Abstract
Targeting of the nuclear prostaglandin receptor peroxisome proliferator-activated receptor δ (PPARδ) by homologous recombination results in placental defects and frequent (>90%) midgestation lethality. Surviving PPARδ−/− mice exhibit a striking reduction in adiposity relative to wild-type levels. This effect is not reproduced in mice harboring an adipose tissue-specific deletion of PPARδ, and thus likely reflects peripheral PPARδ functions in systemic lipid metabolism. Finally, we observe that PPARδ is dispensable for polyp formation in the intestine and colon of APCmin mice, inconsistent with its recently proposed role in the establishment of colorectal tumors. Together, these observations reveal specific roles for PPARδ in embryo development and adipocyte physiology, but not cancer.
Nuclear hormone receptors are ligand-activated transcription factors that regulate multiple physiological processes, including reproduction, development, energy metabolism, and homeostasis (1). Within the nuclear receptor superfamily, peroxisome proliferator-activated receptors (PPAR) α, γ and δ/β comprise a subgroup of three closely homologous genes (2). PPARs have become a major pharmaceutical focus in recent years concomitant with the elucidation of the physiological functions of PPARα and PPARγ in lipid homeostasis and energy metabolism (3). PPARα, the most clinically relevant mediator of the pharmacological effects of peroxisome proliferators, is a transcription factor dedicated to eliminating excess fatty acids by way of catabolism, including the stimulation of hepatic peroxisomes and fatty acid oxidases (4). PPARγ was implicated as a key regulator of adipogenesis (5, 6), as well as in aspects of lipid uptake and efflux in adipocytes and macrophages (7, 8). These two well-studied PPARs regulate lipid homeostasis by nonoverlapping mechanisms: catabolism vs. mobilization. In addition, PPARγ is a high-affinity receptor for the thiazolidinedione class of insulin sensitizers (9, 10), linking lipid metabolism to type II diabetes. The emergence of PPARγ as a central differentiation factor in additional cell types, such as the placental trophoblast (6), further broadens the array of physiological and developmental functions controlled by the PPARs.
Despite a rapid increase in our understanding of PPARα and γ, the identification of PPARδ functions has been lagging behind. Recent studies with a synthetic agonist demonstrated that PPARδ can induce reverse cholesterol transport and rectify lipoprotein profiles and triglyceride levels in obese Rhesus monkeys (11), placing it alongside the remaining PPARs in the regulation of lipid metabolism. More debatable is whether PPARδ is a potential regulator of adipocyte differentiation (12, 13). Apart from metabolism, PPARδ was proposed to be a critical mediator of embryo implantation (14), based on its spatial and temporal expression patterns, and the demonstration that the process requires the naturally occurring PPARδ agonists PGI2 and cPGI (15). Finally, PPARδ was ascribed an oncogenic function after being identified as a direct transcriptional target of β-catenin and as a repression target of the nonsteroidal antiinflammatory drug sulindac, a potent suppressor of colorectal tumors (16). These collective observations implicate PPARδ as a versatile regulator of distinct biological processes including, and extending beyond, lipid metabolism.
We report here the generation of a genetic loss-of-function model for PPARδ, which we use to test existing hypotheses about its function, as well as to identify additional ones. We find that PPARδ has an essential role in placentation, such that its deficiency results in frequent embryonic lethality. Surviving PPARδ null mice, while rare, are generally healthy and fertile, but display more than a 60% reduction in adipose mass. However, this compromise in adiposity is not recapitulated by an adipocyte-specific knockout of PPARδ, suggesting that the phenotype is fat-nonautonomous and may evolve from systemic metabolic perturbations. In addition, we report that PPARδ is not essential for polyp formation in the gut of APCmin mice, although it may have a modest quantitative effect on their growth, reflected in a preferential decrease in the abundance of large polyps in hemizygous and PPARδ null animals. These observations refine and add to the previously proposed roles of the receptor, placing PPARδ alongside the other PPARs, especially PPARγ, in regulating versatile processes, such as placentation, fat homeostasis, and colorectal cancer.
Materials and Methods
PPARδ Gene Targeting.
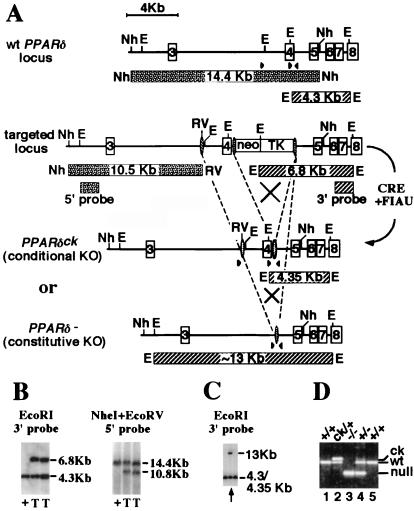
The exon encoding the N-terminal half of the DNA-binding domain of PPARδ was targeted by the CRE-lox methodology (17). A loxP sequence was inserted into the upstream intron, while a neor-TK cassette flanked by two additional loxP sites was introduced into the downstream intron (Fig. 1A). Southern blot analysis of embryonic stem cells transfected with the targeting allele and selected with G418 revealed frequent homologous integration (≈40%) into the PPARδ locus (Fig. 1B). Two correctly targeted embryonic stem clones were subsequently transfected with a cre-recombinase expressing plasmid (18), followed by negative selection with 1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil (FIAU) for daughter clones that lost the neo-TK cassette. One of 48 clones screened (arrow in Fig. 1C) contained cells that had lost the entire segment between the two extreme loxP sites, generating a constitutive loss-of-function allele (PPARδ−). PCR analysis indicated that this clone also harbored a subpopulation of cells in which only the neo-TK cassette was deleted, yielding a conditional knockout allele, where the two loxP sites flanking the targeted exon are intact, amenable to further CRE-mediated deletion (PPARδck; Fig. 1D). Germ-line chimeras derived from this clone transmitted either the PPARδ− or the PPARδck allele to their progeny, and the two allelic pools were maintained separately thereafter (Fig. 1D).
