Abstract
Brucella melitensis is a facultative intracellular bacterial pathogen that causes abortion in goats and sheep and Malta fever in humans. The genome of B. melitensis strain 16M was sequenced and found to contain 3,294,935 bp distributed over two circular chromosomes of 2,117,144 bp and 1,177,787 bp encoding 3,197 ORFs. By using the bioinformatics suite ERGO, 2,487 (78%) ORFs were assigned functions. The origins of replication of the two chromosomes are similar to those of other α-proteobacteria. Housekeeping genes, including those involved in DNA replication, transcription, translation, core metabolism, and cell wall biosynthesis, are distributed on both chromosomes. Type I, II, and III secretion systems are absent, but genes encoding sec-dependent, sec-independent, and flagella-specific type III, type IV, and type V secretion systems as well as adhesins, invasins, and hemolysins were identified. Several features of the B. melitensis genome are similar to those of the symbiotic Sinorhizobium meliloti.
Brucellosis is a major zoonotic disease that causes abortion in wild and domestic animals and Malta or undulant fever in humans. Although the spread of the disease is controlled in developed countries by livestock testing, vaccination, and slaughter programs, brucellosis is a major problem in the Mediterranean region and parts of Asia, Africa, and Latin America, where it causes severe economic losses (1).
The disease is transmitted to humans by consumption of nonpasteurized milk and milk products or by direct contact with infected animals or carcasses (2, 3). On contact, Brucellae penetrate the skin or mucosal membranes and enter the lymph nodes, which become hemorrhagic, resulting in bacteremia, which facilitates dissemination throughout the body. During the early phase of infection, Brucellae invade macrophages, adapt to the acidic environment, and multiply in the vacuolar compartments (4). Like Salmonella, Mycobacterium, and Legionella, Brucella prevents phagosome/lysosome fusion (5, 6). The infection involves many tissue types and organs. Symptoms, which may include fever, chills, headache, pain, fatigue, dementia, and arthritis, are nonspecific. The infection can be treated with combinations of antibiotics such as doxycycline and streptomycin or doxycycline and rifampin (7, 8). Vaccines against Brucellae have varying degrees of success in controlling the disease; however, human vaccines are not available, and the animal vaccines currently in use are pathogenic to humans (2, 9–11).
Based on rRNA sequences, Brucellae belong to the α-2 group of proteobacteria and have close phylogenetic relationships with Agrobacterium, Rickettsia, Rhizobium, and Rhodobacter (12, 13). Most of the α-proteobacteria are intracellular symbionts or pathogens of plants and animals (14). Although there are six recognized species, DNA hybridization studies suggest that Brucellae are part of a monospecific genus. It has been recommended that they be grouped as biovars of the single species Brucella melitensis (15–17). However, each biovar exhibits a preference for specific hosts: B. melitensis infects goats and sheep, Brucella abortus causes abortion in cattle, and Brucella suis infects swine. Brucella canis, Brucella ovis, and Brucella neotomae are pathogenic to dogs, sheep, and desert wood rats, respectively. Brucellae pathogenesis is not well understood (1, 14).
Materials and Methods
DNA Sequencing.
B. melitensis strain 16M (Biotype1, ATCC 23456) was isolated from an infected goat and is pathogenic to humans (18). This strain causes abortion in pregnant goats, pigs, cattle, and sheep and is used to study Brucellosis in the murine animal model. It was grown and molecular manipulations were carried out in a Biosafety Level III laboratory facility. High molecular weight genomic DNA was isolated from strain 16M following standard protocols. The DNA was sheared, size-fractionated, and used to construct libraries in plasmids (2–3-kb inserts in pGEM3), cosmids (30–35-kb inserts using Lorist 6), and λ phages (10–12-kb inserts for λ DASH). Whole-genome shotgun sequencing was performed on about 30,000 plasmids, 3,000 cosmids, and 3,000 phages by using Applied Biosystems 3700 DNA sequencers (Perkin–Elmer). The DNA was assembled by using phred-phrap-consed into 157 contigs in the first sequencing phase. Gaps were closed by primer walking (900 reactions) and by direct chromosomal sequencing (100 reactions). Finally, the genome was assembled into two contigs representing two chromosomes with an average of 9X coverage.
Sequence Analysis.
An Integrated Genomics (Chicago) program was used for ORF calling. Criteria in this program include minimum length of 40 amino acids, alternate starts, and stop sites. The ORFs were entered into the ergo bioinformatics suite for genome annotation and metabolic reconstruction. The predicted ORFs were subjected to two initial rounds of annotation—one automatic and one manual. First, sequence similarities were calculated for all of the ORFs against the sequences in ergo by using the FASTA algorithm. Based on similarity and other criteria, ORFs were assigned to a preexisting orthologous gene(s), which automatically assigns a function to this gene (cluster). Second, an exhaustive manual analysis of every ORF is performed by using sequence similarity tools such as the motif/pattern databases pfam, prosite, prodom, and cogs (19). At this step, a number of tools that exploit the predictive power of gene context were used to predict a function in the absence of adequate sequence similarity. These include algorithms for the identification of chromosomal clusters (20), fusion clusters (21), and regulatory ORF clusters. Once functions were assigned to the ORFs, they were connected to corresponding pathway(s). Finally, metabolic reconstructions were derived by interconnecting the entire set of pathways to the organism under investigation. A third round of manual curation was performed after reconstruction for specific identification of missing functions. The genome sequence has been deposited in GenBank at accession nos. AE008917 and AE008918.
