Abstract
Heritable predisposition to inflammatory bowel disease (IBD) has been demonstrated by epidemiological and genetic analysis. Linkage of IBD to broad regions of chromosome 16 has been established by analysis of multiple populations. NOD2, located on proximal 16q, was recently identified as an IBD gene. As the linkage regions on chromosome 16 are large, we have investigated the possibility that NOD2 is not the only IBD gene located on this chromosome. A high-density experiment using 39 microsatellite markers was performed to identify additional regions of association, and to indicate areas of interest for further investigation. A triple-peaked configuration of the linkage curve with peak logarithm of odds (lod) scores of 2.7, 3.2, and 3.1 was observed on proximal 16p, proximal 16q, and central 16q, respectively. The cohort was stratified by coding individuals carrying the NOD2 single nucleotide polymorphism (SNP)8 and SNP13 “unknown.” Significance at the central peak, corresponding to the genomic location of NOD2, then decreased from 3.2 to 1.2. The maximal lod scores on the proximal p-arm (lod = 2.1) and central q-arm (lod = 2.6) changed only moderately. An exploratory association analysis (TRANSMIT) yielded a strong lead at D16S3068 (P = 0.00028). The region around this marker was further investigated by using anonymous SNPs. An associated haplotype containing three SNPs was identified (peak significance P = 0.00027, IBD phenotype). On stratification based on NOD2 genotype, this significance increased to P = 0.0001. These results confirm the importance of NOD2 and provide evidence for a second IBD gene located on chromosome 16p.
Inflammatory bowel disease [IBD; Mendelian Inheritance in Man (MIM 601458, 266600, 191390)] affects ≈1 of 1,000 individuals in Western countries and has median age of onset in early adulthood (1, 2). The disease is clinically characterized by abdominal pain, chronic diarrhea, rectal bleeding, weight loss, intestinal stenoses, fistulae (3, 4), toxic megacolon, and associated extraintestinal manifestations (5) such as arthritis and uveitis (6). On the basis of clinical and histopathological features, IBD is categorized into two main subtypes, Crohn's disease (CD; MIM 266600) and ulcerative colitis (UC; MIM 191390) (6, 7).
A genetic component in the etiology of IBD has been clearly demonstrated by epidemiological and genetic linkage studies. Epidemiological investigations have consistently shown familial clustering (8) and an increased concordance of the IBD phenotype in monozygotic twins (9, 10). The IBD1 susceptibility region on chromosome 16 was identified by a genomewide linkage analysis (11) and replicated in several independent populations (12–14), including the international IBD consortium (15). It is thus the most widely and consistently replicated region of linkage in IBD. Recently, mutations in the leucine-rich region of the NOD2 gene have been identified as susceptibility factors for CD in a total of four different populations (16–18). The identification of the NOD2 gene as described by Hugot et al. (16) demonstrates the possibility of a systematic positional cloning approach starting from a high-density microsatellite map.
Multipoint linkage analysis of chromosome 16 in our cohort has identified three broad, and only partially overlapping, areas of linkage (Fig. 1). To explain this observation, we postulated that additional IBD genes may be located on chromosome 16 leading to a complex multipoint linkage curve. This hypothesis is supported by the differing locations of linkage peaks in the studies that have detected linkage to chromosome 16 (11–15, 19, 20). We therefore performed a two-phase genetic analysis consisting of a high-density microsatellite mapping experiment of the chromosome 16p linkage region coupled with an association analysis of indicated areas by using genomic single-nucleotide polymorphisms (SNPs). Linkage analyses were performed on both the study population and a stratified set where the phenotypes of individuals with the previously reported NOD2 mutations were removed from the analysis. We report here genetic evidence suggesting that a second IBD gene is located on the proximal region of chromosome 16p.
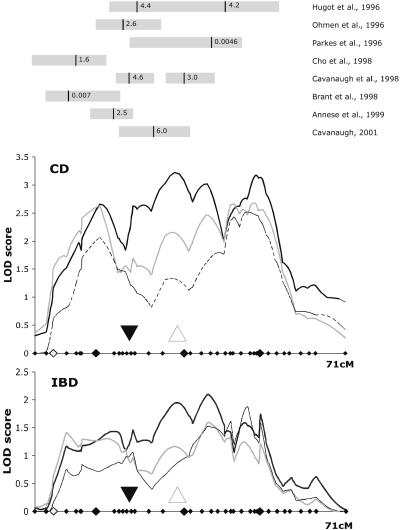
Figure 1.