Figure 1.
PPARδ targeting strategy. (A) (Top to Bottom) The wt, primary targeted allele and the two secondary, CRE-induced recombinant alleles of PPARδ [the conditional knockout allele (PPARδck) and the constitutively null one (PPARδ−)] (see Materials and Methods for details). Expected DNA fragments and their sizes are drawn as patterned bars under the respective genomic structures. Arrowheads indicate approximate location of PCR oligos. Restriction sites are: E, EcoRI; Nh, NheI; RV, EcoRV. (B) Southern blot analysis of homologous integration of the primary targeting construct shows the appropriate genomic alterations both 5′ and 3′ to the homologous recombination site in two of the targeted embryonic stem clones (T), as opposed to wt cells (+). (C) Southern blot analysis of daughter clones after CRE-mediated recombination and 1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil selection of the primary targeted embryonic stem cells. Arrow indicates clone A4, which contains a mixture of cells carrying either the constitutive knockout allele (13-kb fragment) or the PPARδck allele (see D). (D) PCR analysis of mice carrying various PPARδ allele combinations. Bottom band (≈240 bp), null allele; middle band (≈360 bp), wt; top band (≈400 bp), the PPARδck allele. Deduced genotypes are indicated on top.
Genotyping was performed by using a three-oligonucleotide combination as follows: GAGCCGCCTCTCGCCATCCTTTCAG (common, 3′ to the downstream loxP site); GGCGTGGGGATTTGCCTGCTTCA [wild type (wt)-specific, 5′ to the downstream loxP site]; and GGCTGGGTCACAAGAGCTATTGTCTC (null-specific, 5′ to the upstream loxP site). Genomic tail DNA is amplified through 35 cycles of: 94°C at 20 s; 60°C at 30 s; 71.5°C at 70 s. Reaction products of ≈400, ≈360, and ≈240 bp represent the PPARδck, wt, and PPARδ− alleles, respectively.
Induction and Evaluation of Adipocyte-Specific Floxed Allele Recombination.
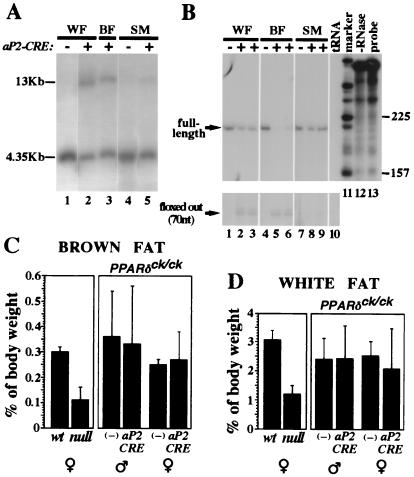
The generation of aP2-CRE transgenic mice and their characterization will be described elsewhere (W.H., Y.B., J.M.O., and R.M.E., unpublished work). These mice express CRE abundantly in brown and white adipose tissue and only marginally in other tissues, such as skeletal muscle, liver, and heart, as judged by Northern blot analysis and a functional test using R26R–ROSA mice (19). The aP2-CRE transgene has been introduced into a PPARδck/ck background, and recombination of the PPARδck allele was assessed in white and brown adipose tissue, as well as skeletal muscle, by Southern blot analysis (See Fig. 4). RNase protection assays to assess the frequency of PPARδ mRNA truncation were performed as described (20), except for the hybridization step, which was carried out by using RPA II hybridization solution (Ambion, Austin, TX).
Figure 4.
