Abstract
Background
CRF01_AE and CRF07_BC are the two most prevalent HIV-1 genotypes in China, and the co-circulation of these two genotypes has led to the continuous generation of CRF_0107 viruses in recent years. However, little is known about the origin and spread of CRF_0107 viruses thus far. This study focused on HIV-1 CRF80_0107, which we previously identified among the MSM population in Beijing and Hebei Province, to explore the demographic distribution, transmission links, and temporal-spatial evolutionary features of the HIV-1 CRF80_0107 strain in China.
Methods
With the partial pol region fragment of the HIV-1 CRF80_0107 subtype standard sequence as a reference, BLAST was used to search for highly similar sequences in the Los Alamos HIV Sequence Database, followed by preliminary subtype identification via COMET. Further phylogenetic and recombination breakpoint analyses were conducted to verify the subtypes and recombination patterns. We also performed a distance-based molecular network analysis to identify potential relationships among different HIV-positive individuals. In addition, spatiotemporal evolutionary dynamics analysis of the candidate CRF80_0107 sequences was performed via a Bayesian approach.
Results
A total of 36 partial pol gene sequences of HIV-1 CRF80_0107 were identified from 2009 to 2018 from 5 provinces in China. Phylogenetic and spatial-temporal dynamics analyses indicated that CRF80_0107 likely originated in Beijing around 2009 and spread to Guangdong Province around 2012. Population dynamics analysis revealed that CRF80_0107 experienced a significant increase in population size from 2009 to 2011 and then stabilized. The study also found that the number of cases in Guangdong Province was second only to that in Beijing and formed 2 relatively independent transmission clusters in the MSM population in Shenzhen, Guangdong Province.
Conclusions
The HIV-1 CRF80_0107 strain has spread to cities beyond its origin, particularly the MSM population in Shenzhen city, Guangdong Province, which is an area with a high incidence of HIV. This highlights the importance of continuous monitoring for the emergence and dynamic changes of novel HIV-1 recombinant viruses and the necessity of implementing effective preventive measures targeting specific populations in particular regions.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-025-10461-0.
Keywords: HIV-1, CRF80_0107, Epidemic, Network, Spatiotemporal dynamics
Introduction
Human immunodeficiency virus (HIV), which causes acquired immune deficiency syndrome (AIDS), is known for its high genetic diversity, which has implications for disease progression, antiretroviral therapy, and vaccine development [1]. The high genetic diversity of HIV mainly arose from its high rates of mutation and recombination due to the error-prone reverse transcriptase enzyme. Recombination occurs when an individual is dually infected or infected by multiple different virus strains and can more rapidly and efficiently alter the genetic makeup of HIV than can mutation [2]. Recombinants can be designated either circulating recombinant forms (CRFs), which are defined as recombinant HIV-1 genomes that have been identified in at least three epidemiologically unrelated individuals, or unique recombinant forms (URFs), which do not show evidence of onward transmission [3, 4]. Currently, 159 HIV-1 CRFs and numerous URFs are deposited in the Los Alamos HIV Sequence Database (https://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html). Although CRF02_AG and CRF01_AE still are the two most prevalent CRFs worldwide, other CRFs (CFRs other than CRF02_AG and CRF01_AE) have increased in prevalence and are predominant in regional epidemics [5, 6]. Therefore, it is highly important to implement surveillance of other CRFs including recently identified CRFs.
China is one of the countries with the highest number of distinct HIV-1 genotypes in the world, and the co-circulation of multiple subtypes has led to the continuous generation of CRFs in China [7]. To date, 60 CRFs have been identified in China, accounting for more than one-third of the total CRFs. First, in the early 1990s, CRF07_BC and CRF08_BC emerged among injecting drug users (IDUs) in Yunnan, a southwestern province of China, and rapidly spread to other regions of China through drug trafficking routes [8]. Subsequently, several CRF_BC viruses were also identified, mainly among IDUs in Yunnan Province. Second, in the early 2000s, the co-circulation of subtypes CRF01_AE and B in the MSM (men who have sex with men) population favour the generation of CRF_01B viruses, among which CRF55_01B rapidly spread throughout the country and became the fifth most prevalent strain in China [9, 10]. In recent years, nearly twenty CRF viruses originating from CRF01_AE and CRF07_BC have been described in China, which calls for more attention to novel CRF_0107 viruses [11, 12]. However, little is known about the origin and spread of CRF_0107 viruses in China thus far.
In 2019, we identified CRF80_0107 among MSM in Beijing and Hebei Province, which was the second CRF_0107 virus identified in China [13]. Recent studies reported the detection of CRF80_0107 either in Hebei province or in Hubei Province, suggesting the ongoing transmission of CRF80_0107 after its generation [14, 15]. To date, only five CRF80_0107 sequences, including three near full-length genome (NFLG) sequences and two partial sequences, have been deposited in the Los Alamos HIV Sequence Database. Notably, CRF80_0107 strains may be misclassified as CRF07_BC strains because the vast majority of pol fragments routinely sequenced in genotypic drug resistance testing belong to CRF07_BC. However, scrutinized recombination analysis and phylogenetic analysis can be used to identify more CRF80_0107 sequences. In this study, we identified 36 HIV-1 partial pol CRF80_0107 sequences (HXB2 positions 2253–3272) from the Los Alamos HIV Sequence Database, investigated the temporal and geographic origins of the CRF80_0107 strains, and explored its epidemiological relationships in different geographic areas and risk groups.