Results and Discussion
General Features of the Genome.
The B. melitensis genome was sequenced by using a shotgun approach. The genome size is 3.29 Mb distributed over two circular chromosomes of 2.12 and 1.18 Mb with a 57% GC content. There are 3,197 predicted ORFs distributed on the two chromosomes. Plasmids were not found. In our annotation scheme, ORF numbers beginning with 1 are on chromosome I and ORFs beginning with 2 are located on chromosome II. The general features of the genome are listed in Table 1. The annotation tools in the ergo suite allowed functional assignments for 2,481 (78%) of the ORFs. Of the remaining 716 ORFs, nearly two-thirds are similar to hypothetical proteins of other organisms and only one-third (228, 7% of the total) did not show any sequence similarity to other known proteins. The ergo suite exploits a number of clustering tools for function predictions, which includes orthologs (2,115 clusters), paralogs (842 ORFs in 235 clusters), and gene clusters. The latter includes chromosomal clusters (1,583 ORFs; ref. 20) and fusion clusters (1,127 ORFs; ref. 21).
Table 1.
General features of the B. melitensis genome
| Genomic features | ||
| No. of chromosomes | 2 | |
| Size of chromosome I | 2,117,144 bp | |
| Size of chromosome II | 1,177,787 bp | |
| DNA total sequence | 3,294,931 bp | |
| DNA coding sequences | 2,874,027 bp (87%) | |
| GC content | 57% | |
| Plasmids | None | |
| General statistics | ||
| Total no. of ORFs | 3,197 | |
| No. of ORFs on chromosome I | 2,059 | |
| No. of ORFs on chromosome II | 1,138 | |
| ORFs with assigned functions | 2,487 | 78% |
| ORFs without assigned function | 710 | 22% |
| ORFs without function or similarity | 228 | 7% |
| ORFs without function, with similarity | 488 | 15% |
| ORFs in ortholog clusters | 2,115 | 66% |
| ORFs in paralog clusters | 842 | 26% |
| No. of paralog clusters | 235 | |
| ORFs in chromosomal clusters | 1,583 | 49% |
| ORFs in possible fusion events | 1,127 | 35% |
| ORFs in fusion events as composites | 282 | 9% |
| ORFs in fusion events as components | 957 | 30% |
Of the 1,127 ORFs that seem to participate in likely fusion events, 282 probably represent composite proteins. These proteins are distributed on both chromosomes and participate in a large number of metabolic processes including secretion, repair, and transposition. The remaining 957 are likely component genes that are fused (composites) in other genomes. The DNA sequences of those have been checked for frame-shift errors and none were found. ORFs that encode proteins with intein sequences were not found.
The B. melitensis genome contains three rRNA operons, two of which are in chromosome I separated by about 200 kb, and the third is in chromosome II. The rRNA structural organization is identical to that of B. suis serovars 1, 2, and 4, consistent with the suggestion that the two chromosomes arose from a single ancestral chromosome by recombination between two rRNA operons (22). All three rRNA operons are organized in the same way: 16S, ile tRNA, ala tRNA, 23S, 5S, and met tRNA. This arrangement is the same in Rhodobacter capsulatus. The closest similarity to the 16S and 23S rRNA sequences of B. melitensis is found in R. capsulatus, Rhodobacter sphaeroides, and Rhodobacter palustris.
Transcription Factors.
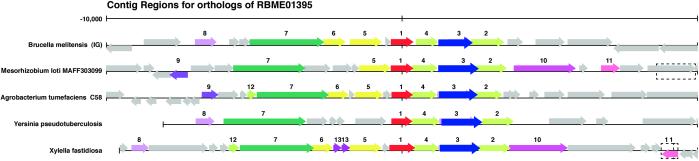
Approximately 193 (6%) ORFs have transcription and translation functions and are distributed on both chromosomes. An ORF similar to the extra-cytoplasmic alternate sigma factor of Pseudomonas aeruginosa is located on chromosome II. Another ORF on chromosome I is similar to virF (AraC family of transcription factor) that perhaps activates the type IV secretion system (See Fig. 1). The operon encoding the large subunits of RNA polymerase is similar to that of other bacteria, containing genes for NusG, four ribosomal protein subunits, and the ββ′ subunits of the RNAP.
Figure 1.