Adipocyte-specific PPARδ knockout does not affect adipose tissue mass. (A) Southern blot analysis of epidydimal white fat pad (WF), interscapular brown fat pad (BF), and thigh skeletal muscle (SM) from mice carrying the PPARδck/ck allele and an aP2-CRE transgene. A 4.35-kb EcoRI fragment represents the nonexcised PPARδck allele, whereas CRE-mediated recombination yields a 13-kb EcoRI fragment (see Fig. 1 and Materials and Methods for further detail). Notice that CRE-dependent conversion into the 13-kb band is substantial in white and brown fat (lanes 2 and 3), and residual in skeletal muscle (lane 5). (B) RNase protection analysis of PPARδ transcript structure in the same mice. CRE-mediated deletion modifies a 210-nt-long protected fragment representing the full-length transcript (see arrow) into a 70-nt-long fragment representing a functionally null transcript devoid of its fourth exon (arrow, Bottom). The assay reveals ≈50% aP2CRE-mediated deletion of the full-length PPARδ mRNA in white fat (lanes 2 and 3 vs. lane 1), >80% in brown fat (lanes 5 and 6 vs. lane 4) and only a marginal one in skeletal muscle (lanes 8 and 9 vs. lane 7). Specificity controls with yeast tRNA and no RNase are shown in lanes 10 and 12, respectively. (C and D) Relative weights of interscapular brown fat pads (C) and epidydimal white fat pads (D) in wt and PPARδ null mice, and in PPARδck/ck mice in the absence (−) or presence of the aP2-CRE transgene. The 2.5- to 3-fold differences in adipose mass between wt and null mice (Left) are not recapitulated by an adipose-specific gene knockout (aP2-CRE vs. −, Right).
Intestinal Polyp Analysis.
Mice were killed after a 15-h fast, and their guts were flushed with 10% formalin for cleaning and initiating tissue fixation. The colons and intestines were subsequently opened along the mesentery and left in formalin until microscopic evaluation. Specimens were stained by dipping in hematoxylin solution and extensive rinsing with PBS, enhancing the contrast between normal and hyperplastic epithelia. Individual polyps were scored and measured by using a stereo microscope (Leica MZ8), with a calibrated eyepiece. Measurements were performed in a double-blind fashion and sampled by a second observer to avoid bias. Standard deviations in matched cohorts were calculated according to Student's t test.
Histology.
Tissue specimens (skin, placenta, polyps) were fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Thin sections were subject to standard hematoxylin and eosin staining. Histological evaluation of intestinal polyps was performed visually by R.B.
Results
Gene Targeting of PPARδ.
We chose a CRE/lox-mediated recombination strategy to disrupt the PPARδ locus (Fig. 1). LoxP sites plus a floxed neo-TK cassette were introduced on both sides of the exon encoding the 5′ half of the DNA-binding domain through homologous recombination. Correctly targeted clones were then transfected with a CRE expression vector (18) and negatively selected against thymidine kinase activity. The procedure yielded two distinct germ-line alleles: a conditional knockout allele (PPARδck), which leaves all PPARδ exons intact and is amenable to CRE-mediated deletion; and a loss-of-function allele (PPARδ−), which excised the exon encoding the DNA-binding domain and resulted in a frameshift of the remainder of the mRNA product. These two alleles were maintained independently after germ-line transmission (see Fig. 1D).
Placental Defects and Frequent Embryonic Lethality in PPARδ Null Mice.
Mice homozygous for the PPARδck allele were obtained from heterozygous crosses at the expected Mendelian frequency (≈25%; data not shown) and exhibited no discernible phenotype. In contrast, homozygous loss of PPARδ caused frequent embryonic lethality, as homozygous null pups were rarely obtained (Table 1). Surviving PPARδ-deficient progeny were markedly runt at term, but typically overcame growth retardation by puberty, although most were still somewhat smaller than their PPARδ-sufficient counterparts. None died postnatally, suggesting that the essential function of the receptor is restricted to the gestational period. Both male and female mutants were fertile.
Table 1.
PPARδ null mouse survival chart
| Stage | No. litters | +/+ | +/− | −/− | Resorbed |
|---|---|---|---|---|---|
| +/− × +/− | |||||
| P21 | 22 | 92 | 185 | 8* (4.5%) | NA |
| E11.5 | 1 | 3 | 3 | 0 | 1 |
| E10.5 | 2 | 4 | 9 | 3 (1†) | 2 |
| E9.5 | 2 | 4 | 12 | 3 | 2 |
| −/− × +/− | |||||
| P21 | 11 | 43 | 6‡ (12%) | NA | |
| E14.5 | 1 | 7 | 1 (7.5%) | 5 | |
| E12.5 | 2 | 9 | 4 (21%) | 6 | |
| E11.5 | 3 | 13 | 4 (15%) | 10 | |
| E10.5 | 4 | 23 | 17 (40%) | 2 | |
| E9.5 | 1 | 5 | 3 (38%) | 0 |
NA, not applicable.
Three of the nulls were born in the same litter.
Dead embryo.
Four of the nulls were born in the same litter.
Survival of PPARδ-deficient mice was relatively even-spread, although not entirely random, and litters with multiple null pups were observed on two different occasions (see Table 1). However, subsequent progeny of the same breeding pairs did not similarly exhibit increased survival. In addition, up to six back-crosses of the original knockout stock (129sv/Jae) against isogenic C57BL/6J breeders did not improve survival rates (data not shown). These observations fail to establish a clear heritable component influencing the survival of PPARδ null mice.