Methods
HIV-1 sequence dataset
To identify additional CRF80_0107 sequences, we utilized a partial pol gene region fragment (HXB2 positions: 2253–3272nt) from the longest reference sequence of CRF80_0107 (Accession number: MH843713, 8986 bp) in the Los Alamos HIV Sequence Database, and employed the Basic Local Alignment Search Tool (BLAST) to search for the 200 most closely related sequences. If two or more sequences presented the same patient code, we retained only one sequence to remove duplicates. To confirm the HIV subtype, we conducted HIV-1 subtyping via the Context-based Modeling for Expeditious Typing (COMET) subtyping tool, followed by phylogenetic analysis as previously described [16–18]. We also confirmed the recombination patterns in the optimized dataset via the Simplot software v3.5 [19]. Finally, we identified 36 HIV-1 CRF80_0107 partial pol (1020 bp) sequences and collected related demographic information, which is publicly available in the Los Alamos HIV Sequence Database.
Transmission network reconstruction
The Tamura-Nei 93 nucleotide substitution model was used to calculate the pairwise genetic distance for all CRF80_0107 partial pol (1020 bp) sequences by Hyphy 2.2.4 software [20, 21]. We then conducted a sensitivity analysis across a reasonable range of genetic distance thresholds from 0.1% to 2.0% to determine the possible genetic linkage between two individuals. The genetic distance before the transmission clusters in a dataset begin to merge and the transmission network loses its accuracy is the optimal cluster formation genetic distance for that dataset, i.e., the genetic distance when the number of clusters is at its maximum [22]. A node represents each individual in the molecular network, and we connected the nodes according to the optimal genetic distances described above and visualized them via Cytoscape 3.10.1 [23].
Maximum likelihood phylogenetic reconstruction
To elucidate the genetic evolutionary relationship of the HIV-1 CRF80_0107 strain in China, we conducted a phylogenetic analysis. First, we identified the most suitable nucleotide substitution model for our data using jModelTest [24]. On the basis of the Akaike information criterion (AIC), the best model for the dataset was the general time reversible model incorporating the gamma distribution for site rate heterogeneity and the invariant site ratio (GTR + G + I). We then utilized PhyML 3.0 to construct the maximum likelihood phylogenetic tree under this nucleotide substitution model [25]. The branch support was inferred from 1000 replicates using standard bootstrap analysis, and clusters with support greater than 900 were considered reliable. Finally, we visualized the final maximum likelihood tree via iTOL [26].
Spatiotemporal dynamics analysis
We checked the dataset for a temporal signal via TempEst v1.5.1, which identifies the correlation of genetic differences between sequences with time (R2 = 0.7386) [27]. We then employed a Bayesian phylogenetic approach to estimate the rate of evolution and the time to the most recent common ancestor (tMRCA) for these data via a GTR + G + I substitution model with an uncorrelated lognormal relaxed clock model and a Bayesian Skygrid coalescent tree prior in BEAST v1.10.4 [28]. Additionally, at least two independent Markov Chain Monte Carlo (MCMC) runs of 5 × 108 simulations with sampling every 10,000 steps were performed. Convergence of the MCMC results was examined in Tracer v1.7.1 with effective sampling size (ESS) > 200 for all parameters considered acceptable [29]. A Bayesian skygrid plot was generated via Tracer v1.7.1 to represent the changes in the viral effective population size over time [30]. Finally, the maximum clade credibility (MCC) tree was generated via TreeAnnotator v1.10.4 after discarding the first 10% as burn-in and visualized via iTOL [31].
We also conducted a Bayesian phylogeographic analysis using the asymmetric discrete traits combined with Bayesian stochastic search variable selection (BSSVS) implemented in BEAST v1.10.4 to identify migration pathways of HIV-1 CRF80_0107 between the different sampling locations [32]. We specified the geographic location of the sequence as a sampling province or municipality. The migration routes were summarized via SPREAD3 v0.9.7.1 and visualized via Adobe Illustration software [33]. Statistical support for migratory routes was determined on the basis of the Bayes factor (BF), and a route with a BF ≥ 3 was considered credible [34].
Results
Identification of CRF80_0107
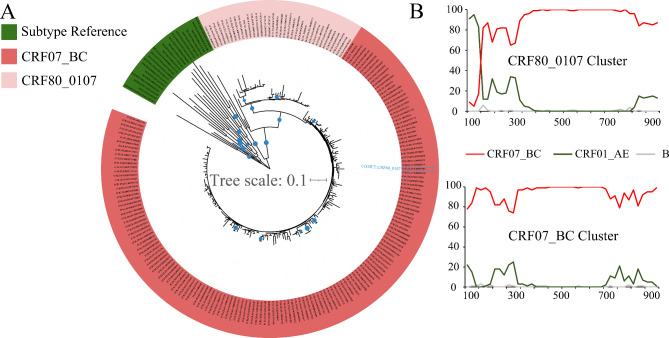
On the basis of the described criteria, a total of 200 HIV-1 partial pol gene (HXB positions: 2253-3272nt) sequences from 2009 to 2018 in China were included in the initial dataset. The subtypes were determined via COMET and phylogenetic analysis. Among these sequences, COMET identified 41 (41/200, 20.50%) sequences as CRF80_0107. However, further phylogenetic analysis revealed that one of the sequences (Accession number: MW956961) initially classified as CRF80_0107 was actually CRF07_BC (Fig. 1A). After further confirming the recombination patterns of the CRF80_0107 branch sequences, the optimized dataset ultimately contained 36 CRF80_0107 sequences (Fig. 1B).