The VirB operon of the type IV secretion system in B. melitensis is shown using the “pinned region” tool from ERGO, which exploits the occurrence of best paired bidirectional hits between ORFs among different organisms. The pinned region for VirB8 (red arrow) shows the type IV secretion cluster from Agrobacterium, Mesorhizobium, Yersinia, and Xylella sp. Identically colored arrows represent similar ORFs. Functions according to numbers are channel protein VirB8 homolog (no. 1), ATPase VirB11 homolog (no. 2), channel protein VirB10 homolog (no. 3), channel protein VirB9 homolog (no. 4), channel protein VirB6 homolog (5), attachment-mediating protein VirB5 homolog (no. 6), ATPase VirB4 homolog (no. 7), attachment-mediating protein VirB1 homolog (no. 8), hypothetical protein (no. 9), ATPase VirD4 homolog (no. 10), DNA recombination protein (no. 11), channel protein VirB3 protein (no. 12), and hypothetical exported protein (no. 13).
Translation Machinery.
B. melitensis encodes a translation apparatus consisting of typical initiation, elongation, and release factors. There is one ORF for each of the elongation factors EF-G, EF-Ts, EF-P, and two ORFs for EF-Tu on chromosome I. The ORF for EF-G and one for EF-Tu flank the ββ′ operon. Two of the peptide chain release factors (RF-2 and RF-3) are located on chromosome I, whereas RF-1 is located on chromosome II. All 33 large and most of the small ribosomal subunits (except S22 and S31, which are absent in most other genomes) are present on chromosome I. Only the S21 subunit gene is on chromosome II.
B. melitensis encodes 18 tRNA synthetases. There are no ORFs for asparagine or glutamine tRNA synthetase, but there are three ORFs for glutamate tRNA synthetase. Among 36 complete bacterial genomes in the ergo database, only 13 contain a gene encoding glutamine tRNA synthetase and 20 have asparagine tRNA synthetase. All of the α-proteobacterial genomes lack both. Three ORFs encode subunits A, B, and C of Glu-tRNA amido transferase, which converts a mis-acylated glutamate on Gln-tRNA to glutamine. The same enzyme probably amidates a mis-acylated aspartic acid on Asn-tRNA to form asparagine. The genes encoding the amido transferase are similar to those found in Mesorhizobium loti, R. sphaeroides, and other members of the α-proteobacteria.
Origins of Replication.
The chromosome I ori is located between ORFs BME2823 and BME2824 with a DnaA binding box identical to that of Caulobacter and Rickettsia and a characteristic low GC skew (55% A + T) (23). In addition, a second putative ori site with two nucleotide differences in the DnaA binding box is located between ORFs BME1001 and BME12059. The ori of chromosome II is located between ORFs BME2001 and BME2118 with a low GC skew (64% A + T) and is similar to that on chromosome I. No plasmid replication origins were found in the genome.
Two ORFs for the replication initiator protein DnaA are located on chromosome I. Genes involved in cell division, such as minC, minD, and topology specificity proteins, are located on chromosome II, whereas other genes such as divJ, ftsE, ftsW, ftsH, ftsQ, ftsX, and ftsZ are on chromosome I. An ftsK gene is located on each chromosome. The gidAB operon, involved in glucose-inhibited cell division, is located on chromosome I, and the chromosome-partitioning operon parAB is located downstream. There is also an ORF for a chromosome segregation protein on chromosome II. DNA polymerases are similar to those of Escherichia coli.
Signal Transduction.
There are 20 ORFs dedicated to signal transduction, 8 of which can be assigned as two-component response regulators. In addition, one ORF encodes a CtrA protein and one sulfur deprivation response regulator is present. Two response regulators of unknown function have been identified. Seven ORFs could be assigned to the sensory transduction histidine kinase family and one to a DivJ family histidine kinase.
Transport.
B. melitensis does not secrete extracellular enzymes involved in carbohydrate degradation. However, specific transporters are present for importing d-fructose, d-mannose, melibiose, sucrose, trehalose, maltose, xylose, and sorbitol. Transport of carbohydrates seems to be accomplished mainly by ABC transporters, although other carbohydrate transporters are also present. ORFs for 17 ABC transporters belonging to families 1 and 2, which are specific for carbohydrate uptake, have been identified (24). Two are similar to the E. coli transport systems for glycerol-3-phosphate and xylose. Another is similar to a Pseudomonas ABC transporter implicated in glucose uptake (25). B. melitensis contains a protein similar to the multiple sugar-binding proteins of Agrobacterium and Azospirillum. These proteins are involved in sugar transport, chemotaxis toward sugars, and regulation of virulence gene expression (26, 27). The B. melitensis gene that codes for this transporter is adjacent to genes involved in galactose metabolism, therefore it may be responsible for galactose uptake. The genomic data suggest that the extracellular sugar-binding domain of one putative carbohydrate ABC transporter is fused to two intracellular ATPase domains. This arrangement has not been described in other bacteria.