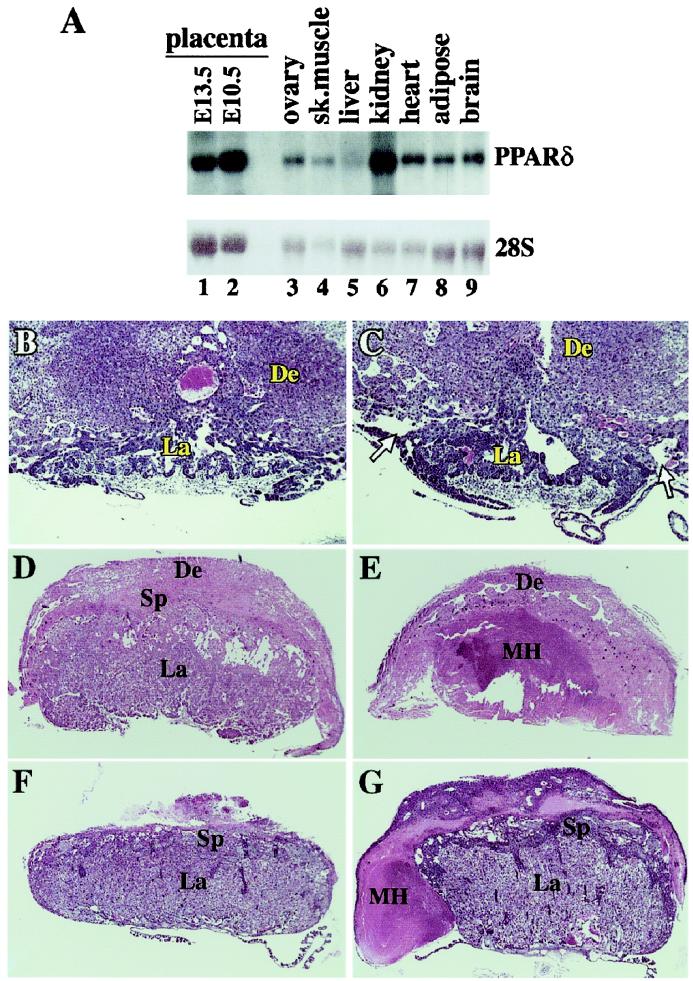
Embryonic lethality and sub-Mendelian ratios of PPARδ null embryos were observed from embryonic day 10.5 (E10.5) onward (Table 1). Mutants surviving beyond that stage were typically smaller than their wt and heterozygous siblings (data not shown). The combination of midterm death and growth restriction pointed to the possibility of defects in extraembryonic tissue. This conjecture was supported by the abundant placental expression of PPARδ (Fig. 2A, lanes 1 and 2), and the fact that null mouse mortality was strictly prenatal.
Figure 2.
Pathologies of PPARδ null placentas. (A) Mouse PPARδ tissue blot. Note the high PPARδ levels in two different stages of placental development (E10.5, E13.5), superior to most tissues, except kidney. (B and C) wt and PPARδ null placentas at E9.5. Arrows in C denote the abnormal detachment of the PPARδ null placenta from the decidua. (D and E) wt and PPARδ null placentas at E12.5. The inner core of the mutant placenta contains a massive maternal hematoma (MH) and is completely devoid of trophoblast cells. (F and G) wt and PPARδ null placentas at E14.5. The mutant placenta is surrounded by a maternal hematoma (MH). De, decidua; Sp, spongiotrophoblast layer; La, placental labyrinth; Th, thrombus. (Magnifications: B and C: ×20; D and E: ×11; F and G: ×7.)
Histological examination of PPARδ null concepti at E9.5, a day before the onset of lethality, revealed that the connections between their placentas and the maternal deciduas are abnormally loose (arrows in Fig. 2C; compare with Fig. 2B). The placental labyrinth, albeit smaller, exhibited a fully differentiated vascular structure, further distinguishing this defect from the one seen in PPARγ null placentas. By E10.5, most PPARδ−/− specimens could not be retrieved without significant detachment of the placenta from the decidua (data not shown), which implies further loosening of the placento-decidual contact upon widening of the gap. By E12.5, three of four PPARδ−/− embryos surviving the major E10.5 lethality point exhibited extensive maternal hemorrhages into the labyrinthine zone (Fig. 2E, MH). Finally, a rare PPARδ null survivor recovered at E14.5 exhibited an attenuated form of the maternal hematoma, in which the thrombus surrounded, rather than infiltrated the labyrinth (compare Fig. 2 E and F). The survival of this embryo to E14.5 suggests a link between the severity of the placental phenotype and embryonic lethality.
PPARδ Deficiency Decreases Adipose Mass.