Fig. 1.
Phylogenetic and Recombination analysis of HIV-1 partial pol sequences of HIV-1 CRF80_0107 in the study. (A) Neighbor-joining tree analysis of HIV-1 CRF80_0107 partial pol gene sequences identified in the present study. The neighbor-joining phylogenetic tree was constructed by combining the reference sequence (all HIV-1 group M, CRF01_AE, and CRF07_BC) with the CRF80_0107 sequence using the Kimura two-parameter model with 1,000 bootstrap replicates. A blue circle on a tree branch represents a branch support value ≥ 0.9; the larger the circle is, the closer the value is to 1.0. The CRF07_BC Cluster is labelled watermelon red, the CRF80_0107 Cluster is light pink, and the other reference sequences (pure subtypes and CRF01_AE) are green. (B) Recombination analysis of HIV-1 CRF80_0107 and CRF07_BC partial pol gene sequences. These HIV-1 CRF80_0107 sequences were compared with those of subtypes B, CRF01_AE, and CRF07_BC, and plots of percentages against nucleotide positions are shown (window size: 200nt; step size: 20nt). Red, CRF07_BC; green, CRF01_AE; gray, subtype B
Demographic details
Most of the sequences analysed in this study represent CRF80_0107 cases from Beijing and Guangdong Provinces (17 cases and 16 cases, respectively). The remaining three cases are from Heilongjiang Province, Hebei Province, and Jiangsu Province. Among these 36 unique cases, 22 were male, one was female, and the rest were unknown. The 23 cases mentioned above included 21 cases of MSM (21/36, 58%, accounting for 91% of the data on known routes of transmission) and 2 cases of heterosexual transmission (HET) (Table 1, Additional Table 1).
Table 1.
Demographic characteristics of HIV-1 CRF80_0107 in this study
| Characteristic | Patients | ||
|---|---|---|---|
| Sex | F | 1 | 2.78% |
| M | 22 | 61.11% | |
| Unknown | 13 | 36.11% | |
| Risk factor | HET | 2 | 5.56% |
| MSM | 21 | 58.33% | |
| Unknown | 13 | 36.11% | |
| Location | Beijing | 17 | 47.22% |
| Guangzhou, Guangdong Province | 1 | 2.78% | |
| Shenzhen, Guangdong Province | 14 | 38.88% | |
| Zhuhai, Guangdong Province | 1 | 2.78% | |
| Hebei | 1 | 2.78% | |
| Heilongjiang | 1 | 2.78% | |
| Jiangsu | 1 | 2.78% | |
| Sample year | 2009 | 1 | 2.78% |
| 2010 | 8 | 22.22% | |
| 2011 | 4 | 11.11% | |
| 2012 | 3 | 8.33% | |
| 2013 | 4 | 11.11% | |
| 2014 | 5 | 13.89% | |
| 2015 | 5 | 13.89% | |
| 2016 | 1 | 2.78% | |
| 2017 | 3 | 8.33% | |
| 2018 | 2 | 5.56% | |
F, female; M, male; HET, heterosexuals; MSM, men who have sex with men
Network transmission analysis of HIV-1 CRF80_0107
We used a genetic distance less than or equal to 0.3% as the cut-off value for constructing a transmission network for the HIV-1 CRF80_0107 strains (Fig. 2A). This helped us identify 3 clusters, encompassing 28 out of 36 (77.78%) sequences (Fig. 2B). The first transmission cluster, named the “Beijing cluster”, contained 16 HIV-1 CRF80_0107 infected individuals from Beijing and one from Shenzhen, Guangdong Province. Of the 16 infected individuals from Beijing, 6 were MSM, and the risk factors for the rest were unknown. Additionally, one infected individual from Shenzhen, Guangdong Province, had risk factors for MSM. The second transmission cluster, named “Guangdong Cluster 1”, consisted of 8 HIV-1 CRF80_0107 infected individuals from Shenzhen, Guangdong Province, and one from Zhuhai, Guangdong Province. Among them, all 8 were MSM, except for one female at risk of heterosexual transmission. The third cluster, named “Guangdong Cluster 2”, is a small cluster consisting of only 2 MSM from Shenzhen City, Guangdong Province. The pairwise genetic distance within all CRF80_0107 sequences in the dataset is generally low (0.008312, 95% confidence interval: 0.007905, 0.008719) (Fig. 2C). Specifically, the Beijing cluster had the lowest mean pairwise genetic distance (P = 0.0086 < 0.05), potentially indicating that CRF80_0107 might originate from a single transmission event. And then, the gradually increasing inner-cluster pairwise genetic distances found in Guangdong clusters might suggest the potentially circulating trend of CRF80_0107 among the MSM group in China.
Fig. 2.