(Top) The approximate locations of the linkage peaks obtained in other positive studies on chromosome 16 (six of the studies have been cited in the text; Ohmen et al. is ref. 39; Parkes et al. is ref. 40). The respective lod scores or P values are given adjacent to the dark bar that indicates a peak location. The gray bars show the approximate location of the intervals with (lodmax = −1). An analysis of chromosome 16 microsatellite linkage to CD (Middle) and IBD (Bottom) is presented. The thick solid line represents the unstratified analysis. The thick gray line and the dashed line represent linkage analyses after the phenotypes of all carriers of the NOD2/SNP13 and NOD2/SNP8 and SNP13 mutation were set to “unknown,” respectively. ▵, The position of the NOD2 gene; ▾, the position of the centromere. The three markers (D16S3145, D16S415, and D16S3106) with the highest MLSs in the CD analysis are marked with large filled diamonds; all other marker positions are marked with small filled diamonds; and ⋄ indicates the position of the locus D16S3068. Linkage analysis was performed by using mapmaker/sibs (as integrated in genehunter 2). cM, centimorgan.
Materials and Methods
Family Ascertainment and Phenotypes.
A family cohort recruited by an international group of IBD investigators at the Charité University Hospital (Berlin), the Department of General Internal Medicine at the Christian-Albrechts-Universtität (Kiel, Germany), and St. Mark's, Guy's, and King's College Hospitals (London), and other European centers as detailed in Acknowledgments was investigated in this study. Informed, written consent was obtained from all study participants. Recruitment protocols were approved by ethics committees at participating centers before commencement of the study. The cohort has been used in previous studies within the collaborative group (12, 21–23). Ascertainment criteria were determined before the initiation of patient collection. Clinical, radiological, and endoscopic (type of lesions, distribution) examinations were required to unequivocally confirm the diagnosis of either UC or CD (24, 25). Histological findings also had to be confirmative or compatible with this diagnosis. In case of uncertainty, the diagnosis of indeterminate colitis was assigned, and the patient was excluded from the study. A venous blood sample was obtained from the affected siblings and their parents, if possible. An overview of the investigated cohort is given in Table 1. A basic cohort of 428 affected sibling pairs was used for initial exploration. For the association analysis in the vicinity of D16S3068, this cohort was extended to 522 affected sibling pairs.
Table 1.
Family cohorts used in the basic study and extended cohort
| Sibship size | Basic
cohort
|
Extended cohort
|
||||||
|---|---|---|---|---|---|---|---|---|
| CD | UC | Mixed (CD/UC) | Total | CD | UC | Mixed (CD/UC) | Total | |
| 2 | 148 (148) | 100 (100) | 48 (48) | 296 (296) | 184 (184) | 116 (116) | 66 (66) | 366 (366) |
| 3 | 14 (42) | 14 (42) | 10 (30) | 38 (114) | 19 (57) | 13 (39) | 14 (42) | 46 (138) |
| 4 | 1 (6) | — | 2 (12) | 3 (18) | 1 (6) | — | 2 (12) | 3 (18) |
| All | 163 (196) | 114 (142) | 60 (90) | 337 (428) | 204 (247) | 129 (155) | 82 (120) | 415 (522) |
Affected siblings pairs are ordered according to sibship size and disease type (mixed refers to sibships with both UC and CD) within the family. The number of sibships and affected sib pairs is given in parentheses in each category. From the extended cohort, 212 families were of German or Dutch descent and 203 were of British descent.
Genotyping.
Genomic DNA was prepared from whole blood by using the Puregene system (Gentra Systems, Minneapolis). Thirty-eight highly polymorphic microsatellite markers on chromosome 16 were genotyped by using fluorescent methods as described (26). Primer sequences were derived from the Genome Data Base (http://www.gdb.org) or the literature (27). In brief, individual DNA samples were arrayed in 96-well microtiter plates and amplified by PCR with the appropriate primers. Product length of microsatellite markers was determined by electrophoresis on denaturing polyacrylamide gels on Applied Biosystems 377 automated DNA sequencers. Allele analyses and individual allele calling were performed as described (26, 28).
SNPs were genotyped by using the TAQMAN technology from Applied Biosystems. This method utilizes the 5′-exonuclease activity of the Taq DNA polymerase in a combination of PCR and competitive hybridization (29, 30). Oligonucleotides and reaction conditions are given in Table 2. PCRs were performed in ABI9700 and Biometra T1 (Göttingen, Germany) thermocyclers, and fluorescence results were determined by using ABI7700 and ABI7900 sequence detectors. The data were managed and checked for Mendelian inheritance errors in an integrated database (31).
Table 2.