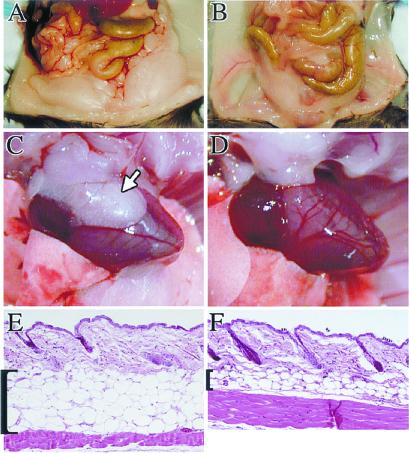
Examination of surviving PPARδ null mice revealed an extremely lean phenotype (Fig. 3 A and B), typified by a 2.5-fold reduction of abdominal fat mass compared with control littermates (e.g., 1.20 ± 0.29% of carcass weight, n = 4, vs. 3.07 ± 0.33%, n = 3, respectively, in females, P < 0.0005; see Fig. 4D). Likewise, the relative mass of interscapular brown fat stores dropped by ≈70% (0.1 ± 0.03% in the null vs. 0.29 ± 0.02% in wt, Fig. 4C; photo not shown). Mesenteric fat, as well as adipose stores associated with internal organs, such as the heart (Fig. 3C, arrow) and kidney, were essentially undeveloped in 4-month-old null animals (Fig. 3D). Microscopic examination revealed no consistent differences between white or brown adipocytes from mutant vs. wt animals (data not shown), suggesting that reduced adiposity may arise from differences in cell number but not size. In contrast, the subcutaneous fat layer, which was also more than 2-fold thinner in null animals, exhibited a combined decrease in both cell number and size (Fig. 3 E and F). Thus, PPARδ deficiency affects all adipose types, although the manifestation of the effect is depot-specific.
Figure 3.
Compromised adipose stores in PPARδ mutants. (A and B) Abdominal fat depots of wt (A) and PPARδ null mice (B), exhibiting a substantial compromise in the amount of fat tissue in a null mouse. (C and D) At 4 months the pericardial white fat depots are routinely found in wt mice (arrow in C) but absent from PPARδ null ones (D). (E and F) Hematoxylin and eosin-stained paraffin sections of skin from the lower back of wt (E) and PPARδ null mice (F). The subcutaneous fat layer is dramatically shrunk in the mutant, reflecting a combined effect of reduced adipocyte number and size. (Magnifications: E and F: ×15.)
PPARδ expression is ubiquitous and its levels in adipose tissue compare with those elsewhere (Fig. 2A, lane 8). Therefore, we wondered whether hypoadiposity of PPARδ null mice reflected the loss of an adipocyte function of the receptor or rather a nonautonomous systemic PPARδ function. To distinguish between these possibilities, we generated mice carrying two copies of the floxed PPARδck allele and an adipose-specific CRE-recombinase transgene, driven by the promoter of the aP2 gene (aP2-CRE, ref. 21; W.H., Y.B., J.M.O., and R.M.E., unpublished work). Genomic DNA and mRNA analyses showed that aP2-CRE deleted ≈50% of PPARδ in the gonadal white fat pad, and >80% of the gene was lost in the interscapular brown fat pad (Fig. 4 A and B). Recombination was adipocyte-specific, as evidenced by its marginal incidence in skeletal muscle (Fig. 4 A and B). Thus, if reduced adiposity was caused by adipocyte-intrinsic functions of PPARδ, its extent in PPARδck/ck/aP2-CRE mice should be proportional to PPARδ loss in this tissue.
Comparison of both epidydimal white and interscapular brown fat pads from 4-month-old PPARδck/ck;TgaP2-CRE/0 mice to those of control PPARδck/ck;Tg0/0 animals revealed no differences between the two cohorts (Fig. 4 C and D). This observation was incompatible with an adipocyte-autonomous effect of PPARδ on adiposity, suggesting that PPARδ controls the process in a systemic fashion. This interpretation ascribes to PPARδ a putative function in lipid homeostasis alongside PPARα and γ.
PPARδ Is Dispensable for Colorectal Polyp Formation.
PPARδ has been recently implicated as a direct target and a potential oncogenic effector of β-catenin in colorectal carcinogenesis (16, 22). To test this hypothesis, we introduced the targeted PPARδ allele into the APCmin mouse strain. This strain is heterozygous for a mutation in the apc (adenomatous polyposis coli) gene, whose tumor suppressor product inhibits β-catenin and its growth-promoting action (23, 24). Upon loss of heterozygosity for apc in cells of the gastrointestinal mucosa, β-catenin activity is deregulated, yielding multiple intestinal neoplasias (min). If PPARδ is a critical transducer of the tumorigenic β-catenin signal, then its loss should substantially reduce, if not eliminate, intestinal polyps in min mice.
Through an extensive breeding effort we were able to obtain three viable PPARδ−/−; APCmin females. Polyp status was assessed in these mice at 4 months, alongside that of matched wt and PPARδ+/− controls. Most importantly, PPARδ null mice harbored conspicuous intestinal and colonic polyps, showing unequivocally that the receptor is not required for polyp formation.