Transmission network analysis of HIV-1 CRF80_0107. (A) Number of clusters and maximum cluster size, as a function of the TN93 distance threshold. The light blue dashed line highlights the genetic distance at 0.003 substitutions/site. (B) Transmission clusters of HIV-1 CRF80_0107. Nodes (circles) represent connected individuals in the overall network, and potential transmission relationships are represented by edges (lines). The different colors of the nodes represent various regions, and the different shapes of the nodes represent different risk factors (ellipse: MSM; rectangle: HET; Triangle shape: Unknown). M: male; F: female; Unknown: -. (C) Distribution of pairwise genetic distances in different transmission clusters of HIV-1 CRF80_0107. Horizontal lines denote the mean with 95% confidence interval (95% CI). Total: the entire CRF80_0107 dataset (Nnode = 36); BJ: Beijing Cluster (Nnode = 17); GD1: Guangdong Cluster1 (Nnode = 9); GD2: Guangdong Cluster2 (Nnode = 2). The Mann-Whitney test was used to compare pairwise genetic distances between 2 clusters (BJ cluster and GD1 cluster) in Fig. 2B (P = 0.0086). GD2 cluster contained only one value (marked with a #) and was not included in the above statistical analysis. *: P < 0.05
Expansion of CRF80_0107 in China
Inferred from the above results of pairwise genetic distance, we further trace the transmission dynamics of CRF80_0107. As shown in the maximum likelihood (ML) tree (Fig. 3A), all CRF80_0107 sequences formed a larger, well-supported monophyletic lineage with a bootstrap value of > 900. The CRF80_0107 sequence from Beijing is not only closer to the root of the evolutionary cluster but also widely distributed in the cluster. The CRF80_0107 sequence branches of the other four provinces are wrapped in the entire Beijing sequence branch, suggesting that Beijing may be the source of CRF80_0107. In addition, the majority of CRF80_0107 sequences from Guangdong Province (12/16, 75%) formed a well-supported, region-specific subcluster (bootstrap value > 900).
Fig. 3.
Maximum likelihood analysis and maximum clade credibility analysis of HIV-1 CRF80_0107. (A) Maximum likelihood phylogenetic tree of HIV-1 CRF80_0107. The maximum likelihood phylogenetic tree was estimated with all CRF80_0107 sequences and several subtype reference sequences via PhyML 3.0 software. The light green background represents all CRF80_0107 sequences, and the light orange background represents CRF80_0107 sequences, which are mainly from Guangdong Province. Except for the black branch, which represents the reference sequence, the other colored branch colors represent different geographic origins. (B) Maximum clade credibility (MCC) tree estimated from HIV-1 partial pol gene sequences of CRF80_0107. Different branch colors represent different geographic sources, different shapes at the end of the branches represent different risk factors, and the shapes are filled with the representative color of the corresponding geographic location. The green and orange stars indicate the tMRCA (95% HPD) for CRF80_0107 in Beijing and Guangdong Province, respectively
Further temporal evolutionary analyses revealed (Fig. 3B) that the origin of CRF80_0107 in China can be traced back to 2008.8 (95% highest posterior density, HPD: 2008.5–2009.0) and identified Beijing as the most likely ancestral location for the HIV-1 CRF80_0107 sub-epidemic. Our study also revealed that the CRF80_0107 strain most likely spread from Beijing to Guangdong Province (Bayes factor, BF = 9.81) around 2012.2 (95% HPD: 2011.3-2012.8). The Bayesian skygrid plot (Fig. 4) showed an exponential increase in the CRF80_0107 effective population size from 2009 to 2011, after which it stabilized. In addition, the analysis of geographic spreading dynamics showed that the well-supported migration occurred from Guangdong Province to Jiangsu Province (BF = 7.95), followed by migration from Beijing to Heilongjiang or Hebei Provinces (all BF>3, Table 2). Currently, CRF80_0107 has been detected in four geographically distant regions of China (Fig. 5): northeastern China (Heilongjiang), northern China (Beijing and Hebei), east China (Jiangsu), and Southern China (Guangdong).
Fig. 4.
Bayesian Skygrid demographic reconstruction of HIV-1 CRF80_0107. The vertical axis shows the effective number of infections (Ne) multiplied by the mean viral generation time (τ). The solid blue line indicates the median effective population size over time; the pink shaded area indicates the 95% highest posterior density (HPD) interval for the effective population size. The first thick solid line indicates the estimated origin date for HIV-1 CRF80_0107(2008.8, 95% HPD: 2008.5–2009)
Table 2.
Inference of the migration events of HIV-1 CRF80_0107 in this study
| From | To | Bayes factor | Posterior probability |
|---|---|---|---|
| Beijing | Guangdong | 9.81 | 0.75 |
| Beijing | Hebei | 3.40 | 0.51 |
| Beijing | Heilongjiang | 4.21 | 0.56 |
| Guangdong | Jiangsu | 7.95 | 0.71 |
Fig. 5.
Inferred geographical migration events of HIV-1 CRF80_0107 strains in China. Visualizing the migration events of HIV-1 CRF80_0107 on a map of China. Points are color-coded by the geographic location of the origin. Viral migration routes with a Bayes factor greater than or equal to 3 are shown, and the routes are color-coded according to the magnitude of the posterior probability value
Discussion
In recent years, the prevalence of novel recombinant strains of HIV has emerged as a significant challenge in global public health. Notably, some recombinant strains, such as CRF01_AE and CRF02_AG, have been confirmed to accelerate disease progression and possess higher replicative capacity than their parental strains [6, 35, 36]. China is also one of the countries with the highest prevalence of HIV recombinant strains, where CRF01_AE, CRF07_BC, CRF08_BC, and subtype B were previously dominant [10]. Nowadays, China has entered an era of prevalent HIV-1 s-generation recombinant strains, such as the CRF55_01B strain [37]. According to the latest national HIV molecular epidemiological survey, novel CRF and URF strains account for up to 3.5% and 5.0%, respectively, covering almost all provinces and municipalities in China. Among them, recombinants with CRF01_AE and CRF07_BC as parental strains are the most common and are transmitted primarily among MSM [38]. HIV-1 recombinant strains often enhance biological adaptability, evade host immune responses, and produce dual or multiple drug-resistant variants, posing significant challenges to prevention, monitoring, drug resistance, treatment, and the broad development of vaccines [39–41]. In this context, conducting in-depth research on the epidemiology and transmission dynamics of specific recombinant strains, such as the 0107 recombinant strain, is particularly important.