Several types of channels and transporters are involved in adaptation to changes in osmolarity (28). These include aquaporins, mechanosensitive channels, transporters for the uptake of osmoprotectants, and probable ion-specific channels. B. melitensis contains an ORF that is similar to the aquaporin of B. abortus (29). Three putative ABC transporters are present for osmoprotectants, but members of the betaine/carnitine/choline transporter (BCCT) family were not found. There are no potassium channel proteins, but there are two ORFs with similarity to chloride channels.
Resistance to heavy metals is accomplished by a number of transporters. Two members of the cation diffusion facilitator (CDF) family and three P-type ATPases that are involved in metal efflux are found. There are two ABC transporters similar to those involved in drug efflux and two specific to the sodium-dependent multidrug efflux pump NorM, similar to that found in Vibrio parahaemolyticus. Three homologs of E. coli multidrug-resistance proteins A and B and seven proteins related to the acriflavine-resistance pumps were observed. The common outer membrane component TolC required for several efflux pumps is present. Several ORFs with similarities to the quaternary ammonium compound resistance protein QacF of Enterobacter aerogenes and the multiple antibiotic-resistance protein MarC of E. coli were found. Seven penicillin-binding proteins, three of which belong to the PBP 1A family, three to the PBP 6 precursor family, and one to the PBP2 family, were observed. The penicillin acyclase gene is located on chromosome II, whereas all of the penicillin-binding protein genes are located on chromosome I.
Catabolism.
Among the polyols, only glycerol and erythritol degradation pathways are present. The genes that encode glycerol kinase and glycerol-3 phosphate dehydrogenase have been identified, although no ORF for the glycerol uptake facilitator protein was found. Erythritol is a key sugar produced by the pregnant host, and Brucella can use it. The erythritol pathway is shown in Fig. 2. The first three enzymes are encoded by the eri operon, which also includes a fourth gene encoding a repressor for the operon. The entire operon is present in B. abortus and parts of it are in M. loti. There are no erythritol-specific transporters such as those found in Rhizobium, but both glycerol and erythritol can be taken up by several putative polyol ABC transporters.
Figure 2.
The erythritol uptake and utilization pathway in B. melitensis, reconstructed from the genome sequence. BME nos. in the boxes correspond to ORFs in the genome.
With the exception of the glycogen cycle, all pathways of central carbohydrate metabolism are present in B. melitensis. The enzymes of the oxidative and nonoxidative branches of the pentose phosphate pathway (PPP) are intact with the presence of an ORF for transketolase on each chromosome. All of the ORFs for enzymes of the Entner–Doudoroff pathway are present. One ORF codes for a 4-hydroxy-2-oxoglutarate/2-dehydro-3-deoxyphosphogluconate aldolase fusion protein. B. melitensis also has a methyl glyoxal bypass pathway for the synthesis of pyruvate. There are fermentation pathways that produce acetate, formate, lactate, ethanol, and butanol.
The enzymes required for the pyruvate-malate cycle and anaplerotic reactions, such as NAD-dependent malic enzymes and phosphoenolpyruvate (PEP) carboxylase, are present. Similarly, conversion of oxaloacetate to pyruvate can be catalyzed by NADP-dependent malic enzyme and by pyruvate carboxylase. In addition, there is a pyruvate phosphate dikinase, which converts pyruvate to the intermediate PEP.
Fatty Acid Metabolism.
B. melitensis is able to synthesize and metabolize saturated and unsaturated fatty acids, as well as other major lipid classes such as lipid A, phospholipids, and isoprenoids. Synthesis of isoprenoids occurs via the deoxy-xylulose-phosphate pathway, which has a bifunctional enzyme for 4-diphosphocytidyl-2-C-methyl-d-erythritol synthase and 2-C-methyl-d-erythritol-2, 4-cyclodiphosphate synthase, similar to those found in Rhodopseudomonas, Mesorhizobium, Treponema, and Helicobacter. An ORF for deoxyxylulose-5-phosphate reductoisomerase was not found, thus B. melitensis probably uses an alternate enzyme to produce 2-C-methyl-d-erythritol-4-phosphate for the synthesis of isoprenoids. B melitensis possesses all of the genes required for the biosynthesis of the typical bacterial phospholipids, phosphatidylglycerol, cardiolipin, phosphatidylserine, and phosphatidylethanolamine. Additionally, B. melitensis seems to be capable of synthesizing phosphatidylcholine, a phospholipid common in eukaryotes but rare in bacteria.
Nitrogen Metabolism.
Brucella does not fix atmospheric nitrogen but it encodes enzymes to oxidize nitrogen compounds, such as copper-containing nitrate reductase, a nitrous oxide–nitric oxide reductase complex, and nitrous oxide reductase. The respiratory nitrate reductase complex consists of four subunits encoded by a single operon. In addition, there is a nitrite extrusion protein to move nitrite from the cytosol to the periplasm, where the nitrite reductase is located. Thus, it should be possible for B. melitensis to grow anaerobically by respiration of nitrate.
Sulfur Metabolism.