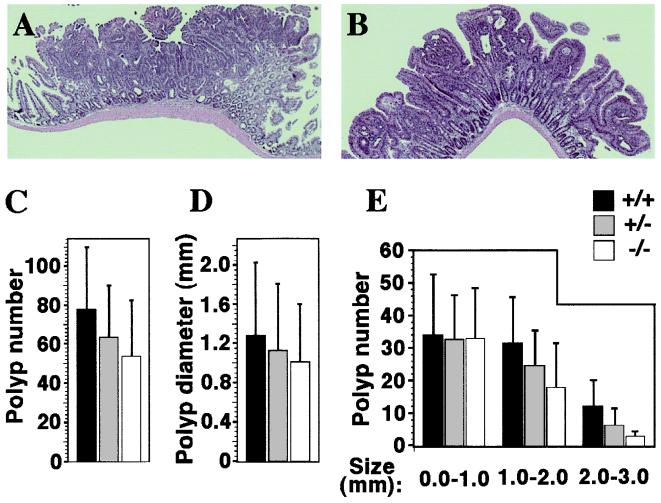
We next turned to histological and quantitative comparisons of polyps between wt, heterozygous, and null animals. We concentrated on polyps in the small intestine, because differences in the number and size of colonic polyps could not be reliably assessed in this small cohort because of their typical small number. Histological evaluation of 12 intestinal polyps from PPARδ+/+ and nine from PPARδ−/−; APCmin mice identified all of them as low-grade, noninvasive tubular adenomas (Fig. 5 A and B), excluding major effects of PPARδ on the cellular phenotype of the tumor. The average number of intestinal polyps was not significantly different between PPARδ+/+, PPARδ+/−, and PPARδ−/−; APCmin mice (Fig. 5C). Similarly, loss of PPARδ did not elicit a statistically significant change in the median size of intestinal polyps (Fig. 5D). Detailed size distribution analysis revealed a decrease in the abundance of large polyps (>1.0 mm diameter) upon PPARδ gene dosage reduction (Fig. 5E), which was further pronounced at the largest polyp size category (>2.0 mm diameter; Fig. 5E). In contrast, the number of small polyps (<1.0 mm in diameter) was essentially identical in all PPARδ genotype groups. Although below statistical significance in our small cohort, this trend could indicate that polyps with varying degrees of PPARδ gene knockout grow slower and are therefore less likely to attain a large size. Thus, PPARδ is qualitatively dispensable for the tumorigenic process, although we cannot rule out the possibility that it influences the pace of polyp growth.
Figure 5.
Intestinal polyp analysis in PPARδ-deficient APCmin mice. Histology of representative polyps from wt (A) and PPARδ−/− (B) APCmin mutants. Both adenomas display a similar, benign, tubular phenotype, regardless of the genetic status of PPARδ. (Magnification: ×16.) (C) Average polyp numbers in APCmin females carrying PPARδ+/+ (black bars; 77.6 ± 32.3), PPARδ+/− (gray bars; 63.6 ± 26.4), and PPARδ−/− genotypes (white bars; 54.0 ± 28.7). Differences between these values are statistically insignificant. (D) Median polyp diameters in PPARδ+/+ (1.28 ± 0.75 mm), PPARδ+/− (1.13 ± 0.68 mm), and PPARδ−/− (1.01 ± 0.59 mm). Differences between these values are statistically insignificant. (E) Intestinal polyp size distribution. Polyps in each of the three PPARδ genotypes were classified into three size ranges (0–1.0 mm; 1.0–2.0 mm; 2.0–3.0 mm). Note that wt PPARδ allele dosage is directly related to the incidence of larger polyps (>1.0 mm). However, differences are still statistically insignificant according to Student's t test.
Discussion
PPARδ Function During Embryonic Development.
We report here that PPARδ deficiency is lethal to over 90% of embryos. This observation is similar to an earlier study using a different PPARδ targeting configuration (25), with two critical differences. First, the lethal phenotype of our knockout variant appears to occur earlier during gestation (E10.5 vs. E18.5). Second, in contrast to Peters et al. (25), we identified neither a heritable component nor a specific genetic background that alleviates this lethality. The difference between the two mutant strains may arise from the different targeting strategies. Whereas our knockout configuration eliminates almost the entire PPARδ gene product (see Fig. 1 and Materials and Methods), the one generated by Peters et al. only truncates the C-terminal 60 aa. Conceivably, this limited deletion may fail to abolish PPARδ activity in its entirety, because from a structural perspective, the truncated receptor should retain DNA-binding activity, and possibly heterodimerization with retinoid-X receptor. Such residual functions may suffice for increased embryonic survival and a higher proportion of live births.
PPARδ has been implicated as a mediator of prostacyclin cPGI and PGI2 functions in embryo implantation (14). However, the uncompromised fertility of PPARδ null females, and the proper Mendelian distribution of PPARδ−/− postimplantation embryos up to E9.5, do not support this contention. Rather, they unambiguously demonstrate that implantation can proceed in the complete absence of either maternal or embryonic PPARδ. Thus, either PPARδ is not the exclusive molecular target of prostacyclins in this process, or prostacyclins are essential only when the receptor is around. The latter situation is theoretically possible, if unliganded PPARδ actively represses implantation, with prostacyclins providing a temporal cue to relieve this inhibition; such derepression would become dispensable if PPARδ was missing altogether.
We show here that PPARδ is essential for placentation. PPARδ−/− embryos start dying in parallel to the appearance of an abnormal gap in the placento-decidual interface. Most survivors of the first wave of mortality succumb to subsequent maternal hemorrhages. Conceivably, to survive to parturition, embryos have to evade these structural mishaps to some extent. This seems to be the case with the embryo shown in Fig. 2G, which at E14.5 exhibited a thrombus that had not invaded the placental labyrinth. This phenotypic variant suggests that embryonic survival may reflect an incomplete penetrance of the placental defects.