This study conducted a comprehensive analysis of partial pol gene sequences of HIV-1 CRF80_0107 from China between 2009 and 2018 in the Los Alamos HIV Sequence Database, focusing on its demographic distribution, transmission relationships, and spatial-temporal evolutionary patterns. Our findings revealed the epidemiological characteristics and transmission dynamics of the HIV-1 CRF80_0107 subtype in China, particularly between two first-tier major cities with significant geographical distances (Beijing and Shenzhen).
First, this study indicates that Beijing is the most likely ancestral origin of HIV-1 CRF80_0107 and has the highest proportion of cases (17/36, 47.22%). Since the emergence of HIV-1 in Beijing, various subtypes have been reported [42, 43]. Currently, the main prevalent subtypes are still dominated by the parental strains CRF01_AE and CRF07_BC of CRF80_0107 [44, 45]. As the capital of China, Beijing has higher population mobility and more complex social network than that in Hebei Province, offering more fertile ground for the occurrence and transmission of recombinant strains [46–48]. It is therefore not be difficult to interpret that CRF80_0107 mainly formed a transmission network among MSM in Beijing. Although the risk factors for some infected individuals in the Beijing transmission cluster are still unclear, a large proportion of cases in the overall CRF80_0107 dataset occur within the MSM population. Moreover, the Beijing transmission cluster included MSM from Shenzhen, Guangdong Province, suggesting that MSM may still constitute a high-risk group for HIV-1CRF80_0107 infection in Beijing. Therefore, intervention measures targeting MSM populations infected with the CRF80_0107 strain in Beijing should be prioritized in prevention and control strategies to effectively curb the cross-regional transmission of this strain.
Shenzhen, another first-tier city in China, not only faces a more complex HIV epidemic but also has emerged as a significant hotspot for the origin and transmission of HIV-1 recombinant strains [49–51]. Previous studies have confirmed that the CRF07_BC strain in Shenzhen has reversed its status since 2012, surpassing CRF01_AE as the most prevalent HIV-1 strain [52]. Furthermore, the novel second-generation recombinant strain CRF55_01B originated and spread along railway lines to various parts of the country [53, 54]. This study revealed that CRF80_0107, which originated in Beijing around 2009, spread to Guangdong Province within just approximately three years, potentially facilitated by the developed and convenient railway network. Furthermore, the number of CRF80_0107 cases in Guangdong Province ranks second only to that in Beijing (N = 16), and specific phylogenetic clusters have formed, indicating the presence of independent transmission chains in the region. Further transmission network analysis confirmed that CRF80_0107 formed several relatively independent transmission clusters among the MSM population in Shenzhen, Guangdong. However, individuals infected with the novel recombinant strain of HIV in the MSM population in Shenzhen have higher viral loads and lower CD4 + T-cell counts, further increasing the risk of virus transmission [55]. Additionally, virus transmission within this population may be more concealed and rapid, increasing the difficulty of prevention and control [49]. Therefore, HIV prevention and control efforts targeting the MSM population in Shenzhen should be more detailed and thorough.
In addition to these main findings mentioned above, the Bayesian skygrid plot indicates that the effective population size of CRF80_0107 increased exponentially from 2009 to 2011 and then stabilized, which may reflect the effectiveness of prevention and control measures implemented as well as changes in the transmission characteristics of the virus itself [56]. We also found two cases of heterosexual transmission, suggesting that the transmission route of CRF80_0107 is not exclusively limited to the MSM population and carries the risk of transmission to the general population. The sequences used for calculating these results are mostly from partial pol region, with only three NFLG sequences. As pol region has a lower mutation rate than gag and env, it may affect the estimation of the true origin time of CRF80_0107. However, previous studies have attempted to trace the transmission dynamics of HIV-1 CRF55_01B using the pol sequences [57]. Moreover, the pol fragments were routinely used in genotypic drug resistance testing and often submitted to the HIV database. To include as many potential CRF80_0107 as possible, it would be a better choice to use pol sequences for analyses based on HIV database. Even so, only 36 samples were finally included in this study, and the small sample size may limit geographic representation. Despite these limitations, our study further revealed the cross-regional transmission dynamics of CRF80_0107 in China, including migration from Beijing to Heilongjiang Province and from Guangdong Province to Jiangsu Province. It requires us continue monitoring the prevalence of CRF80_0107, and include more NFLG sequences in our future work.