Results of the exploratory association analysis using microsatellite markers in the broad chromosome 16 linkage region
| Marker | IBD phenotype | CD phenotype |
|---|---|---|
| D16S420 | 0.042084 | 0.108538 |
| D16S401 | 0.207753 | 0.197653 |
| D16S3133 | 0.130597 | 0.055268 |
| D16S3068 | 0.000281 | 0.007916 |
| D16S3116 | 0.202301 | 0.265602 |
| D16S3131 | 0.064864 | 0.246106 |
| D16S296 | 0.472402 | 0.12474 |
| D16S3100 | 0.328345 | 0.183747 |
| D16S3093 | 0.174147 | 0.364688 |
| D16S3145 | 0.165404 | 0.222713 |
| D16S298 | 0.049384 | 0.127025 |
| D16S3022 | 0.017771 | 0.041568 |
| SPN | 0.146076 | 0.074623 |
| D16S409 | 0.101376 | 0.067997 |
| D16S411 | 0.072485 | 0.228119 |
| D16S261 | 0.36953 | 0.384983 |
| D16S416 | 0.209683 | 0.523343 |
| D16S415 | 0.182533 | 0.033321 |
| D16S3032 | 0.012341 | 0.478398 |
| D16S408 | 0.256492 | 0.398304 |
| D16S3057 | 0.011381 | 0.003971 |
| D16S3089 | 0.315063 | 0.03513 |
| D16S3132 | 0.014381 | 0.564171 |
| D16S514 | 0.131528 | 0.072258 |
| D16S3143 | 0.037327 | 0.756801 |
| D16S503 | 0.104004 | 0.060076 |
| D16S3019 | 0.006044 | 0.03252 |
| D16S496 | 0.028046 | 0.03326 |
| D16S3095 | 0.197467 | 0.49872 |
| D16S3106 | 0.038197 | 0.066823 |
| D16S3066 | 0.376165 | 0.136382 |
| D16S3115 | 0.175036 | 0.074596 |
| D16S515 | 0.162787 | 0.013197 |
| D16S3125 | 0.03327 | 0.466642 |
| D16S518 | 0.036965 | 0.119995 |
| D16S3049 | 0.017347 | 0.137686 |
| D16S516 | 0.097475 | 0.307015 |
| D16S511 | 0.045429 | 0.003309 |
| D16S520 | 0.042084 | 0.108538 |
Markers that have been used in the genome scan are marked in italics. For each marker, nominal P values obtained from the transmit analysis are given. Significances for the marker D16S3068 that was selected for further analysis are given in bold print.
Genomic Sequencing.
Library construction from the bacterial artificial clone (BAC) clone, shotgun sequencing, and assembly of the BAC sequence were performed according to standard protocols as described (32).
Mutation Detection.
SNPs were established by genomic resequencing with primers based on the database sequences (see Results). Sequences were masked by using REPEATMASKER (http://repeatmasker.genome.washington.edu) before primer design. The BigDye chemistry (Applied Biosystems) was used according to the manufacturer's recommendations and analyzed on ABI310 and ABI3700 automated sequencers.
Statistical Analysis.
Genetic analyses were conducted by using the two standard diagnostic categories CD and UC as described. The combined IBD phenotype represented all affected individuals with either UC or CD as a unified phenotype for analysis.
To facilitate inclusion of markers from different genetic maps (e.g., CEPH and CHLC), marker order and distances were calculated by using genotype data from the 337 pedigrees of the basic cohort. The genetic map was constructed by using the automated mapping program MULTIMAP v. 2.0 (33). Linkage analyses were performed by using the lod statistics of MAPMAKER/SIBS as implemented in GENEHUNTER 2. This analysis estimates the allele sharing in affected sibling pairs and compares it to the sharing distribution expected under H0 (no linkage). Family-based linkage and association statistics were calculated by using the TRANSMIT program (34, 35) with the robust variance estimator option. TRANSMIT implements a likelihood-based transmission disequilibrium test (TDT) test that is specifically designed to be applicable in general pedigree data. Significance measurements were verified within TRANSMIT using 10,000 bootstrap samples. Intermarker disequilibrium was estimated by using the program EH (36). EH implements the estimation maximization algorithm for determination of haplotypes from unrelated individuals.
Results
Microsatellite Saturation Experiment.
Thirty-nine microsatellite markers, spanning the interval from D16S420 to D16S520, were genotyped in a basic cohort of 428 affected sibling pairs with IBD. The average marker density was one marker every 1.8 centimorgan. Linkage analyses using the categories of IBD, CD, and UC were performed. A peak multipoint lod score (MLS) of 3.2 was observed near the marker D16S415 for CD disease category. Overall, a triple-peaked configuration of the linkage curve in the CD phenotype was observed with additional peaks near D16S3145 on 16p (MLS 2.7) and near D16S496 more telomeric on 16q (MLS 3.1). The MLS curve for the CD phenotype is depicted in Fig. 1 Middle. For the combined IBD phenotype, an MLS of 2.1 was observed at the marker D16S3057 (Fig. 1 Bottom). The UC analysis yielded maximum MLS values of 0.6 and 0.5 on the q and p arms, respectively (graph not shown).
Stratification on NOD2 Genotypes.