Placentation is a complex tissue remodeling process, balancing trophoblast invasion and differentiation, decidual response, proteolysis of the extracellular matrix (ECM), and vascular development (26). The histological appearance of PPARδ−/− placentas shares striking similarities with those of compound keratin deficiencies (27, 28). This finding raises the possibility that breakdown of the placento-decidual interface in PPARδ null concepti may reflect an imbalance in ECM remodeling, caused by either compromised matrix build-up or excessive proteolysis.
PPARδ and Adiposity.
The closest homologues of PPARδ, namely PPARα and PPARγ, are both established regulators of lipid homeostasis (3). It is therefore reasonable to assume that PPARδ should have a related function. This idea is supported in the broadest sense by the ability of PPARδ to bind and moderately respond to many of the fatty acids that also activate PPARα and γ (2, 15). In addition, PPARδ can up-regulate ABCA1 expression and cholesterol efflux in multiple cell types (11), and the expression of liver fatty-acid binding protein in the gut (29). Furthermore, administration of a synthetic PPARδ-selective agonist to primates with adult-onset obesity restores their high density lipoprotein cholesterol levels along with a parallel decrease in plasma triglycerides (11). In obese-diabetic db/db mice, PPARδ agonists induce a modest increase in total cholesterol and a significant reduction in adipocyte lipoprotein lipase levels (30). These studies provide compelling evidence that, in the adult, PPARδ acts within the context of lipid and lipoprotein metabolism.
In support of this notion, we report here that PPARδ null mice display a dramatic reduction in adiposity. This effect is registered uniformly in all types of fat tissue, including gonadal, mesenteric, brown, and subcutaneous stores, all of which exhibit an ≈3-fold decrease.
Strikingly, we find that this effect of PPARδ deficiency is adipocyte-nonautonomous and cannot be reconstructed by adipocyte-specific PPARδ deficiency. Thus, reduced adiposity of PPARδ null mice reflects a response of the tissue to an exogenous stimulus, rather than an intrinsic function of the receptor within the fat cell. However, an examination of systemic lipid and lipoprotein profiles did not detect significant changes in the levels of either total cholesterol, high density lipoprotein cholesterol, triglycerides, or free fatty acids in the plasma of fasted PPARδ null mice, relative to control littermates (data not shown). These results suggest that PPARδ might not impact basal lipid homeostasis, and therefore that its contribution to adiposity is either through an unrelated route, or during active phases of the feeding cycle.
PPARδ and Colorectal Cancer.
PPARδ was recently implicated as a direct transcriptional target of β-catenin and a critical, sulindac-sensitive factor in the development of gastrointestinal neoplasias (16). This hypothesis was further reinforced by the demonstration that colon cancer cells in which PPARδ has been knocked out fail to form tumors in nude mice, where their wt counterparts readily thrive (22). However, we observe here that PPARδ is clearly dispensable for both the formation and elaboration of intestinal and colonic tumors. We did not measure the potential effect of PPARδ-specific stimuli, such as pharmacological or dietary activation, which may impact tumor progression in the presence of PPARδ.
While clearly nonessential, PPARδ may emerge from our quantitative analysis as a potential modifier of intestinal adenomas. We observed a selective and gradual reduction in the number of larger polyps upon loss of each wt PPARδ allele. This trend may provide an indication that PPARδ contributes quantitatively toward maximal polyp growth, such that in its absence fewer polyps exceed a diameter of 1 mm.
Conclusion.
We describe here the phenotypic analysis of complete and tissue-specific PPARδ null mice. These genetic platforms reveal that PPARδ acts at two temporally distinct phases. First, during early development the receptor regulates placentation and is consequently essential for the survival of most embryos. Second, in adult mice it comprises a nonautonomous determinant of adiposity, providing a plausible link to lipid metabolism, and pointing to PPARδ as a potential drug target candidate in the treatment of metabolic disorders. Finally, our analyses warrant reassessment of two previously proposed functions of this receptor: embryo implantation and colon cancer. We observe that PPARδ is broadly dispensable for both processes. However, we cannot exclude that it may fine-tune these events in conjunction with certain pharmacological or dietary stimuli.
This study fits nicely with the notion that the PPAR family comprises a triad of related receptors controlling lipid homeostasis, among other functions. Intriguingly, placental pathologies further broaden the functional links of PPARδ to PPARγ, which in addition to its established role in adipose tissue, regulates trophoblast differentiation (6). However, unlike PPARγ mutants, defects in PPARδ null placentas disrupt the placental-decidual interface and do not affect differentiation of the labyrinthine trophoblast, clearly distinguishing between the placental functions of either PPAR. Thus, while conceivably related, PPAR functions are nevertheless nonredundant.
Acknowledgments
We thank B. Dominguiez for blastocyst injection, M. Lieberman for whole animal photography, M. Lawrence for expert histology, and E. Stevens for administrative assistance. Y.B. was supported by a European Molecular Biology Organization long-term fellowship and the Charles and Anna Stern Foundation. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute for Biological Sciences and March of Dimes Chair in Molecular and Developmental Biology. This work was supported by the Howard Hughes Medical Institute.