Conclusion
In summary, this study reports that the HIV-1 CRF80_0107 strain originated in Beijing around 2009 and was subsequently introduced into Shenzhen, Guangdong Province, an area with a high prevalence of HIV-1 epidemics and recombination, around 2012, where it formed several relatively independent transmission clusters among the MSM population in Shenzhen. This is another example of the increasing complexity of the domestic North-South HIV-1 epidemic. Using CRF80_0107 as an example, our study emphasized the emergence of recombinant CRF80_0107 strains in high-risk populations in other cities for early detection, prevention, and control. We place greater emphasis on the importance of today’s large first-tier cities in the prevalence and transmission of novel recombinant strains of HIV-1 and the importance of continuous and accurate molecular surveillance of novel recombinant strains of HIV in MSM at risk in large cities.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank all participants and peer workers for their collaboration during the study.
Abbreviations
- AIC
Akaike information criterion
- AIDS
Acquired immune deficiency syndrome
- BF
Bayes factor
- BLAST
Basic Local Alignment Search Tool
- BSSVS
Bayesian stochastic search variable selection
- COMET
The Context-based Modeling for Expeditious Typing
- CRFs
Circulating recombinant forms
- ESS
Effective sampling size
- HET
Heterosexual
- HIV
Human immunodeficiency virus
- IDUs
Injecting drug users
- MCC
Maximum clade credibility
- MCMC
Markov Chain Monte Carlo
- ML
Maximum likelihood
- MSM
Men who have sex with men
- NFLG
Near full-length genome
- tMRCA
The most recent common ancestor
- URFs
Unique recombinant forms
- 95% HPD
95% highest posterior density
Author contributions
Yongjian Liu, Hongling Wen, and Lin Li designed the study. Xiaorui Wang participated in the process of organizing the sequence data, genotype determination, and phylogenetic analyses. Bo Zhu performed the phylodynamic analysis. Hanping Li and Jingwan Han collected the demographic data of CRF80_0107. Xiaolin Wang and Lei Jia performed the network analysis. Xiaorui Wang produced the illustrations and wrote the manuscript. Bohan Zhang, Jingyun Li and Linding Wang edited the manuscript. All authors read and approved the final manuscript.
Funding
This project was supported by the National Natural Science Foundation of China (NSFC: 82373642, 82173583), and the National Key Research and Development Program of China (2022YFC2305202, 2022YFC2304903).
Data availability
Data is provided within the manuscript or supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Clinical trial number
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yongjian Liu, Email: yongjian325@sina.com.
Hongling Wen, Email: wenhongling@sdu.edu.cn.
Lin Li, Email: dearwood@sina.com.
References
- 1.Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med. 2008;358(15):1590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Kuyl AC, Cornelissen M. Identifying HIV-1 dual infections. Retrovirology. 2007;4:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, Gao F, Hahn BH, Kalish ML, Kuiken C, et al. HIV-1 nomenclature proposal. Science. 2000;288(5463):55–6. [DOI] [PubMed] [Google Scholar]
- 4.Zeng H, Sun B, Yang R. The nomenclature of a new HIV circulating recombinant form should be cautious. AIDS. 2013;27(16):2663–4. [DOI] [PubMed] [Google Scholar]
- 5.Williams A, Menon S, Crowe M, Agarwal N, Biccler J, Bbosa N, Ssemwanga D, Adungo F, Moecklinghoff C, Macartney M et al. Geographic and population distributions of HIV-1 and HIV-2 circulating subtypes: a systematic literature review and meta-analysis (2010–2021). The Journal of infectious diseases 2023. [DOI] [PMC free article] [PubMed]
- 6.Hemelaar J, Elangovan R, Yun J, Dickson-Tetteh L, Fleminger I, Kirtley S, Williams B, Gouws-Williams E, Ghys PD. Global and regional molecular epidemiology of HIV-1, 1990–2015: a systematic review, global survey, and trend analysis. Lancet Infect Dis. 2019;19(2):143–55. [DOI] [PubMed] [Google Scholar]
- 7.Hemelaar J, Loganathan S, Elangovan R, Yun J, Dickson-Tetteh L, Kirtley S. Country Level Diversity of the HIV-1 pandemic between 1990 and 2015. J Virol. 2020;95(2). [DOI] [PMC free article] [PubMed]
- 8.Tee KK, Pybus OG, Li X-J, Han X, Shang H, Kamarulzaman A, Takebe Y. Temporal and spatial dynamics of human immunodeficiency virus type 1 circulating recombinant forms 08_BC and 07_BC in Asia. J Virol. 2008;82(18):9206–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J, Cai W, Zheng C, Yang Z, Xin R, Li G, Wang X, Chen L, Zhong P, Zhang C. Origin and outbreak of HIV-1 CRF55_01B among MSM in Shenzhen, China. J Acquir Immune Defic Syndr. 2014;66(3):e65–67. [DOI] [PubMed] [Google Scholar]
- 10.Vrancken B, Zhao B, Li X, Han X, Liu H, Zhao J, Zhong P, Lin Y, Zai J, Liu M et al. Comparative Circulation Dynamics of the five main HIV types in China. J Virol. 2020;94(23). [DOI] [PMC free article] [PubMed]