Three SNPs (SNP8, SNP12, and SNP13, using the nomenclature from Hugot et al.) that were independently associated with CD in the paper by Hugot et al. (16) were genotyped in the basic cohort. As shown in previous studies (16–18), two of these mutations were significantly associated with the CD phenotype on TRANSMIT TDT analysis (P = 0.00013 and 0.0001 for SNP8 and SNP13, respectively) in the present family sample. SNP12 yielded only marginal significance with a P value of 0.014. On analysis in the group with the IBD phenotype, only SNP13 yielded a significant result at the 1% significance level (P = 0.04, 0.19 and 0.0005 for SNP8, SNP12, and SNP13, respectively). The three SNPs were not in significant linkage disequilibrium (LD) (χ2 < 2.5, df = 3). As SNP8 and SNP13 contribute significantly (at the P = 0.01 level) to the CD risk in this cohort, we stratified the cohort in a two-stage process by removing the phenotypes of all individuals carrying a mutation at SNP13 and SNP8. There is some evidence for pairwise LD between SNP8 and SNP13 (χ2 = 7.2, df = 2, P = 0.03). The removal of phenotypes for carriers of either mutation is therefore conservative with regard to stratification. For this stratification, the phenotypes of all individuals carrying the respective mutations were coded as “unknown” in the linkage files. As a result of this stratification, 166 phenotypes in the SNP13 and 370 phenotypes in the combined stratification cohort were removed from the analysis.
The results of the linkage analysis in the stratification cohorts are shown in Fig. 1. The evidence for linkage drops steeply at the location of the NOD2 gene (from lod 3.2 to 1.2 in CD). The two peaks on the proximal p-arm and the more distal q-arm change moderately: the peak lod score drops from 2.7 to 2.1 on the proximal p arm and from 3.1 to 2.6 on the q arm in the CD phenotype. Interestingly, a minor increase of significance on the q-arm peak from 1.7 to 1.9 is observed in the IBD phenotype.
Exploratory Association Analysis.
An exploratory association analysis on the microsatellite genotypes was performed in the CD and IBD phenotype groups by using TRANSMIT in the basic cohort. Table 2 shows the nominal P values from this exploratory analysis for all markers. The most significant result was obtained at marker D16S3068 with a P value of 0.00028 for the IBD and 0.0079 for the CD phenotype. The genomic vicinity of this marker was therefore selected for further investigation.
Investigation of the Association Lead at D16S3068.
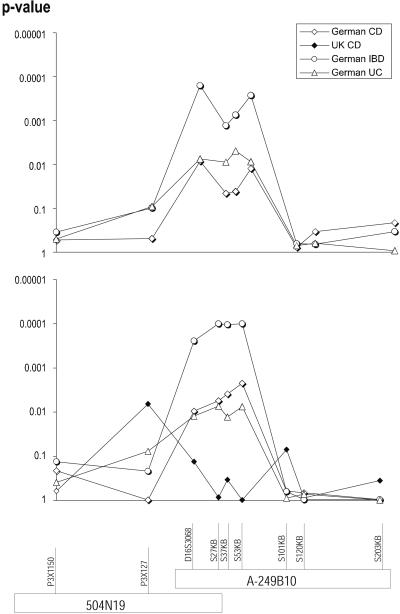
The microsatellite marker D16S3068 is located on the BAC CIT-A-249B10 (GenBank accession no. AC002288). Because D16S3068 is located 3 kb from the end of this BAC, the CIT-B (California Instiute of Technology Human BAC library B: CIT978SK) library was screened by using PCR pools. Several positive BACs were identified. When PCR mapping was used, the BAC clone covering the most additional genomic distance was identified as 504N19 and sequenced (GenBank accession no. AF265340). SNPs were identified on both BACs by using direct genomic resequencing in 24 affected individuals with IBD. Eight previously undiscovered SNPs were identified at nucleotide positions 27 kb, 37 kb, 53 kb, 101 kb, 120 kb, and 203 kb on A-249B10 (coordinates based on GenBank sequence AC002288) and at positions 61824 (P3X 127), 99363 (P3X 1150) on 504N19. Positions and primers used for genotyping are given in Table 3. These SNPs and the microsatellite D16S3068 were genotyped in the extended cohort. The analysis was performed in a population-specific manner, separating the United Kingdom and the Germany/Netherlands part of the study cohort (see Table 1). The strongest evidence of association was observed for SNPs 27 kb, 37 kb, and 53 kb for the IBD phenotype in the German part of the cohort. Among the SNPs analyzed, a peak nominal single point TRANSMIT result of P = 0.00027 was observed for the A-249B10-53 kb SNP (444 observed versus 413 expected transmissions). On analysis of the subphenotypes, UC yielded higher significances (maximum significance P = 0.0049 at 37 kb) than CD in German patients (maximum significance P = 0.012 at 53 kb). The P values obtained in the German population are shown in Fig. 2 Upper. All results in the United Kingdom cohort were insignificant with a peak P value of 0.018 in the CD phenotype for A-249B10/101 kb (data not shown).
Table 3.