Abbreviations
- APC
adenomatous polyposis coli
- PPAR
peroxisome proliferator-activated receptor
- wt
wild type
- E(n)
embryonic day
- min
multiple intestinal neoplasias
References
- 1.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kliewer S A, Forman B M, Blumberg B, Ong E S, Borgmeyer U, Mangelsdorf D J, Umesono K, Evans R M. Proc Natl Acad Sci USA. 1994;91:7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kliewer S A, Xu H E, Lambert M H, Willson T M. Recent Prog Horm Res. 2001;56:239–263. doi: 10.1210/rp.56.1.239. [DOI] [PubMed] [Google Scholar]
- 4.Pineda Torra I, Gervois P, Staels B. Curr Opin Lipidol. 1999;10:151–159. doi: 10.1097/00041433-199904000-00009. [DOI] [PubMed] [Google Scholar]
- 5.Tontonoz P, Hu E, Spiegelman B M. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 6.Barak Y, Nelson M C, Ong E S, Jones Y Z, Ruiz-Lozano P, Chien K R, Koder A, Evans R M. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 7.Tontonoz P, Nagy L, Alvarez J G, Thomazy V A, Evans R M. Cell. 1998;93:241–252. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- 8.Chawla A, Boisvert W A, Lee C H, Laffitte B A, Barak Y, Joseph S B, Liao D, Nagy L, Edwards P A, Curtiss L K, et al. Mol Cell. 2001;7:161–171. doi: 10.1016/s1097-2765(01)00164-2. [DOI] [PubMed] [Google Scholar]
- 9.Lehmann J M, Moore L B, Smith-Oliver T A, Wilkison W O, Willson T M, Kliewer S A. J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 10.Forman B M, Tontonoz P, Chen J, Brun R P, Spiegelman B M, Evans R M. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 11.Oliver W R, Jr, Shenk J L, Snaith M R, Russell C S, Plunket K D, Bodkin N L, Lewis M C, Winegar D A, Sznaidman M L, Lambert M H, et al. Proc Natl Acad Sci USA. 2001;98:5306–5311. doi: 10.1073/pnas.091021198. . (First Published April 17, 2001; 10.1073/pnas.091021198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bastie C, Holst D, Gaillard D, Jehl-Pietri C, Grimaldi P A. J Biol Chem. 1999;274:21920–21922. doi: 10.1074/jbc.274.31.21920. [DOI] [PubMed] [Google Scholar]
- 13.Brun R P, Tontonoz P, Forman B M, Ellis R, Chen J, Evans R M, Spiegelman B M. Genes Dev. 1996;10:974–984. doi: 10.1101/gad.10.8.974. [DOI] [PubMed] [Google Scholar]
- 14.Lim H, Gupta R A, Ma W G, Paria B C, Moller D E, Morrow J D, DuBois R N, Trzaskos J M, Dey S K. Genes Dev. 1999;13:1561–1574. doi: 10.1101/gad.13.12.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forman B M, Chen J, Evans R M. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He T C, Chan T A, Vogelstein B, Kinzler K W. Cell. 1999;99:335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauer B. Methods. 1998;14:381–392. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- 18.O'Gorman S, Dagenais N A, Qian M, Marchuk Y. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soriano P. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 20.Barak Y, Gottlieb E, Juven-Gershon T, Oren M. Genes Dev. 1994;8:1739–1749. doi: 10.1101/gad.8.15.1739. [DOI] [PubMed] [Google Scholar]
- 21.Ross S R, Graves R A, Spiegelman B M. Genes Dev. 1993;7:1318–1324. doi: 10.1101/gad.7.7b.1318. [DOI] [PubMed] [Google Scholar]
- 22.Park B H, Vogelstein B, Kinzler K W. Proc Natl Acad Sci USA. 2001;98:2598–2603. doi: 10.1073/pnas.051630998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su L K, Kinzler K W, Vogelstein B, Preisinger A C, Moser A R, Luongo C, Gould K A, Dove W F. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 24.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 25.Peters J M, Lee S S, Li W, Ward J M, Gavrilova O, Everett C, Reitman M L, Hudson L D, Gonzalez F J. Mol Cell Biol. 2000;20:5119–5128. doi: 10.1128/mcb.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cross J C, Werb Z, Fisher S J. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- 27.Tamai Y, Ishikawa T, Bosl M R, Mori M, Nozaki M, Baribault H, Oshima R G, Taketo M M. J Cell Biol. 2000;151:563–572. doi: 10.1083/jcb.151.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hesse M, Franz T, Tamai Y, Taketo M M, Magin T M. EMBO J. 2000;19:5060–5070. doi: 10.1093/emboj/19.19.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirier H, Niot I, Monnot M C, Braissant O, Meunier-Durmort C, Costet P, Pineau T, Wahli W, Willson T M, Besnard P. Biochem J. 2001;355:481–488. doi: 10.1042/0264-6021:3550481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leibowitz M D, Fievet C, Hennuyer N, Peinado-Onsurbe J, Duez H, Bergera J, Cullinan C A, Sparrow C P, Baffic J, Berger G D, et al. FEBS Lett. 2000;473:333–336. doi: 10.1016/s0014-5793(00)01554-4. [DOI] [PubMed] [Google Scholar]