- 11.Li Y, Feng Y, Li F, Xue Z, Hu J, Xing H, Ruan Y, Shao Y. Genome sequence of a Novel HIV-1 circulating recombinant form (CRF79_0107) identified from Shanxi, China. AIDS Res Hum Retroviruses. 2017;33(10):1056–60. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y-Q, Li Q-H, Li E-L, Wang Y-R, Tang Z-Y, Gao X, Lu R-R, Liu S-Y, Chen X-H, Wang F-X, et al. Identification of a novel HIV-1 second-generation circulating recombinant form (CRF136_0107) among MSM in China. AIDS. 2023;37(8):F19–23. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Pei Z, Li H, Han J, Li T, Li J, Liu Y, Li L. Characterization of a Novel HIV-1 circulating recombinant form (CRF80_0107) among men who have sex with men in China. AIDS Res Hum Retroviruses. 2019;35(4):419–23. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Luo Y, Li Y, Zhang Y, Wu W, Peng H, Han L, Chen Y, Ruan L, Yang R. Genetic diversity, complicated recombination, and deteriorating Drug Resistance among HIV-1-Infected individuals in Wuhan, China. AIDS Res Hum Retroviruses. 2021;37(3):246–51. [DOI] [PubMed] [Google Scholar]
- 15.Fan W, Wang X, Zhang Y, Meng J, Su M, Yang X, Shi H, Shi P, Lu X. Prevalence of resistance mutations associated with integrase inhibitors in therapy-naive HIV-positive patients in Baoding, Hebei Province, China. Front Genet. 2022;13:975397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Struck D, Lawyer G, Ternes AM, Schmit JC, Bercoff DP. COMET: adaptive context-based modeling for ultrafast HIV-1 subtype identification. Nucleic Acids Res. 2014;42(18):e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Zhang Y, Liu Y, Li H, Jia L, Han J, Li T, Wang X, Li J, Wen H et al. Phylogenetic Analysis of Sequences in the HIV Database Revealed Multiple Potential Circulating Recombinant Forms in China. AIDS research and human retroviruses: 2021. [DOI] [PubMed]
- 19.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73(1):152–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–26. [DOI] [PubMed] [Google Scholar]
- 21.Wertheim JO, Kosakovsky Pond SL. Purifying selection can obscure the ancient age of viral lineages. Mol Biol Evol. 2011;28(12):3355–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D, Wu J, Zhang Y, Shen Y, Dai S, Wang X, Xing H, Lin J, Han J, Li J, et al. Genetic characterization of HIV-1 epidemic in Anhui Province, China. Virol J. 2020;17(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25(7):1253–6. [DOI] [PubMed] [Google Scholar]
- 25.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–21. [DOI] [PubMed] [Google Scholar]
- 26.Letunic I, Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill V, Baele G. Bayesian estimation of Past Population dynamics in BEAST 1.10 using the Skygrid Coalescent Model. Mol Biol Evol. 2019;36(11):2620–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4(1):vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in bayesian phylogenetics using Tracer 1.7. Syst Biol. 2018;67(5):901–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gill MS, Lemey P, Faria NR, Rambaut A, Shapiro B, Suchard MA. Improving bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol Biol Evol. 2013;30(3):713–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dellicour S, Gill MS, Faria NR, Rambaut A, Pybus OG, Suchard MA, Lemey P. Relax, keep walking - A practical guide to continuous phylogeographic inference with BEAST. Mol Biol Evol. 2021;38(8):3486–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Comput Biol. 2009;5(9):e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bielejec F, Baele G, Vrancken B, Suchard MA, Rambaut A, Lemey P. SpreaD3: interactive visualization of Spatiotemporal history and trait evolutionary processes. Mol Biol Evol. 2016;33(8):2167–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ssemwanga D, Bbosa N, Nsubuga RN, Ssekagiri A, Kapaata A, Nannyonjo M, Nassolo F, Karabarinde A, Mugisha J, Seeley J et al. The Molecular Epidemiology and Transmission Dynamics of HIV Type 1 in a General Population Cohort in Uganda. Viruses 2020, 12(11). [DOI] [PMC free article] [PubMed]
- 35.Konings FAJ, Burda ST, Urbanski MM, Zhong P, Nadas A, Nyambi PN. Human immunodeficiency virus type 1 (HIV-1) circulating recombinant form 02_AG (CRF02_AG) has a higher in vitro replicative capacity than its parental subtypes a and G. J Med Virol. 2006;78(5):523–34. [DOI] [PubMed] [Google Scholar]
- 36.Li X, Xue Y, Zhou L, Lin Y, Yu X, Wang X, Zhen X, Zhang W, Ning Z, Yue Q, et al. Evidence that HIV-1 CRF01_AE is associated with low CD4 + T cell count and CXCR4 co-receptor usage in recently infected young men who have sex with men (MSM) in Shanghai, China. PLoS ONE. 2014;9(2):e89462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He N. Research Progress in the epidemiology of HIV/AIDS in China. China CDC Wkly. 2021;3(48):1022–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He S, Song W, Guo G, Li Q, An M, Zhao B, Gao Y, Tian W, Wang L, Shang H, et al. Multiple CRF01_AE/CRF07_BC recombinants enhanced the HIV-1 Epidemic Complexity among MSM in Shenyang City, Northeast China. Front Microbiol. 2022;13:855049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Njai HF, Gali Y, Vanham G, Clybergh C, Jennes W, Vidal N, Butel C, Mpoudi-Ngolle E, Peeters M, Ariën KK. The predominance of human immunodeficiency virus type 1 (HIV-1) circulating recombinant form 02 (CRF02_AG) in West Central Africa may be related to its replicative fitness. Retrovirology. 