ABC transporters carry out the uptake of sulfur in the form of sulfate or thiosulfate, but a thiosulfate ATP-binding protein is not present. Sulfate is incorporated into adenosine phosphosulphate by a sulfate adenylyltransferase. Interestingly, the adenylyltransferase also has an adenyl kinase domain. The phosphorylated adenosine phosphosulphate is reduced to sulfite by a specific phosphoadenosine phosphate reductase (APS reductase) complex and thioredoxin. Thiosulphate is converted to sulfite by a thiosulfate sulfur transferase, and eventually the sulfite is converted to hydrogen sulfide.
Cofactor Synthesis.
B. melitensis is not able to synthesize NAD de novo because of the absence of quinolinate synthetase and nicotinate-nucleotide pyrophosphorylase. Nicotinamidase, nicotinate phosphoribosyltransferase, nicotinate-nucleotide adenylyltransferase, and glutamine-hydrolyzing NAD synthase can synthesize NAD if niacin is present in the growth medium. Pathways for synthesis of other coenzymes and cofactors such as riboflavin, tetrahydrofolate, molybdenum cofactor, molybdopterin guanine dinucleotide, lipoate, biotin, thiamin, CoA, siroheme, protoheme, cobalamin, ubiquinone, and pyridoxine are present.
Electron Transfer.
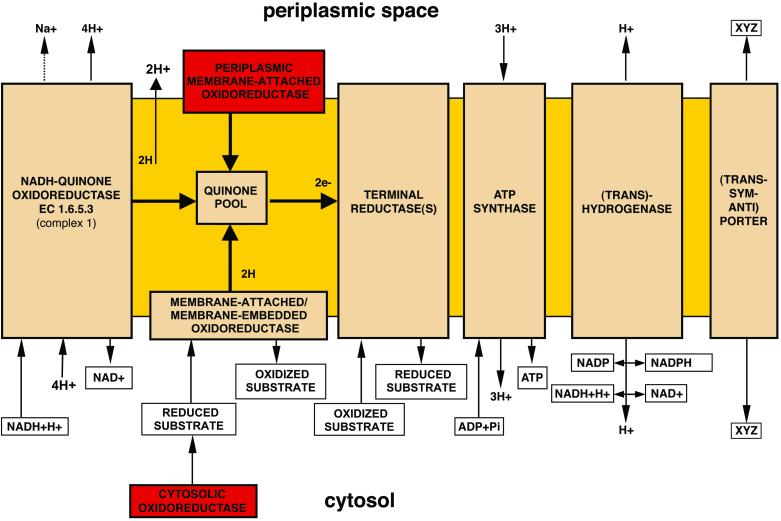
Analysis of the genome indicates that B. melitensis has the capability to adapt to either an aerobic or anaerobic environment. It can switch between different terminal oxidases depending on the oxygen concentration. A cytochrome o ubiquinol oxidase serves as a terminal electron donor to oxygen under normal oxygen concentration. The same function is carried out by cytochrome cbb3 oxidase under low oxygen concentration. During microaerobic conditions, B. melitensis can use the cytochrome bd quinol oxidase for electron transfer to oxygen from the membrane quinone pool. All other key members of the aerobic respiratory complexes are present. Complete sets of ORFs for all of the subunits for complex I (NADH-quinone oxidoreductase), complex II (succinate dehydrogenase), and complex III (ubiquinol-cytochrome c reductase) as well as the soluble electron-transporting cytochromes c2 and c552 are present. Also found are genes that encode enzymes such as d- and l-lactate dehydrogenases, electron transfer flavoprotein-quinone oxidoreductase, glycerol-3-phosphate dehydrogenase [NAD(P)+], and malate:quinone oxidoreductase. These enzymes supply electrons and protons for the aerobic respiratory chain and oxidative phosphorylation pathways.
Under anaerobic conditions, Brucella can use enzymes such as formate dehydrogenase and d-lactate dehydrogenase for electron transfer (Fig. 3). Furthermore, there is a complete system for reduction of nitrate to nitrogen and sulfate to sulfide. B. melitensis has an H+-transporting ATP synthase, NAD(P) trans-hydrogenase, and Na+/H+ and K+/H+ antiporters to maintain the proton gradient and for effective oxidative phosphorylation. Arsenate reductase, which could use a wide range of metal oxides, is also present.
Figure 3.
Reconstruction of possible anaerobic respiratory pathways in B. melitensis, based on the genome sequence. Red boxes indicate that these systems are absent from the genome.
Flagellar Genes.
B. melitensis is nonmotile but it has several flagellar operons. Unlike the enteric bacteria, it does not have a master class I operon (flhDC) but has several ORFs of flagellar class II and class III genes. B. melitensis has all of the components of the flagellar basal body structure except for FlgA, which is necessary for the flagellar P-ring formation. Genes for the flagellar specific sigma factor (FliA) and the anti-sigma factor (FlgM) are absent. ORFs that encode the flagellar L-ring, P-ring proteins, flagellar hook protein (FlgE), and the hook-length control protein are also absent. Among the flagellar class III genes, the motor genes motBCD, motA, structural flagellin (fliC), and two flagella hook-associated proteins are present. B. melitensis lacks all of the genes that are necessary for chemotaxis (Che) and all of the methyl-accepting proteins (MCPs).