SNP markers analyzed in the study
| SNP | Nucleotide position | Primers | Probes |
|---|---|---|---|
| 504N19-P3X127 | 61824 | F: TTTTTGTACATCTTAATCTTGCG | FAM: CCTTCAGGACAAAAcGATTATTGTAAATCTAATACTT |
| R: AAATCCCTTTAGAAAAGGTAATCA | TET: CCTTCAGGACAAAAtGATTATTGTAAATCTAATACTTT | ||
| 504N19-P3X1150 | 99363 | F: AGCAGGGAGATACA | FAM: AAAGAATCATGTATTTaAATTTTCAGTTAATTCAAATCTGT |
| R: CTTTACTCATTTGTTGGCCTATTTGAAGT | TET: AAAAGAATCATGTATTTgAATTTTCAGTTAATTCAAATCT | ||
| A-249B10-27kb | 27461 | F: CATTTTCCTGTCATTGAGTAT | FAM: TTCCTGTTTATTTATTATcATTTTATGGAGAGTGTTGGTACAGAAA |
| R: TTGACTGGTTGATCACATAAATTCATTTTAGACTTT | TET: TTCCTGTTTATTTATTATgATTTTATGGAGAGTGTTGGTACAGAAA | ||
| A-249B10-37kb | 37391 | F: CTGTTTTTTGAATCTAGCTCACGG | FAM: TGGGAAAGACTTCAATAaGTTTACATTCCTAGATTCTCC |
| R: AATAGCGACCTTTGGAGGATATACTC | TET: TGGGAAAGACTTCAATAtGTTTACATTCCTAGATTCTCC | ||
| A-249B10-53kb | 53534 | F: ACAAATGTGAAAACTGAAGGCAA | FAM: ACTTATTGAGGcACCCAGTTGACACCCATG |
| R: CTAGGATCTGTGGCTCATTCCAT | TET: ACTTATTGAGGtACCCAGTTGACACCCATGGAA | ||
| A-249B10-101kb | 101056 | F: AGGCAGTGGCCAATTATTACTCA | FAM: CCTCCTTCTTAATGCcGGCAAAGATCATTTT |
| R: GAGGATACACTGATGGAGTATTTGGA | TET: CCTCCTTCTTAATGCtGGCAAAGATCATTTTTG | ||
| A-249B10-120kb | 120688 | F: GGTTCTCCAAGTCCAGCTTTGTT | FAM: CAGCTCAGATTCCACATAAGTTTGTCAGGCC (+4 insertion) |
| R: CATTCTGAAACTCAAAGGCT | TET: CAGCTCAGATTCCACATTTGTCAGGCC | ||
| A-249B10-203kb | 203064 | F: GATGCTTTTGGTTTTATATTCATAGGAA | FAM: AAAGAAAAAGGGACgGGGCAGTAAAGGAGA |
| R: AATATGTTTAAGAACTTGGCAAACAACT | TET: AAAGAAAAAGGGACaGGGCAGTAAAGGAGACA |
The primers and probes used for SNP detection in the taqman assays are given. F, forward; R, reverse. Lowercase letters indicate the position of the mutated nucleotide; FAM and TET indicate the fluorescent dye used for probe labeling. The position refers to the genomic sequences of the BACs as deposited in GenBank.
Figure 2.
Association analysis (transmission distortion as calculated by TRANSMIT) of markers in the vicinity of the association signal in the vicinity of D16S3068. (Upper) The unstratified analysis of the German part of the extended cohort (no significant results were obtained in the United Kingdom part of the cohort, which have thus been omitted for clarity). (Lower) The results of the analysis of the NOD2-stratfied genotypes. The key of phenotypes and populations is given in Upper. The marker names and the corresponding physical map using the local BAC clones are shown below the association graphs.
To investigate the influence of NOD2 genotype on this association finding, we stratified the cohorts on NOD2 genotype. Phenotypes of individuals carrying either the NOD2–SNP8 or –SNP13 mutation were set to unknown. Thereby, 345 and 204 phenotypes were removed in the German and United Kingdom cohorts, respectively. After this stratification, the significance levels in the Germany/Netherlands part of the cohort increased to 0.0001 for the SNP at 53 kb (293 observed versus 264 expected transmissions) in the IBD phenotype. In the subphenotype analysis, CD significances peaked at P = 0.002 at 53 kb in this population. The UC association evidence dropped to 0.0076 at the 53 kb SNP (127 versus 115 transmissions). The three SNPs at 27 kb, 37 kb, and 53 kb are in high pairwise LD (χ2 > 250), as revealed by an eh analysis of the CD patients in the German cohort. These three markers thus form the associated haplotype as also shown by very similar association significances in the single marker TRANSMIT analysis. The United Kingdom part of the cohort yielded a peak significance of 0.0065 at the P3X 127 SNP in the CD phenotype group (95 versus 82 transmissions) on stratification. A graphical representation of the local map and the results after stratification are shown in the Fig. 2 Lower.