2006;3:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Streeck H, Li B, Poon AF, Schneidewind A, Gladden AD, Power KA, Daskalakis D, Bazner S, Zuniga R, Brander C, et al. Immune-driven recombination and loss of control after HIV superinfection. J Exp Med. 2008;205(8):1789–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikolaitchik O, Keele B, Gorelick R, Alvord WG, Mazurov D, Pathak VK, Hu WS. High recombination potential of subtype a HIV-1. Virology. 2015;484:334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Su B, Zhang Y, Jia L, Li H, Li Z, Han J, Zhang T, Li T, Wu H, et al. Brief report: onward transmission of multiple HIV-1 unique recombinant forms among men who have sex with men in Beijing, China. J Acquir Immune Defic Syndr. 2019;81(1):1–4. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z, Dai L, Jiang Y, Feng K, Liu L, Xia W, Yu F, Yao J, Xing W, Sun L, et al. Transmission network characteristics based on env and gag sequences from MSM during acute HIV-1 infection in Beijing, China. Arch Virol. 2017;162(11):3329–38. [DOI] [PubMed] [Google Scholar]
- 44.Shi YZ, Huang HH, Wang XH, Song B, Jiang TJ, Yu MR, Wang ZR, Li RT, Jiao YM, Su X et al. Retrospective study on genetic diversity and drug resistance among people living with HIV at an AIDS Clinic in Beijing. Pharmaceuticals (Basel Switzerland) 2024, 17(1). [DOI] [PMC free article] [PubMed]
- 45.Li R, Song C, Chen D, Li C, Hao Y, Zeng H, Han J, Zhao H. Prevalence of transmitted drug resistance among ART-naïve HIV-infected individuals, Beijing, 2015–2018. J Glob Antimicrob Resist. 2022;28:241–8. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, Vermund SH, Ruan Y, Liu H, Zhang C, Yin L, Shao Y, Qian HZ. HIV testing and sexual risks among migrant men who have sex with men: findings from a large cross-sectional study in Beijing, China. AIDS Care. 2018;30(1):86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Z, Xu J, Liu E, Mao Y, Xiao Y, Sun X, Liu Y, Jiang Y, McGoogan JM, Dou Z, et al. HIV and Syphilis prevalence among men who have sex with men: a cross-sectional survey of 61 cities in China. Clin Infect Diseases: Official Publication Infect Dis Soc Am. 2013;57(2):298–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi J, Zhang D, Fu X, Li C, Meng S, Dai M, Liu H, Sun J. High risks of HIV transmission for men who have sex with men–a comparison of risk factors of HIV infection among MSM associated with recruitment channels in 15 cities of China. PLoS ONE. 2015;10(4):e0121267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.An M, Zheng C, Chen L, Li H, Zhang Y, Gan Y, Zhao B, Zhang H, Han X, Zhao J et al. Sustained spread of HIV-1 CRF55_01B in its place of initial origin: Dynamics and hotspots. The Journal of infectious diseases 2024. [DOI] [PubMed]
- 50.Wang X, Zhao J, Li X, Li H, Zhang Y, Liu Y, Chen L, Zheng C, Jia L, Han J et al. Identification of a novel HIV-1 second-generation circulating recombinant forms CRF109_0107 in China. The Journal of infection 2020. [DOI] [PubMed]
- 51.Li M, Zhou J, Zhang K, Yuan Y, Zhao J, Cui M, Yin D, Wen Z, Chen Z, Li L, et al. Characteristics of genotype, drug resistance, and molecular transmission network among newly diagnosed HIV-1 infections in Shenzhen, China. J Med Virol. 2023;95(7):e28973. [DOI] [PubMed] [Google Scholar]
- 52.Zhang D, Zheng C, Li H, Li H, Liu Y, Wang X, Jia L, Chen L, Yang Z, Gan Y, et al. Molecular surveillance of HIV-1 newly diagnosed infections in Shenzhen, China from 2011 to 2018. J Infect. 2021;83(1):76–83. [DOI] [PubMed] [Google Scholar]
- 53.Han X, Takebe Y, Zhang W, An M, Zhao B, Hu Q, Xu J, Wu H, Wu J, Lu L, et al. A large-scale survey of CRF55_01B from Men-Who-Have-Sex-with-Men in China: implying the Evolutionary History and Public Health Impact. Sci Rep. 2015;5:18147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gan M, Zheng S, Hao J, Ruan Y, Liao L, Shao Y, Feng Y, Xing H. The prevalence of CRF55_01B among HIV-1 strain and its connection with traffic development in China. Emerg Microbes Infections. 2021;10(1):256–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei L, Li H, Lv X, Zheng C, Li G, Yang Z, Chen L, Han X, Zou H, Gao Y, et al. Impact of HIV-1 CRF55_01B infection on the evolution of CD4 count and plasma HIV RNA load in men who have sex with men prior to antiretroviral therapy. Retrovirology. 2021;18(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu QH, Qian HZ, Li JM, Leuba SI, Chu ZX, Turner D, Ding HB, Jiang YJ, Vermund SH, Xu JJ, et al. Assisted Partner Notification and Uptake of HIV Testing among men who have sex with men: a Randomized Controlled Trial in China. Lancet Reg Health West Pac. 2021;12:100171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zai J, Liu H, Lu Z, Chaillon A, Smith D, Li Y, Li X. Tracing the transmission dynamics of HIV-1 CRF55_01B. Sci Rep. 2020;10(1):5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is provided within the manuscript or supplementary information files.