Secretion Systems.
B. melitensis does not seem to secrete proteins into the external medium or inject them into the host. It lacks typical type I, II, and, III secretion systems. However, it has genes for sec-dependent, sec-independent, and flagellar-specific type III, IV, and V secretion systems. The sec-dependent pathway ORFs for protein translocation such as SecA, SecE, and SecY have been identified. Two ORFs comprise SecD/SecF fusion proteins, unlike the situation in B. abortus in which SecD and SecF are individual ORFs. Proteins involved in maturation and release of secreted protein such as signal peptidase, lipoprotein signal peptidase, and the signal recognition particle FfH/SRP54, and the receptor (FtsY) are present. The twin arginine motif translocation (TAT) pathway accomplishes the secretion of folded cofactor-containing proteins across the membrane. Of the five known Tat proteins (A–E), TatA and TatC are present. General chaperones such as GroES and GroEL are also present.
Unlike Yersinia or Salmonella, B. melitensis does not have a distinct type III secretion system. However, a set of 12 ORFs located on chromosome II code for a type IV secretion system. Of these, 11 genes are homologous to the B. abortus- and B. suis-type IV secretion system (30) and to that of Agrobacterium tumefaciens (31). These include 3 attachment-mediating proteins, VirB1, -2, and -5, and 6 channel proteins, VirB 3, 6, 7, 8, 9, and 10 (Fig. 1). The virF transcriptional activator is present on chromosome I. There are no ORFs encoding the VirE family of proteins. The nature of the macromolecules that are translocated by this system in B. melitensis is not known. Three ORFs for type V secreted proteins, such as extracellular protease, have been identified. Each includes a protease domain and an adhesin domain belonging to the autotransporter protein family.
Transposable Elements, Insertion Sequence (IS) Elements, and Phage.
There are 41 ORFs assigned to transposases and 30 possible IS elements (22 on chromosome I and 8 on chromosome II). Seven of the IS elements are about 840-bp-long and belong to the class IS711. There are eight IS elements with similarity to those found in Mesorhizobium, which form three distinct subgroups.
Of 41 transposases, there are two identical transposases on chromosome II. There is another group of seven with identical amino acid sequence, of which only one is located on chromosome II. On chromosome II, the genes that encode mannosyltransferase and hemagglutinins are flanked by transposable elements. There are no intact lysogenic phages in the chromosomes; however, there are 16 ORFs located on chromosome I that are related to various phage proteins. An anti-repressor (ant) gene has been identified close to an IS element in addition to a phage-related DNA-binding protein. Ten ORFs are clustered together, located on chromosome I, corresponding to phage portal, DNA packaging, tail, phage host specificity, and other phage-related proteins. This cluster seems to comprise the sole cryptic phage element in the B. melitensis genome.
Virulence.
The outer membrane protein (Omp) genes of B. melitensis are located on chromosome I. The Omps are classified into three major groups. Group 1 is a 94-kDa Omp. Group 2 has two members, Omp2a with a molecular mass of 41 kDa and Omp2b with a molecular mass of 43 kDa. Group 3 consists of Omp25 and Omp31. Group 1 Omp is a minor component of the cell envelope. Omp31 is on the outer cell membrane and is associated with the peptidoglycan. Omp31 forms oligomers that are resistant to SDS at low temperature. Omp25 is an outer membrane structural protein that is highly conserved in all Brucellae. It is associated with both lipopolysaccharide and peptidoglycan. B. melitensis also encodes an OmpA protein, which is homologous to that of E. coli. Other minor outer membrane proteins such as OmpE, which associates with lipoproteins, are also found. All these Omps are associated with virulence (32).
B. melitensis also encodes several other types of virulence factors. There are three types of structurally unrelated hemolysins, namely a hemolysin, an alpha-hemolysin, and hemolysin III. The hemolysin (rRNA methylase) and a membrane-associated hemolysin III are predicted to be pore-formers. Three ORFs that code for adhesion proteins (AidA-I precursors) have been identified. One additional ORF is significantly homologous to the C-terminal region of the AidA-I precursor. An invasin (InvA) precursor, homologous to cell surface proteins of M. loti and Burkholderia cepacia, has also been identified. This protein is also weakly homologous to an invasin of Yersinia pseudotuberculosis. The invasins of enteric bacteria facilitate entry into the host cell by binding to β integrins.