Discussion
Genetic linkage of chromosome 16 to CD was established by a genomewide linkage study in 1996. This susceptibility region has since been confirmed in many independent studies, making this the most robust linkage region observed for IBD. In fact, this linkage region may be the most consistent non-HLA linkage observed for any complex genetic trait. The recent identification and verification of NOD2 as an IBD gene in four independent population samples has clearly demonstrated the ability of positional cloning to identify IBD susceptibility genes, and confirmed the importance of chromosome-16 linkage results. The NOD2 discovery also raises new questions regarding the chromosome-16 linkage areas, and provides new tools for interrogation of these regions. One of the most intriguing possibilities raised by continued genetic analysis of chromosome 16 is the suggestion that additional susceptibility genes may reside on this chromosome. In part, this question is raised by the substantially different locations of the linkage peaks reported in different studies (11–15, 19, 20).
To investigate the possibility of additional chromosome-16 IBD genes, we performed a systematic microsatellite mapping experiment to both clearly define linkage regions in which undiscovered IBD genes may be located, and to derive association leads for gene identification. This experiment was initiated in our laboratory before the NOD2 gene identification, and in the absence of NOD2 genotype information.
To define initial regions of interest, 38 microsatellite markers were genotyped in an exploratory cohort of 428 affected sibling pairs. The peak linkage result occurs near NOD2, using the strict CD phenotype classification. This curve spans the intervals implicated in previous studies and may indicate that this relatively diverse cohort contains many of the different susceptibility loci on chromosome 16, thus producing the complex linkage result. On stratification on NOD2 genotype, the central linkage peak, corresponding to the location of NOD2, drops substantially in significance from lod 3.2 to lod 1.2, indicating a major influence of NOD2 on allele sharing in this region. The relatively unchanged flanking peaks suggest the existence of additional susceptibility genes. This would point to a total of two NOD2-independent risk loci for IBD on chromosome 16. Nonparametric linkage analysis as performed in this study has limited fine mapping power. However, an additional linkage located 20 centimorgans away from NOD2 cannot be explained on the basis of methodological imprecision alone (37, 38).
Analysis of the STR data by using association tests yielded a strong lead at D16S3068 (P = 0.00028 for all IBD and 0.0079 for CD). D16S3068 is located on a sequenced BAC clone (A-249B10), but available flanking sequence was limited. To facilitate identification of novel genes and new SNP markers for this region, we isolated and completely sequenced a neighboring BAC (504P11). Three newly established SNP loci, along with D16S3068, showed a transmission distortion with the IBD phenotype. Interestingly, this distortion was restricted to the German subset of the study cohort. On stratification based on NOD genotype (SNP8 and SNP13), this association signal increased in significance. This increase is interesting, as a significant proportion of the sample set was removed from the analysis by conditioning the sample based on NOD2. In case of a neutral finding, a substantial decrease in significance would have been expected as 370 phenotypes were removed from the analysis. In addition, the significances in the subphenotypes shifted toward CD, a result consistent with the predominant CD linkage signal on this chromosome. The three SNPs, 27 kb, 37 kb, and 53 kb, and the microsatellite D16S3068 define the associated haplotype. This haplotype is very frequent, pointing either to a common, low-impact susceptibility factor or to another, yet unidentified haplotype with a lower population frequency, which is superimposed on the frequent haplotype. In the unstratified United Kingdom cohort, no evidence for association to this genomic region was detected. After stratification on NOD2 genotype, a peak association of P = 0.0065 was observed in the CD phenotype group. This result suggests the presence of a second population specific susceptibility haplotype, which is unmasked by stratification. The degree of association detected in the United Kingdom part of the population is lower than in the German families. A different mutational spectrum or a population-specific haplotype history at this locus could account for this finding. Both population-specific haplotypes may point to the same gene.
There are no obvious genes annotated on the sequence surrounding D16S3068. Therefore, the molecular basis of the observed genetic association is not yet clear. The lack of a consistent gene model in this region may also be caused by the relatively short sequence available for gene prediction.
In summary, we present in this report evidence for the existence of susceptibility loci in addition to NOD2 on chromosome 16. We furthermore demonstrate an associated population-specific transmission distortion indicating a putative susceptibility gene on chromosome 16p and have localized this signal to a small pericentromeric region. Finally, we have shown that stratification on NOD2 genotype leads to increased significances in both analyzed populations. In total, these data might help to discover additional IBD susceptibility genes in this region and further our genetic understanding of this disorder.
