372:679–683. doi: 10.1038/372679a0. [DOI] [PubMed] [Google Scholar]
- 18.Parr B A, McMahon A P. Nature (London) 1995;374:350–353. doi: 10.1038/374350a0. [DOI] [PubMed] [Google Scholar]
- 19.Monkley S J, Delaney S J, Pennisi D J, Christiansen J H, Wainwright B J. Development (Cambridge, UK) 1996;122:3343–3353. doi: 10.1242/dev.122.11.3343. [DOI] [PubMed] [Google Scholar]
- 20.Ikeya M, Lee S M K, Johnson J E, McMahon A P, Takada S. Nature (London) 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 21.Dorsky R I, Moon R T, Raible D W. Nature (London) 1998;396:370–372. doi: 10.1038/24620. [DOI] [PubMed] [Google Scholar]
- 22.Akazawa H, Komuro I, Sugitani Y, Yazaki Y, Nagai R, Noda T. Genes Cells. 2000;5:499–513. doi: 10.1046/j.1365-2443.2000.00339.x. [DOI] [PubMed] [Google Scholar]
- 23.Yamauchi Y, Abe K, Mantani A, Hitoshi Y, Suzuki M, Osuzu F, Kuratani S, Yamamura K. Dev Biol. 1999;212:191–203. doi: 10.1006/dbio.1999.9323. [DOI] [PubMed] [Google Scholar]
- 24.Sakai K, Miyazaki J. Biochem Biophys Res Commun. 1997;237:318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- 25.LeDouarin N M, Ziller C, Couly G F. Dev Biol. 1993;159:24–49. doi: 10.1006/dbio.1993.1219. [DOI] [PubMed] [Google Scholar]
- 26.LeDouarin N M, Dupin E, Ziller C. Curr Opin Genet Dev. 1994;4:685–695. doi: 10.1016/0959-437x(94)90135-p. [DOI] [PubMed] [Google Scholar]
- 27.Kirby K L, Waldo K L. Circ Res. 1995;77:211–215. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- 28.Olson E N, Strivastava D. Science. 1996;272:671–676. doi: 10.1126/science.272.5262.671. [DOI] [PubMed] [Google Scholar]
- 29.Alini M, Matsui Y, Dodge D R, Poole A R. Calcif Tissue Int. 1992;50:327–335. doi: 10.1007/BF00301630. [DOI] [PubMed] [Google Scholar]
- 30.Fujisawa-Sehara A, Nabeshima Y, Hosoda Y, Obinata T, Nabeshima Y. J Biol Chem. 1990;265:15219–15223. [PubMed] [Google Scholar]
- 31.Anderson D J. Trends Genet. 1997;13:276–280. doi: 10.1016/s0168-9525(97)01187-6. [DOI] [PubMed] [Google Scholar]
- 32.Damalas A, Ben-Ze'ev A, Simcha I, Shtutman M, Fernando J, Leal M, Zhurinsky J, Geiger B, Oren M. EMBO J. 1999;18:3054–3063. doi: 10.1093/emboj/18.11.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 34.Gondo Y, Nakamura K, Nakao K, Sasaoka T, Ito K, Kimura M, Katsuki M. Biochem Biophys Res Commun. 1994;202:830–837. doi: 10.1006/bbrc.1994.2005. [DOI] [PubMed] [Google Scholar]
- 35.Ahmed Y, Hayashi S, Levine A, Wieschaus E. Cell. 1998;93:1171–1182. doi: 10.1016/s0092-8674(00)81461-0. [DOI] [PubMed] [Google Scholar]
- 36.Kaplan K B, Burds A A, Swedlow J R, Bekir S S, Sorger P K, Nathke I B. Nat Cell Biol. 2001;3:429–432. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- 37.Fodde R, Kuipers J, Rosenberg C, Smits R, Kielman M, Gasper C, van Es J H, Breukel C, Wiegant J, Giles R H, et al. Nat Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]
- 38.Basu J, Bousbaa H, Logarinho E, Li Z, Williams B C, Lopes C, Sunkel C E, Goldberg M L. J Cell Biol. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobles M, Liberal V, Scott M L, Benezra R, Sorger P K. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]