Other virulence-associated genes that have been identified are acvB and mviN. The AcvB protein is homologous to the VirJ protein of A. tumefaciens, required for the transfer of the Ti-plasmid (33). MviN is a membrane-bound protein similar to one found in Salmonella that is necessary for virulence in mice (34). Another ORF encodes the virulence-associated protein E that is involved in symbiosis of Bradyrhizobium japonicum (35). Two orthologs show similarity to a B. abortus ORF, which has been shown to be required for virulence (36). Pathways for the synthesis of lipid IV and UDP-N-acetylmuramoyl pentapeptide biosynthesis are present. Other pathways such as those involved in KDO2 lipid and acetylglucosaminyl-acetylmuramoyl-pentapeptidyl diphosphoundecaprenol syntheses are absent. This absence results in a (bacterial) lipopolysaccharide (LPS) on B. melitensis that is much less potent than the macrophage-activation LPS of Salmonella. The entCE and entBA operons on chromosome I are involved in the synthesis of the siderophore-2,3-dihydrobenzoic acid. Another ORF, which is divergently transcribed from the entCE operon, encodes enterobactin synthetase (entD). The role of the ent operon in iron uptake or virulence in B. melitensis is not known. This operon does not seem to play a role in virulence in B. abortus (37).
B. melitensis contains the response regulator NodW, which is the equivalent of the Bradyrhizobium NolA, and histidine kinase (NodV) homologs necessary for host specificity. Intracellular survival requires the adaptive acid tolerance regulatory protein ActR; its orthologs are found in Bradyrhizobium. Under alkaline conditions, Brucella adapts by use of the phaABCDEFG operon. This operon encodes proteins involved in proton uptake energized by potassium efflux. The PhaA and PhaB proteins are fused. The heme-catalase gene katA, which is located on chromosome II, lacks the signal sequence necessary for its export. It is likely to be involved in protecting the bacterium from hydrogen peroxide toxicity during intracellular multiplication. There are two superoxide dismutases for scavenging superoxides produced by the host, an Fe-Mn-SOD located on chromosome I and a Cu-Zn SOD located on chromosome II (see Appendix, which is published as supporting information on the PNAS web site, www.pnas.org).
Conclusions
The complete genomic sequence of B. melitensis has provided significant information on the basic nature of the organism including its core metabolism, secretion, adhesion, transporters, and cell-wall features. B. melitensis, like other bacteria such as Vibrio cholerae, A. tumefaciens, and Sinorhizobium meliloti, has multiple chromosomes; hence, chromosome segregation and partitioning should be interesting to study. Identification of the nature of the molecules secreted by the type IV secretion system may help in understanding pathogenesis. The genome has also revealed several cases of fusion proteins in B. melitensis. Genomic analysis leads to several unanswered questions: Are flagellar genes cryptic or are some of them expressed in the host? Did B. melitensis lose motility and convert the flagellar proteins to a suprainjecting needle complex or a secretion apparatus to deliver toxins? Which genes are involved when B. melitensis invades and multiplies in diverse hosts? Which genes are required to abort fetuses? Identifying these genes will require microarray and proteomic approaches enabled by the genome sequence. Finally, the response of B. melitensis to its host environments, featuring low pH and nitrous oxide, as well as its ability to evade the host immune response, will provide a major challenge for future studies.
Supplementary Material
Acknowledgments
We thank Karl Reich, Dieter Soll, and Rodolfo Ugalde, as well as the sequencing and assembly teams at Integrated Genomics, for assistance and suggestions. We thank E. Peter Greenberg and Mitchell Sogin for useful reviews of the manuscript. The project was funded by Department of Energy Grant DE-FG02-00ER62773.
Abbreviation
- IS
insertion element
Footnotes
References
- 1.Corbel M J. Emerg Infect Dis. 1997;3:213–221. doi: 10.3201/eid0302.970219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young E J. Rev Infect Dis. 1983;5:821–842. doi: 10.1093/clinids/5.5.821. [DOI] [PubMed] [Google Scholar]
- 3.Young E J. Clin Infect Dis. 1995;21:283–289. doi: 10.1093/clinids/21.2.283. [DOI] [PubMed] [Google Scholar]
- 4.Porte F, Liautard J P, Kohler S. Infect Immun. 1999;67:4041–4047. doi: 10.1128/iai.67.8.4041-4047.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldwin C L, Winter A J. Immunol Ser. 1994;60:363–380. [PubMed] [Google Scholar]