Acknowledgments
We thank the clinicians that have played a leading role in the patient recruitment: John Lennard-Jones, Sander van Deventer, Andrew J. S. MacPherson, John C. W. Lee, Susanna Nikolaus, and Jochen Grebe. We thank the cooperating physicians, IBD patients, and their families for participating in this study. The cooperation of the German Crohn's and Colitis Foundation (DCCV e. V.), Prof. Raedler (Hamburg), Prof. Kruis (Köln), Dr. Theuer (Heilbronn); Dr. Meckler (Gedern); Prof. Lochs, Dr. Wedel, and T. Herrmann (Berlin); Dr. Herchenbach (Recklinghausen); Prof. Scheurlen (Würzburg); Dr. Demharter (Augsburg); Dr. Simon (Munich); Dr. Purrmann (Moers); Dr. Jessen (Kiel); Dr. Zehnter (Dortmund); Dr. Lübke and Dr. Weismüller (Koblenz); Dr. Eiche (Denkendorf); Dr. Schönfelder (Aachen); Prof. Fleig (Halle) all in Germany; and Dr. Hodgson, Dr. Sanderson, Dr. Pounder, and Dr. Forgacs (London); Dr. Bird (Maidstone); Dr. Hines (Haywards-Heath); Dr. Cairns and Dr. Ireland (Brighton); Dr. Barrison (St. Albans); and Dr. Smith-Lang (Sidcup) in the United Kingdom, is gratefully acknowledged. We also acknowledge the expert technical help from Kathy King, Anja Hasselmeyer, Tanja Wesse, Tam Ho Kim, and Birte Köpke. This work was supported in the United Kingdom by The Wellcome Trust, The Generation Trust, the National Association for Colitis and Crohn's disease, Crohn's in Childhood Research Association, and the Sir Halley Stewart Trust. In Germany, support was from the Deutsche Forschungsgemeinschaft (For 423), a Training and Mobility of Research Network Grant of the European Union (ERB-4061-PL-97-0389), a competence network Chronisch-entzündliche Darmerkrankungen, the German Human Genome Project, and the National Genome Research Network (all funded by the German Federal Department for Research and Education).
Footnotes
Abbreviations: IBD, inflammatory bowel disease; CD, Crohn's disease; UC, ulcerative colitis; SNP, single-nucleotide polymorphisms; lod, logarithm of odds; BAC, bacterial artificial clone; MLS, multipoint lod score.
References
- 1.Shivananda S, Lennard Jones J, Logan R, Fear N, Price A, Carpenter L, van Blankenstein M. Gut. 1996;39:690–697. doi: 10.1136/gut.39.5.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Probert C S, Jayanthi V, Rampton D S, Mayberry J F. Int J Colorectal Dis. 1996;11:25–28. doi: 10.1007/BF00418851. [DOI] [PubMed] [Google Scholar]
- 3.Yamamoto T, Keighley M R. Int J Colorectal Dis. 2000;15:211–215. doi: 10.1007/s003840000233. [DOI] [PubMed] [Google Scholar]
- 4.Sabir N, Sungurtekin U, Erdem E, Nessar M. Int J Colorectal Dis. 2000;15:317–322. doi: 10.1007/s003840000251. [DOI] [PubMed] [Google Scholar]
- 5.Voigt E, Griga T, Tromm A, Henschel M G, Vorgerd M, May B. Int J Colorectal Dis. 1999;14:304–307. doi: 10.1007/s003840050234. [DOI] [PubMed] [Google Scholar]
- 6.Podolsky D K. N Engl J Med. 1991;325:928–937. doi: 10.1056/NEJM199109263251306. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton S R. Int J Colorectal Dis. 1987;2:113–117. doi: 10.1007/BF01647703. [DOI] [PubMed] [Google Scholar]
- 8.Orholm M, Munkholm P, Langholz E, Nielsen O H, Sorensen I A, Binder V. N Engl J Med. 1991;324:84–88. doi: 10.1056/NEJM199101103240203. [DOI] [PubMed] [Google Scholar]
- 9.Tysk C, Lindberg E, Jarnerot G, Floderus Myrhed B. Gut. 1988;29:990–996. doi: 10.1136/gut.29.7.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson N P, Driscoll R, Pounder R E, Wakefield A J. Br Med J. 1996;312:95–96. doi: 10.1136/bmj.312.7023.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugot J P, Laurentpuig P, Gower-Rousseau C, Olson J M, Lee J C, Beaugerie L, Naom I, Dupas J L, Vangossum A, Orholm M, et al. Nature (London) 1996;379:821–823. doi: 10.1038/379821a0. [DOI] [PubMed] [Google Scholar]
- 12.Curran M E, Lau K F, Hampe J, Schreiber S, Bridger S, MacPherson A J S, Cardon L R, Sakul H, Harris T J R, Stokkers P, et al. Gastroenterology. 1998;115:1066–1071. doi: 10.1016/s0016-5085(98)70075-7. [DOI] [PubMed] [Google Scholar]
- 13.Brant S R, Fu Y, Fields C T, Baltazar R, Ravenhill G, Pickles M R, Rohal P M, Mann J, Kirschner B S, Jabs E W, et al. Gastroenterology. 1998;115:1056–1061. doi: 10.1016/s0016-5085(98)70073-3. [DOI] [PubMed] [Google Scholar]