- 6.Pizarro-Cerda J, Meresse S, Parton R G, van der Goot G, Sola-Landa A, Lopez-Goni I, Moreno E, Gorvel J P. Infect Immun. 1998;66:5711–5724. doi: 10.1128/iai.66.12.5711-5724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicoletti P. Adv Biotechnol Processes. 1990;13:147–168. [PubMed] [Google Scholar]
- 8.Garin-Bastuji B, Blasco J M, Grayon M, Verger J M. Vet Res. 1998;29:255–274. [PubMed] [Google Scholar]
- 9.Meyer M E, Morgan W J. Int J Syst Bacteriol. 1973;23:135–141. [Google Scholar]
- 10.Bertrand A. Presse Med. 1994;23:1128–1131. [PubMed] [Google Scholar]
- 11.Solera J, Martinez-Alfaro E, Espinosa A. Drugs. 1997;53:245–256. doi: 10.2165/00003495-199753020-00005. [DOI] [PubMed] [Google Scholar]
- 12.Ke D, Boissinot M, Huletsky A, Picard F J, Frenette J, Ouellette M, Roy P H, Bergeron M G. J Bacteriol. 2000;182:6913–6920. doi: 10.1128/jb.182.24.6913-6920.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreno E, Stackebrandt E, Dorsch M, Wolters J, Busch M, Mayer H. J Bacteriol. 1990;172:3569–3576. doi: 10.1128/jb.172.7.3569-3576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ugalde R A. Microbes Infect. 1999;1:1211–1219. doi: 10.1016/s1286-4579(99)00240-3. [DOI] [PubMed] [Google Scholar]
- 15.Cheville N F. Ann NY Acad Sci. 2000;916:147–153. doi: 10.1111/j.1749-6632.2000.tb05285.x. [DOI] [PubMed] [Google Scholar]
- 16.Verger J M, Grimont F, Grimont P A, Grayon M. Int J Syst Bacteriol. 1985;35:292–295. [Google Scholar]
- 17.Gandara B, Merino A L, Rogel M A, Martinez-Romero E. J Clin Microbiol. 2001;39:235–240. doi: 10.1128/JCM.39.1.235-240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Vallve S, Romeu A, Palau J. Genome Res. 2000;10:1719–1725. doi: 10.1101/gr.130000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kyrpides N C, Ouzounis C A, Iliopoulos I, Vonstein V, Overbeek R. Nucleic Acids Res. 2000;28:4573–4576. doi: 10.1093/nar/28.22.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Overbeek R, Fonstein M, D'Souza M, Pusch G D, Maltsev N. Proc Natl Acad Sci USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enright A J, Iliopoulis I, Kyrpides N C, Ouzounis C A. Nature (London) 1999;6757:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 22.Jumas-Bilak E, Michaux-Charachon S, Bourg G, O'Callaghan D, Ramuz M. Mol Microbiol. 1998;27:99–106. doi: 10.1046/j.1365-2958.1998.00661.x. [DOI] [PubMed] [Google Scholar]
- 23.Brassinga A K, Siam R, Marczynski G T. J Bacteriol. 2001;183:1824–1829. doi: 10.1128/JB.183.5.1824-1829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saier M H., Jr Microbiology. 2000;146:1775–1795. doi: 10.1099/00221287-146-8-1775. [DOI] [PubMed] [Google Scholar]
- 25.Adewoye L O, Worobec E A. Gene. 2000;253:323–330. doi: 10.1016/s0378-1119(00)00285-7. [DOI] [PubMed] [Google Scholar]
- 26.Kemner J M, Liang X, Nester E W. J Bacteriol. 1997;179:2452–2458. doi: 10.1128/jb.179.7.2452-2458.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Bastelaere E, Lambrecht M, Vermeiren H, Van Dommelen A, Keijers V, Proost P, Vanderleyden J. Mol Microbiol. 1999;32:703–714. doi: 10.1046/j.1365-2958.1999.01384.x. [DOI] [PubMed] [Google Scholar]
- 28.Wood J M. Microbiol Mol Biol Rev. 1999;63:230–262. doi: 10.1128/mmbr.63.1.230-262.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez M C, Froger A, Rolland J P, Thomas D, Aguero J, Delamarche C, Garcia-Lobo J M. Microbiology. 2000;146:3251–3257. doi: 10.1099/00221287-146-12-3251. [DOI] [PubMed] [Google Scholar]
- 30.Sieira R, Comerci D J, Sanchez D O, Ugalde R A. J Bacteriol. 2000;182:4849–4855. doi: 10.1128/jb.182.17.4849-4855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stachel S E, Nester E W. EMBO J. 1986;5:1445–1454. doi: 10.1002/j.1460-2075.1986.tb04381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verstreate D R, Creasy M T, Caveney N T, Baldwin C L, Blab M W, Winter A J. Infect Immun. 1982;35:979–989. doi: 10.1128/iai.35.3.979-989.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan S Q, Jin S, Boulton M I, Hawes M, Gordon M P, Nester E W. Mol Microbiol. 1995;17:259–269. doi: 10.1111/j.1365-2958.1995.mmi_17020259.x. [DOI] [PubMed] [Google Scholar]
- 34.Katsukaka K, Okada T, Yokoseki T, Iino T. Gene. 1994;1:49–54. doi: 10.1016/0378-1119(94)90603-3. [DOI] [PubMed] [Google Scholar]
- 35.Gottfert M, Rothlisberger S, Kundig C, Beck C, Marty R, Hennecke H. J Bacteriol. 2001;183:1405–1412. doi: 10.1128/JB.183.4.1405-1412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Endley S, McMurray D, Ficht T A. J Bacteriol. 2001;183:2454–2462. doi: 10.1128/JB.183.8.2454-2462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bellaire B H, Elzer P H, Baldwin C, Roop R M. Infect Immun. 1999;67:2615–2618. doi: 10.1128/iai.67.5.2615-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.