- 14.Annese V, Latiano A, Bovio P, Forabosco P, Piepoli A, Lombardi G, Andreoli A, Astegiano M, Gionchetti P, Riegler G, et al. Eur J Hum Genet. 1999;7:567–573. doi: 10.1038/sj.ejhg.5200328. [DOI] [PubMed] [Google Scholar]
- 15.Cavanaugh J. Am J Hum Genet. 2001;68:1165–1171. doi: 10.1086/320119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hugot J P, Chamaillard M, Zouali H, Lesage S, Cezard J P, Belaiche J, Almer S, Tysk C, O'Morain C A, Gassull M, et al. Nature (London) 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 17.Ogura Y, Bonen D K, Inohara N, Nicolae D L, Chen F F, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr R H, et al. Nature (London) 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 18.Hampe J, Cuthbert A, Croucher P J P, Mirza M M, Mascheretti S, Fisher S, Frenzel H, King K, Hasselmeyer A, MacPherson A J S, et al. Lancet. 2001;357:1925–1928. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 19.Cavanaugh J A, Callen D F, Wilson S R, Stanford P M, Sraml M E, Gorska M, Crawford J, Whitmore S A, Shlegel C, Foote S, et al. Ann Hum Genet. 1998;62:291–298. doi: 10.1046/j.1469-1809.1998.6240291.x. [DOI] [PubMed] [Google Scholar]
- 20.Cho J H, Nicolae D L, Gold L H, Fields C T, LaBuda M C, Rohal P M, Pickles M R, Qin L, Fu Y, Mann J S, et al. Proc Natl Acad Sci USA. 1998;95:7502–7507. doi: 10.1073/pnas.95.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hampe J, Schreiber S, Shaw S H, Lau K F, Bridger S, MacPherson A J S, Cardon L R, Sakul H, Harris T J R, Buckler A, et al. Am J Hum Genet. 1999;64:808–816. doi: 10.1086/302294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olavesen, M. G., Hampe, J., Mirza, M. M., Saiz, R., Lewis, C. M., Bridger, S., Teare, D., Easton, D. F., Herrmann, T., Scott, G., et al. (2000) Immunogenetics, in press. [DOI] [PubMed]
- 23.Hampe J, Hermann B, Bridger S, MacPherson A J S, Mathew C G, Schreiber S. Int J Colorectal Dis. 1998;13:260–263. doi: 10.1007/s003840050173. [DOI] [PubMed] [Google Scholar]
- 24.Lennard-Jones J E. Scand J Gastroenterol Suppl. 1989;170:2–6. doi: 10.3109/00365528909091339. [DOI] [PubMed] [Google Scholar]
- 25.Truelove S C, Pena A S. Gut. 1976;17:192–201. doi: 10.1136/gut.17.3.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall J, Nanthakumar E. In: Current Protocols in Human Genetics. Boyle A L, editor. Vol. 2. New York: Wiley; 1997. pp. 2.8.1–2.8.19. [Google Scholar]
- 27.Partanen J, Koskimes S. Scand J Immunol. 1988;28:313–316. doi: 10.1111/j.1365-3083.1988.tb01454.x. [DOI] [PubMed] [Google Scholar]
- 28.Idury R M, Cardon L R. Genome Res. 1997;7:1104–1109. doi: 10.1101/gr.7.11.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holland P M, Abramson R D, Watson R, Gelfand D H. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak K J, Flood S J, Marmaro J, Giusti W, Deetz K. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 31.Hampe, J., Wollstein, A., Lu, T., Frevel, H. J., Will, M. & Schreiber, S. (2001) Bioinformatics, in press. [DOI] [PubMed]
- 32.Kioschis P, Wiemann S, Heiss N S, Francis F, Gotz C, Poustka A, Taudien S, Platzer M, Wiehe T, Beckmann G, et al. Genomics. 1998;54:256–266. doi: 10.1006/geno.1998.5560. [DOI] [PubMed] [Google Scholar]
- 33.Matise T C, Perlin M, Chakravarti A. Nat Genet. 1994;6:384–390. doi: 10.1038/ng0494-384. [DOI] [PubMed] [Google Scholar]
- 34.Clayton D. Am J Hum Genet. 1999;65:1170–1177. doi: 10.1086/302577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clayton D, Jones H. Am J Hum Genet. 1999;65:1161–1169. doi: 10.1086/302566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terwilliger J D, Ott J. Handbook of Human Genetic Linkage. Baltimore: Johns Hopkins Univ. Press; 1994. pp. 245–250. [Google Scholar]
- 37.Kruglyak L, Lander E S. Am J Hum Genet. 1996;58:1092–1093. [PMC free article] [PubMed] [Google Scholar]
- 38.Hampe J, Wienker T, Nürnberg P, Schreiber S. Hum Hered. 2000;50:91–101. doi: 10.1159/000022896. [DOI] [PubMed] [Google Scholar]
- 39.Ohmen J D, Yang H Y, Yamamoto K K, Zhao H Y, Ma Y, Bentley L G, Huang Z, Gerwehr S, Pressman S, McElree C, et al. Hum Mol Genet. 1996;5:1679–1683. doi: 10.1093/hmg/5.10.1679. [DOI] [PubMed] [Google Scholar]
- 40.Parkes M, Satsangi J, Lathrop G M, Bell J I, Jewell D P. Lancet. 1996;348:1588. doi: 10.1016/S0140-6736(05)66204-6. (lett.). [DOI] [PubMed] [Google Scholar]